Abstract
One of the most powerful techniques for attributing functions to genes in uni- and multicellular organisms is comprehensive analysis of mutant traits. In this study, systematic and quantitative analyses of mutant traits are achieved in the budding yeast Saccharomyces cerevisiae by investigating morphological phenotypes. Analysis of fluorescent microscopic images of triple-stained cells makes it possible to treat morphological variations as quantitative traits. Deletion of nearly half of the yeast genes not essential for growth affects these morphological traits. Similar morphological phenotypes are caused by deletions of functionally related genes, enabling a functional assignment of a locus to a specific cellular pathway. The high-dimensional phenotypic analysis of defined yeast mutant strains provides another step toward attributing gene function to all of the genes in the yeast genome.
Keywords: cell morphology, functional genomics, high-dimensional phenotyping, phenome
One of the ultimate goals of genetics is to reveal relationships between gene function and phenotypic traits. Comprehensive analysis of mutant traits is a very powerful technique for attributing functions to genes in uni- and multicellular organisms. In the budding yeast Saccharomyces cerevisiae, a complete set of mutants, each of which carries a precise deletion of one yeast ORF, has been systematically constructed (1). By using these mutant strains combined with microarray and robot technology, genome-wide analyses of various mutant traits, including general growth rate, fitness under a particular condition, and sensitivity to drugs, has been reported (reviewed in ref. 2).
Cell morphology becomes an attractive target for comprehensive analysis, because more powerful methods for fluorescent microscopic imaging analysis in biological research have been emerging after development of high-resolution microscopes and specific fluorescent dyes. Yeast cell morphology reflects various cellular events, including progression through the cell cycle, establishment of cell polarity, and regulation of cell size control. Previous genome-wide studies of yeast morphology were focused on a specific morphology, such as cell size, cell shape, or bud site pattern (3-6), and therefore extracted limited information. Because morphological traits are often judged “by eye,” it has remained difficult to obtain quantitative and reproducible results.
We recently developed an image-processing system that automatically processes digital cell images of each yeast cell (7, 8) to obtain quantitative morphological data of yeast mutant cells. Mannoprotein (as a cell wall component marker), the actin cytoskeleton, and nuclear DNA are specifically stained simultaneously. Cells are then photographed, and fluorescence images are automatically processed. The obtained images of all yeast mutants and data-mining functions are available at our Saccharomyces cerevisiae Morphological Database (SCMD) web site (8, 9).
In this study, we employ high-dimensional and quantitative phenotyping of yeast mutants. This large-scale phenotyping represents the comprehensive detailed analyses of the SCMD morphological data set. Our quantitative and high-dimensional analysis is an improvement of previously published, rather qualitative analyses (10-16). Similar phenotypes are caused by deletions of functionally related genes, enabling a functional assignment of a locus to a specific cellular pathway.
Materials and Methods
Strains, Molecular Biology, and Functional Assays. A set of haploid S. cerevisiae MATa deletion strains (4,786 strains) was obtained from the European Saccharomyces cerevisiae Archive for Functional Analysis (EUROSCARF). For obtaining fks1ts data, YOC1085 (MATa fks1-1144 fks2Δ) and its isogenic control strain YOC1001 were used. Each strain was grown in yeast extract/peptone/dextrose medium, and logarithmic-phase cells were fixed. To obtain fluorescent images of the cell-surface mannoprotein, actin cytoskeleton, and nuclear DNA, cells were triply stained with fluorescein isothiocyanate-Con A, rhodaminephalloidin, and 4′,6-diamidino-2-phenylindole, respectively. Four deletion strains (yal016w/tpd3, ylr131c/ace2, yor290c/snf2, and ylr425w/tus1) could not be processed because of aggregation. Quantitative data were obtained from a total of 4,782 strains. We omitted 62 strains for further analysis because corresponding ORFs have been reported to be merged or deleted according to the updated version of the S288C sequence. Deletion strains of 4,718 ORFs were used. Disruption of the corresponding genes was verified by PCR for 81 randomly sampled strains. Standard molecular biological techniques were used (17).
