Abstract
Cell cycle checkpoints are essential for maintaining genomic integrity. Human topoisomerase II binding protein 1 (TopBP1) shares sequence similarity with budding yeast Dpb11, fission yeast Rad4/Cut5, and Xenopus Cut5, all of which are required for DNA replication and cell cycle checkpoints. Indeed, we have shown that human TopBP1 participates in the activation of replication checkpoint and DNA damage checkpoints, following hydroxyurea treatment and ionizing radiation. In this study, we address the physiological function of TopBP1 in S phase by using small interfering RNA. In the absence of exogenous DNA damage, TopBP1 is recruited to replicating chromatin. However, TopBP1 does not appear to be essential for DNA replication. TopBP1-deficient cells have increased H2AX phosphorylation and ATM-Chk 2 activation, suggesting the accumulation of DNA double-strand breaks in the absence of TopBP1. This leads to formation of gaps and breaks at fragile sites, 4N accumulation, and aberrant cell division. We propose that the cellular function of TopBP1 is to monitor ongoing DNA replication. By ensuring proper DNA replication, TopBP1 plays a critical role in the maintenance of genomic stability during normal S phase as well as following genotoxic stress.
Genomic stability in eukaryotic cells is maintained by multiple checkpoint mechanisms which coordinate cell cycle progression and other processes including transcription, apoptosis, and repair (15). These networks involve many proteins that relay the signal of DNA damage, faulty DNA replication, or aberrant chromosome segregation to downstream effectors.
In mammals, ATM (ataxia-telangiectasia mutated) and ATR (ATM and rad3 related), members of the phosphatidylinositol 3-kinase-related family of proteins, play critical roles as checkpoint regulators (1). ATM phosphorylates and activates downstream effectors such as checkpoint kinase 2 (Chk2) in response to ionizing irradiation (5). On the other hand, ATR detects incompletely replicated or UV-damaged DNA and promotes phosphorylation-dependent activation of Chk1 (14, 16, 18, 50). In addition to ATM and ATR, the Rad17-replication factor C clamp loader, the Rad9-Rad1-Hus1 sliding clamp, and Mre11-Rad50-Nbs1 complexes have all been implicated as sensors of DNA lesions (39, 52). Some of these proteins not only participate in checkpoint control but also function during normal DNA replication. These include ATR/ATM kinase, the Rad17-replication factor C complex, the Rad9-Rad1-Hus1 complex, the single-strand DNA binding protein replication protein A (RPA), the DNA helicases BLM (for Bloom's syndrome protein) and WRN (for Werner's syndrome protein), and topoisomerase binding protein 1 (TopBP1) (4, 12, 19, 27, 35, 43, 52).
TopBP1 was initially identified as a DNA topoisomerase II β-interacting protein (47). Human TopBP1 possess eight BRCA1 carboxyl-terminal (BRCT) domains, a motif which was first described at the C terminus of the breast cancer susceptibility gene product, BRCA1, and is conserved in many proteins related to cell cycle checkpoint and DNA damage response (8). TopBP1 shares sequence homology with Saccharomyces cerevisiae Dpb11, Schizosaccharomyces pombe Rad4/Cut5, Drosophila melanogaster Mus101, and Xenopus Cut5. All these homologs are believed to participate in DNA replication and DNA damage checkpoints.
Budding yeast Dpb11 containing four BRCT domains assembles on replication origins in a Cdc45-dependent manner and plays a role in loading DNA polymerases α and ɛ (21, 38, 42). In the presence of incomplete replication, Dpb11 mutants still progress into mitosis, suggesting that Dpb11 is needed for the activation of replication checkpoint. Dpb11 mutants have an increased rate of genome rearrangements, indicating that one of the Dpb11 functions is to prevent spontaneous genome rearrangements that arise from replication errors (24). Dpb11 mutants are sensitive to hydroxyurea and UV irradiation. In addition, Dpb11 is required for Rad53 activation in response to DNA replication blocks. These data suggest that Dpb11 acts in the DNA damage checkpoint pathway (2, 42). Similarly, fission yeast Rad4/Cut5 is required for Cdc45 loading during normal DNA replication (11), as well as replication checkpoint and DNA damage checkpoint controls (20, 22, 33, 34, 41). In higher eukaryotes, the mutant of the mutagen sensitive 101 (mus101) gene of Drosophila melanogaster encoding seven BRCT domains shows defects in DNA synthesis, chromosome instability, and hypersensitivity to DNA damage (7, 45). Xenopus Cut5 (also known as Mus101) contains eight BRCT domains and is required for the recruitment of Cdc45 to origins of DNA replication (40). In the presence of stalled replication forks, Cut5 facilitates ATR chromatin binding and polymerase α chromatin association (26).