Estimation of Mutants. Statistical tests are not applicable to our data set because we have only one set of data for each deletion strain; thus, we defined morphological normality of a deletion strain as the probability that data from wild-type cells would have measurements outside the range of the deletion strain. We estimated the distribution of wild-type data by transformation and discarded 247 parameters by a normality test because the estimated normal distributions for these parameters were not reliable. Assuming that the transformed wild-type distribution follows the estimated normal distribution in the remaining 254 parameters, we estimated the number of mutants by counting deletion strains that show abnormal morphology in at least 1 parameter. The detailed method is described in Supporting Text, which is published as supporting information on the PNAS web site.
Gene Ontology (GO) Analysis. We downloaded the GO-gene associations and the complete list of yeast ORFs annotated with GO terms from the Saccharomyces genome database as of Jan. 27, 2005.
Functional Prediction. For the “DNA recombinational repair” group, we selected 14 well defined mutant strains with a deletion in a gene locus affecting DNA repair. For the “cell wall biosynthesis” group, we selected a group of 42 mutant strains characterized by a reduced content of glucan in the cell wall (data not shown). To automatically predict the genes having the query functional module of interest from high-dimensional morphological data, we first selected morphological parameters that characterize the query functional module by using classified clustering (18). The classified clustering allows us to extract the most discriminative combination of parameters between the mutants that are known to be related to the query functional module and the other mutants in terms of interclass variance, a common statistical measure for classifying high-dimensional data. Focusing on the dimensions selected, we then separated the periphery of the mutants relevant to the query functional module from the others by using a support vector machine (19). The mutants in the periphery are predicted to be part of the query functional module.
URLs. The Saccharomyces genome database is available at www.yeastgenome.org, the GO database at www.geneontology.org, and the Saccharomyces cerevisiae Morphological Database (SCMD) at http://scmd.gi.k.u-tokyo.ac.jp.
Results and Discussion
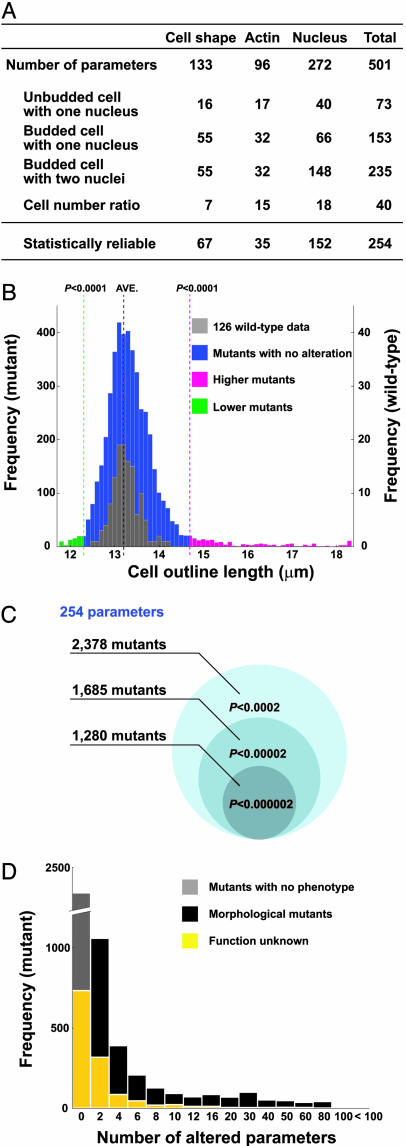
Systematic Identification of Yeast Morphological Mutants. To systematically identify yeast morphological mutants, we first defined parameters that accurately reflect yeast cellular morphology, then we used those parameters to find mutants whose morphology was significantly different from wild-type cells. We defined 501 quantitative parameters representing information about cell shape (visualized by cell wall staining), the actin cytoskeleton, and nuclear morphology of cells at a specific stage of the cell cycle (Fig. 1A; see also Table 1 and Fig. 5, which are published as supporting information on the PNAS web site). Of these 501 parameters, 254 were statistically reliable after power transformation of wild-type data (see Supporting Text). Of the 4,718 haploid mutants of the systematically constructed gene deletion collection (1), a total of 2,378 mutant strains exhibit differences from wild-type cells in at least one of the 254 morphological parameters with normal distribution (Fig. 1 B and C, P < 0.0001 for one side). This estimation indicates that individual deletion of nearly half of the nonessential genes in the genome affects cellular morphology. Two hundred and forty-seven of the 254 parameters identify differences in at least one mutant strain (see Table 2, which is published as supporting information on the PNAS web site). Among the 2,378 morphological mutants identified, 544 carry a deletion in a gene of unknown function (Fig. 1D and Table 3, which is published as supporting information on the PNAS web site), indicating that our high-dimensional analytical approach significantly advances the phenotyping of the yeast mutant collection significantly. The statistical analysis of all yeast mutant strains analyzed, as well as the Java-based calmorph program, will be provided on request.
Fig. 1.
Comprehensive analysis of morphological phenotypes in yeast. (A) Classification of parameters. A subset of parameters used to characterize individual cells is shown. Among all of the 501 parameters measured, 254 revealed statistically reliable data (see Supporting Text). Thresholded at a correlation coefficient value of 0.9, 175 of 254 parameters were considered to be independent (see Fig. 6, which is published as supporting information on the PNAS web site). (B) Data sets were obtained from 126 independent wild-type samples and compared with all data sets from the 4,718 mutant strains. As an example, the distribution of the average of outline length of cells with no bud and a single nucleus is shown. (C) Number of morphological mutants distinct from wild type at the P value for both sides indicated. Deletion strains whose morphological normality P is less than or equal to the threshold in at least one parameter are counted as morphological mutants (see Supporting Text for details). (D) Frequency of the number of parameters altered in the strains analyzed. The number of deletion strains in each category with the GO term “biological process unknown” assigned to the corresponding locus is shown in yellow.
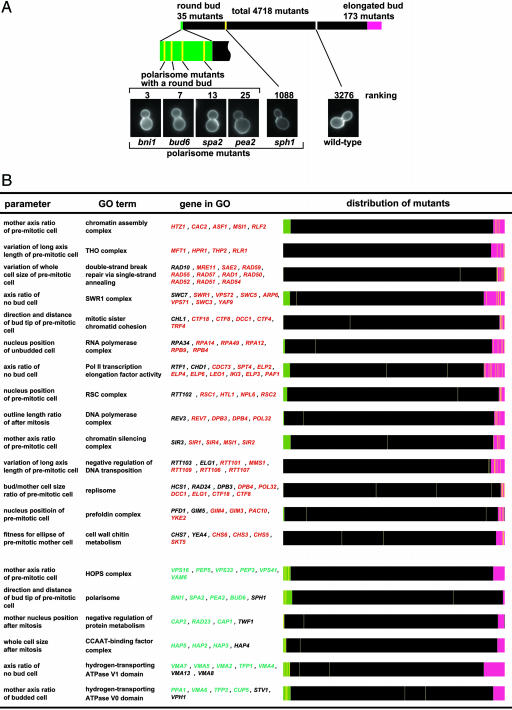
Relationship Between Morphology and Gene Function. One of the ultimate goals of genetics is to reveal relationships between gene function and phenotypic traits. With the high-dimensional phenotypic traits, we demonstrate the close relationship between cellular morphology of the mutant strains and the functions of the deleted loci. As an example, we describe the analysis of polarisome function. The polarisome (Fig. 2A) is a protein complex that helps to determine cell polarity (20). Accordingly, four of five polarisome mutant strains (as provided by the GO consortium) display a round bud phenotype. The frequency of the round bud phenotype occurs by chance with a probability of 1.51 × 10-8 (binomial test, Fig. 2 A), indicating that a round bud is a characteristic trait of polarisome perturbation. A similar correlation is observed in 902 cases among a total of 368,808 combinations (1,452 GO groups × 254 parameters, P < 0.01, Bonferroni-corrected) (Fig. 2B; see also Tables 4 and 5, which are published as supporting information on the PNAS web site). Particular morphological phenotypes are enriched in 260 of the 1,452 GO groups analyzed. This finding implies that, in the case of the genes belonging to these annotated groups, the phenotype of a mutant strain deficient in such a locus can be predicted.