Human TopBP1 has been suggested to be involved in DNA replication and checkpoint control. TopBP1 physically interacts with DNA polymerase ɛ. The addition of an antibody against TopBP1 inhibits DNA synthesis in vitro, suggesting that TopBP1 may be required for normal DNA replication (19). In response to ionizing radiation, TopBP1 is phosphorylated by ATM (48), implying a role of TopBP1 in the DNA damage checkpoint. The role of TopBP1 in checkpoint control is directly demonstrated by a later study using TopBP1 antisense oligonucleotides, showing that ionizing radiation-induced G2/M checkpoint and Chk1 phosphorylation is partially abrogated in the absence of TopBP1 (46).
While it is clear that human TopBP1 participates in the DNA damage checkpoint, the exact role of TopBP1 during normal S-phase progression is not fully understood. The S phase is a period of increased genomic instability as DNA is unpacked and exposed to numerous intrinsic and exogenous replication stress. Therefore, a system monitoring proper DNA replication is pivotal for protecting cells against genomic instability. In this study, we show that TopBP1 is required for Chk1 activation in response to a stalled replication fork. Furthermore, we find that TopBP1 is a chromatin-associated protein during DNA replication. TopBP1 probably monitors the integrity of DNA replication and protects cells from genomic instability during normal cell cycle progression.
MATERIALS AND METHODS
Antibodies.
Rabbit anti-TopBP1 antibody was raised by immunizing rabbits with GST-TopBP1 containing amino acids 979 to 1435. Anti-TopBP1 antibody was affinity purified using the AminoLink Plus Immobilization and Purification kit (Pierce). The phospho-specific antibodies to pT68 of Chk2 and to phosphorylated H2AX (γ-H2AX) were generated as described previously (28, 44). The anti-Chk2pT68 antibody and anti-γ-H2AX were purified by affinity chromatography using the phosphopeptide linked to agarose beads (SulfoLink kit; Pierce) according to the manufacturer's instructions. The anti-Chk1 antibody and phospho-specific antibody to pS317 of Chk1 were purchased from Cell Signaling Technology. The phospho-specific antibody to pS1981 of ATM was obtained from Rockland, Inc.; anti-5-bromo-2′-deoxyuridine (anti-BrdU) antibody was obtained from BD Biosciences; and anti-origin recognition complex 2 (anti-ORC2) was obtained from Santa Cruz Biotechnology.
Cell culture and synchronization.
All cell lines were obtained from the American Tissue Culture Collection and maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum at 37°C in 5% (vol/vol) CO2. Synchronization of cells at mitosis was achieved by nocodazole treatment. In brief, cells were cultured with 0.1-μg/ml nocodazole for 14 h and were harvested by a shake-off procedure. The mitotic cells were released and allowed to grow in serum-containing media.
Cell fractionation.
Whole cell extracts were prepared by resuspension of cells in Laemmli buffer, followed by sonication for 15 s. Preparation of chromatin-bound proteins was performed as described by Mendez and Stillman (23). A total of ∼3 × 106 cells were washed with phosphate-buffered saline (PBS), resuspended in 300 μl of solution A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, 10 mM NaF, 1 mM Na2VO3, and protease inhibitors) containing 0.1% of Triton X-100, and incubated on ice for 5 min. Cytoplasmic proteins were separated from nuclei (P1) by low-speed centrifugation (1,300 × g for 4 min; 4°C). Isolated nuclei were washed once with solution A and then lysed in 300 μl of solution B (3 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol, and protease inhibitors) on ice for 10 min. Soluble nuclear proteins were separated from insoluble chromatin (P2) by centrifugation (1,700 × g for 4 min; 4°C). Isolated chromatin was washed once with solution B and centrifuged at 10,000 × g for 1 min. The final chromatin (P3) was resuspended in 300 μl of Laemmli buffer and sonicated for 15 s. To release chromatin-bound proteins, nuclei (P1) were resuspended in solution A supplemented with 1 mM CaCl2 and 50 U of micrococcal nuclease (Sigma). After 1 min of incubation at 37°C, the nuclease reaction was stopped by the addition of 1 mM EGTA. Nuclei were collected by low-speed centrifugation, lysed, and fractionated as above.
siRNA.
TopBP1 and control small interfering RNA (siRNA) were synthesized by Dharmacon, Inc. The siRNA duplexes were 21 bp as follows: control siRNA sense strand, 5′-UUCAAUAAAUUCUUGAGGUdTdT (where dT is deoxyribosylthymine); TopBP1 siRNA sense strand, 5′-CUCACCUUAUUGCAGGAGAdTdT; BRCA1 siRNA sense strand, 5′-GGAACCUGUCUCCACAAAGdTdT and 5′-UCACAGUGUCCUUUAUGUAdTdT. Transfection was performed twice 24 h apart with 200 nM of siRNA with Oligofectamine reagent according to the manufacturer's instructions (Invitrogen).