Fig. 2.
Phenotypic analysis of mutant strains and correlation to cellular functions. (A) Polarisome mutants are enriched in the mutant class with altered bud morphology; 4,718 mutants were sorted by roundness of bud. Mutant strains that deviate from wild-type phenotype are shown in magenta or green. The positions of polarisome mutant strains and wild type are indicated with yellow and white lines, respectively. A round bud is a common feature for polarisome perturbation. (B) Cellular functions (as defined by GO terms) that are affected in subsets of mutant strains displaying specific morphological phenotypes. Mutant strains significantly different from wild-type cells are shown in green and magenta, respectively. Genes annotated to the GO term but that exhibit wild-type measurements are in black text. The relative positions of mutant strains with deletions in the loci given are indicated by yellow lines.
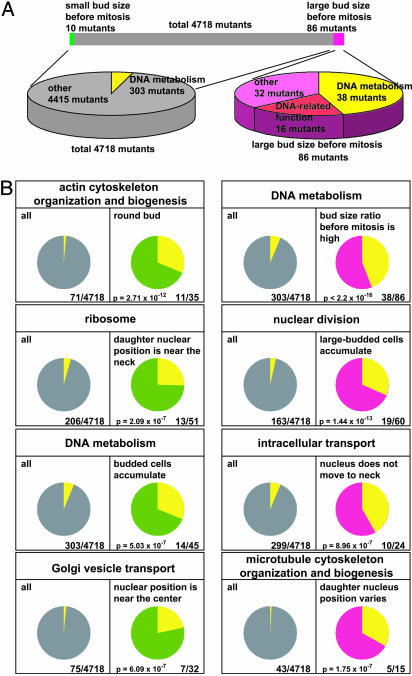
A specific phenotype can be assigned to a specific function. Fig. 3A shows the example of the collection of strains displaying the phenotype “large bud size before mitosis.” Of 86 strains within this group, 39 mutants carry a deletion of a gene annotated with “DNA metabolism” (P < 2.2 × 10-16 by chance); most of these loci are involved in DNA-damage response (21-26). Among the remaining 48 mutant strains with this phenotype, 16 corresponding loci are annotated to other DNA-related functions (27-29). Therefore, we conclude that mutant strains defective in DNA damage response represent the majority within the phenotypic class “large bud size before mitosis.” Likewise, for 109 of 254 morphological traits, the enrichment of mutant strains with deletions in loci with a specific annotation is observed (binomial test, P < 0.01, Bonferroni-corrected) (Fig. 3B; see also Tables 6 and 7, which are published as supporting information on the PNAS web site). Therefore, the function of a given gene can be predicted based on the morphological phenotype of the corresponding mutant strain.
Fig. 3.
Enrichment of defined mutant classes within a defined set of morphological phenotypes. (A) Mutant strains affected in DNA metabolism and DNA-related functions are enriched in strains displaying a large bud size before mitosis. Distributions of the mutants with a large bud size before mitosis are shown in magenta. The pie chart on the right represents the ratio of mutants with defects in DNA-related function within the morphological group “large bud size before mitosis.” Genes associated with the GO term “DNA metabolism” and genes involved in DNA-related function are shown in yellow and red, respectively. The pie chart on the left represents the ratio of genes associated with the GO term “DNA metabolism” to total nonessential mutants. (B) Mutants affected in specific cellular functions are enriched in mutant class displaying the defined phenotype. The cellular function is given above each statistical analysis. The enrichment is observed at indicated probability by chance. (Left) The diagram of each analysis displays the frequency of the mutant strains assigned to the given cellular function. (Right) The diagram displays the frequency of such defined strains within strains with the indicated morphological phenotype.
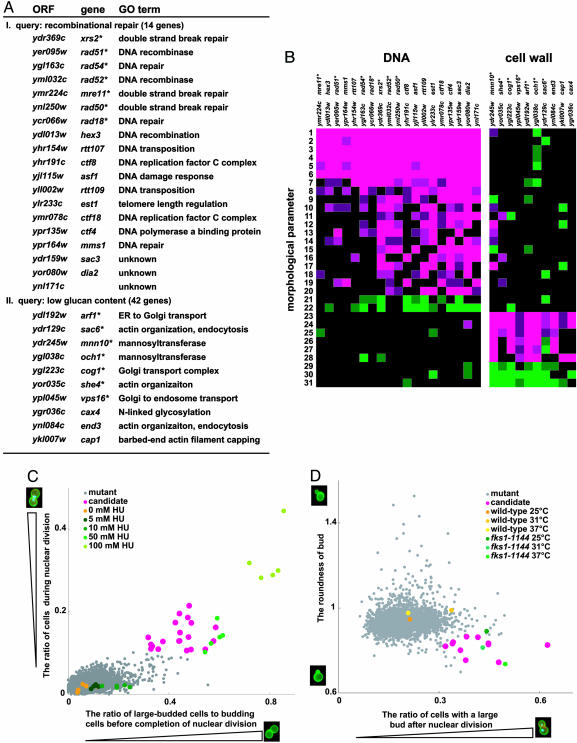
Function Prediction of the Gene from Morphological Data. The validity of gene function prediction is improved by applying two or three phenotypic parameters for classification. We have chosen a set of deletion strains as a query group, by automatically selecting multiple phenotypic parameters that discriminate best between the strains of the query group and the rest of the strain collection. These parameters are then used to identify mutant strains in the whole collection that correspond to these phenotypes. We applied this approach to two groups of strains. One group is characterized as deficient in “DNA recombinational repair,” the other as impaired in “cell wall biosynthesis.” For the “DNA recombinational repair” group, we selected 14 well defined mutant strains with a deletion in a gene locus affecting DNA repair. This group of strains is characterized best by the parameters “ratio of cells in nuclear division” and “ratio of large-budded cells to budding cells before completion of nuclear division,” indicating that cells that arrest or delay at mitosis accumulated in these mutant strains. By using these query terms, a set of 19 mutant strains was identified in the collection, and 7 of these strains were included in the group of 14 strains that defined the query parameters (Fig. 4A). Importantly, the group of the 19 identified strains reveals additional common phenotypic characteristics as visualized by the presence of large contiguous patches of color representing similar morphological features (Fig. 4B). Wild-type cells treated with hydroxyurea, depleting the nucleoside triphosphate pool and inhibiting DNA synthesis, show morphological phenotypes similar to those of the group of the 19 mutant strains identified (Fig. 4C). Interestingly, all candidate strains identified show increased sensitivity to the DNA-damaging reagent hydroxyurea or methyl methanesulfonate (see Table 8, which is published as supporting information on the PNAS web site, and Supporting Text). Among these candidates, we further characterized SAC3 and DIA2, whose functions in DNA repair are unknown, suggesting involvement of these genes in DNA repair (see Fig. 7, which is published as supporting information on the PNAS web site, and Supporting Text).
Fig. 4.
Functional classification of morphological mutants. (A) The descriptive morphological phenotype of a set of 14 recombinational repair mutant strains (I) and 42 mutant strains with low glucan content (II) was defined and used as a query to identify mutant strains that display the appropriate phenotype. Nineteen (for query I) and 10 (for query II) candidate strains were identified. The locus deleted in these strains, the gene name, and the putative function (as given by the GO term) is given. A * indicates mutant strains used as a query. (B) Color-coded representation of the morphological phenotypes displayed by these mutant strains. Morphological difference from wild type is shown for each mutant such that the magnitude is indicated by the intensity of the colors displayed. The brightest colors represent a significant difference (P < 0.00001) for one side. Magenta and green indicate that the mutant has a significantly higher or lower value than that of the wild type, respectively. The numerals in the vertical axis represent the following morphological parameters: 1, C12-2_A1B; 2, A7-2_A1B; 3, C117_A1B; 4, C118_A1B; 5, DCV114_A1B; 6, C125_A1B; 7, D118_A1B; 8, CCV115_C; 9, C118_C; 10, DCV112_C; 11, D109_C; 12, D125_C; 13, CCV104_C; 14, DCV151_C; 15, C117_C; 16, DCV106_C; 17, D103_C; 18, A108; 19, DCV145_C; 20, DCV146_C; 21, A107_A1B; 22, C123; 23, D202; 24, D213; 25, D216; 26, A110; 27, A119; 28, D110_A1B; 29, D207; 30, C106_A1B; and 31, D214. A precise parameter description is shown in Fig. 5 and Table 1. (C and D) Scatter plots of indicated mutant phenotypes of each strain analyzed. The candidate strains identified by using the specified group phenotypes are highlighted. The phenotype of wild-type strains treated with hydroxyurea (HU) is shown in C. (D) The phenotype of wild-type and fks1-1144 mutant cells grown at the temperature indicated is shown.
The similarity between the inhibitor-induced morphological phenotype and the morphological characteristics of the functionally defined set of mutant strains suggests that morphological phenotyping can be used to address the target area of a given chemical. The chemical-induced phenotype can be used to identify mutant strains with similar phenotypes. Potentially, the chemical targets the function(s) that are affected by the gene deletions present in the identified mutant strains.
Next, we selected a group of mutant strains characterized by a reduced content of glucan in the cell wall. This group reveals the identifying parameters “the ratio of cells with large bud after nuclear division” and “roundness of bud.” When these parameters were used, 10 strains were identified, and 7 of these strains belonged to the original query group (Fig. 4A). The phenotype used for the characterization of these strains suggests that cytokinesis or cell separation is incomplete in these mutants. Nine of the 10 strains are sensitive to one of four cell wall damaging drugs (calcofluor white, caffeine, echinocandin B, or SDS) and have altered polysaccharide content in cell wall fractions. Transmission electron microscopic analysis reveals altered cell wall architecture in 8 of 10 strains (see Fig. 8, which is published as supporting information on the PNAS web site, and Supporting Text). A similar morphological phenotype is obtained in an fks1-1144 mutant strain when grown under semipermissive conditions (Fig. 4D). This mutant allele affects the essential 1,3-β-glucan synthase activity and results in a temperature-sensitive phenotype (30). Our results indicate that cell wall integrity is impaired in the identified mutant strains, resulting in very similar morphological phenotypes.
In summary, our quantitative, high-dimensional phenotype analysis reveals that deletions of about half of the nonessential genes in yeast affect cell morphology. Therefore, our approach reveals a phenotype for ≈500 yeast mutant strains with a deletion in a locus of unknown function. Importantly, morphological phenotypes strongly correlate with gene function. We propose to use morphological phenotyping of yeast mutant strains to assign potential functions to unknown genes. In addition, chemically induced morphological phenotypes of yeast can be used to rapidly identify cellular targets of drugs.
Supplementary Material
Acknowledgments
We thank Tamao Goto, Yuka Kitamura, and Emi Shimoi for technical assistance; Kara Dolinski and Fugaku Aoki for critical reading of the manuscript; and the members of the Ohya and Morishita groups at the University of Tokyo for stimulating discussions. This work was supported by the Institute for Bioinformatics and Research and Development of the Japan Science and Technology Corporation.
Author contributions: Y.O. and S. Morishita designed research; M.Y., F.S., A.S., T.F., S.I., S.O., G.S., M.W., A.H., N.F., S.T., S. Muramatsu, and S.N. performed research; J.-P.L., J.M.F., M.A., H.A., and K.S. contributed new reagents/analytic tools; J.S., Y.N., M.O., and H.S. analyzed data; Y.O. and S.N. wrote the paper; J.S. predicted relationship between morphology and gene function; M.Y. designed experiments and performed biological analysis; F.S. collected morphological data of mutants; Y.N. estimated morphological mutants; and T.L.S. constructed and managed database web site.
Conflict of interest statement: No conflicts declared.
Abbreviation: GO, gene ontology.
References
- 1.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 2.Bader, G. D., Heilbut, A., Andrews, B., Tyers, M., Hughes, T. & Boone, C. (2003) Trends Cell Biol. 13, 344-356. [DOI] [PubMed] [Google Scholar]
- 3.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- 4.Jorgensen, P., Nishikawa, J. L., Breitkreutz, B. J. & Tyers, M. (2002) Science 297, 395-400. [DOI] [PubMed] [Google Scholar]
- 5.Zhang, J., Schneider, C., Ottmers, L., Rodriguez, R., Day, A., Markwardt, J. & Schneider, B. L. (2002) Curr. Biol. 12, 1992-2001. [DOI] [PubMed] [Google Scholar]
- 6.Ni, L. & Snyder, M. (2001) Mol. Biol. Cell 12, 2147-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohtani, M., Saka, A., Sano, F., Ohya, Y. & Morishita, S. (2004) J. Bioinform. Comput. Biol. 1, 695-709. [DOI] [PubMed] [Google Scholar]
- 8.Saito, T. L., Ohtani, M., Sawai, H., Sano, F., Saka, A., Watanabe, D., Yukawa, M., Ohya, Y. & Morishita, S. (2004) Nucleic Acids Res. 32, D319-D322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito, T. L., Sese, J., Nakatani, Y., Sano, F., Yukawa, M., Ohya, Y. & Morishita, S. (2005) Nucleic Acids Res. 33, W753-W757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie, K. R., Weng, S., Balakrishnan, R., Costanzo, M. C., Dolinski, K., Dwight, S. S., Engel, S. R., Feierbach, B., Fisk, D. G., Hirschman, J. E., et al. (2004) Nucleic Acids Res. 32, D311-D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guldener, U., Munsterkotter, M., Kastenmuller, G., Strack, N., van Helden, J., Lemer, C., Richelles, J., Wodak, S. J., Garcia-Martinez, J., Perez-Ortin, J. E., et al. (2005) Nucleic Acids Res. 33, D364-D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S. & O'Shea, E. K. (2003) Nature 425, 686-691. [DOI] [PubMed] [Google Scholar]
- 14.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. (2000) Nature 403, 623-627. [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M. & Sakaki, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180-183. [DOI] [PubMed] [Google Scholar]
- 17.Burke, D., Dawson, D. & Stearns, T. (2000) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual (Cold Spring Harbor Lab. Press, Woodbury, New York).
- 18.Sese, J., Kurokawa, Y., Monden, M., Kato, K. & Morishita, S. (2004) Bioinformatics 20, 3137-3145. [DOI] [PubMed] [Google Scholar]
- 19.Vapnik, V. (1995) The Nature of the Statistical Learning Theory (Springer, Berlin).
- 20.Pruyne, D. & Bretscher, A. (2000) J. Cell Sci. 113, 365-375. [DOI] [PubMed] [Google Scholar]
- 21.Broomfield, S., Hryciw, T. & Xiao, W. (2001) Mutat. Res. 486, 167-184. [DOI] [PubMed] [Google Scholar]
- 22.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouprina, N., Kroll, E., Bannikov, V., Bliskovsky, V., Gizatullin, R., Kirillov, A., Shestopalov, B., Zakharyev, V., Hieter, P., Spencer, F., et al. (1992) Mol. Cell. Biol. 12, 5736-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullen, J. R., Kaliraman, V., Ibrahim, S. S. & Brill, S. J. (2001) Genetics 157, 103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scholes, D. T., Banerjee, M., Bowen, B. & Curcio, M. J. (2001) Genetics 159, 1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hryciw, T., Tang, M., Fontanie, T. & Xiao, W. (2002) Mol. Genet. Genomics 266, 848-857. [DOI] [PubMed] [Google Scholar]
- 27.Ramey, C. J., Howar, S., Adkins, M., Linger, J., Spicer, J. & Tyler, J. K. (2004) Mol. Cell. Biol. 24, 10313-10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taggart, A. K. & Zakian, V. A. (2003) Curr. Opin. Cell Biol. 15, 275-280. [DOI] [PubMed] [Google Scholar]
- 29.Wintersberger, U., Kuhne, C. & Karwan, A. (1995) Yeast 11, 929-944. [DOI] [PubMed] [Google Scholar]
- 30.Dijkgraaf, G. J., Abe, M., Ohya, Y. & Bussey, H. (2002) Yeast 19, 671-690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.