Immunofluorescence staining.
Cells grown on coverslips were fixed with 3% paraformaldehyde at room temperature for 10 min. After permeabilization with 0.5% Triton X-100 for 5 min, cells were blocked with 5% goat serum-1% bovine serum albumin for 30 min and incubated with primary antibodies recognizing BrdU, TopBP1, γ-H2AX, ATM, or phosphohistone H3 at room temperature for 1 h. After being washed out with PBS, the cells were incubated with secondary antibodies, fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG), rhodamine-conjugated goat anti-rabbit IgG, or rhodamine-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc.) at room temperature for 1 h. Nuclei were counterstained with 4′6-diamidino-2-phenylindole (DAPI). After a final wash with PBS, coverslips were mounted with glycerin containing paraphenylenediamine. For costaining of PCNA with either TopBP1 or γ-H2AX, cells were permeabilized with methanol:acetone (1:1) at −20°C for 30 min and then incubated sequentially with primary and secondary antibodies.
Fragile-site analysis.
Fragile sites were induced by exposure of HeLa cells to 0.4 μM aphidicolin for 24 h. Cells were harvested for chromosome preparation by standard conditions of 45 min of Colcemid treatment (50 ng/ml), followed by 20 min incubation in 0.075 M KCl at 37°C. Cells were fixed by multiple changes of Carnoy fixative (3:1 methanol:acetic acid) and were dropped onto slides. Slides were baked overnight at 60°C before Giemsa banding, according to the trypsin digestion procedure. Metaphase spreads were scored for gaps and breaks (9). Common fragile sites were determined following the list in Richards (30).
RESULTS
TopBP1 is a replication-dependent chromatin binding protein.
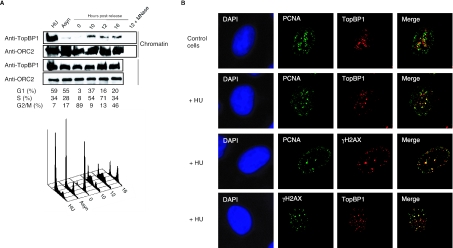
In this study, we were interested in identifying the physiological function of TopBP1 in the absence of DNA damage. Given the roles of TopBP1 homologs in other model organisms (8, 11, 19, 21, 38, 42, 45), we first examined whether TopBP1 would associate with replicating chromatin in S phase. HCT116 colon carcinoma cells were synchronized at prometaphase with nocodazole and then released into the next cell cycle. Cell cycle distribution was confirmed by fluorescence-activated cell sorter analysis. As shown in Fig. 1A, while ORC2, a subunit of origin recognition complex, remained bound to chromatin in all cell cycle phases including mitosis (31), TopBP1 was only detected in the chromatin-containing fractions after cells left mitosis and entered S phase. Treatment of the S phase-chromatin fractions with micrococcal nuclease released TopBP1 (Fig. 1A), demonstrating that TopBP1 indeed associates with DNA during S phase. Furthermore, in the presence of replication stress induced by hydroxyurea, TopBP1 was enriched in the chromatin-containing fraction (Fig. 1A), in agreement with a role of TopBP1 in replication checkpoint control (19). These observations suggest that TopBP1 binds to replicating chromatin independently of DNA damage and is robustly recruited to the damaged DNA.
FIG. 1.
TopBP1 associates with chromatin in S-phase cells. (A) HCT116 cells were synchronized in mitosis with nocodazole treatment for 14 h and then released in fresh medium without nocodazole. Cells were collected at 0, 10, 12, or 16 h later. HCT116 cells were also treated with 1 mM hydroxyurea (HU) for 14 h. Cell cycle distributions were determined by flow cytometry. Chromatin-containing fractions were blotted with indicated antibodies. Asyn, asynchronized; MNase, micrococcal nuclease. (B) HeLa cells treated with or without hydroxyurea (HU) were coimmunostained with the indicated antibodies.
Next, we sought to examine whether or not TopBP1 is recruited to replication forks during S phase. However, as previously reported (19), TopBP1 foci in S-phase cells were distinct from PCNA foci, a marker of the sites of DNA synthesis (Fig. 1B). This indicates that although TopBP1 associates with chromatin, it is not directly linked with the ongoing replication forks during normal S phase. Also similar to the previous report showing that TopBP1, BRCA1, and PCNA colocalize at stalled replication forks (19), it was observed that TopBP1 foci colocalized with most of PCNA foci and γ-H2AX foci following hydroxyurea treatment, supporting that TopBP1 is recruited to the sites of stalled replication forks following replication stress.
TopBP1 deficiency leads to the accumulation of cells with 4N DNA content.
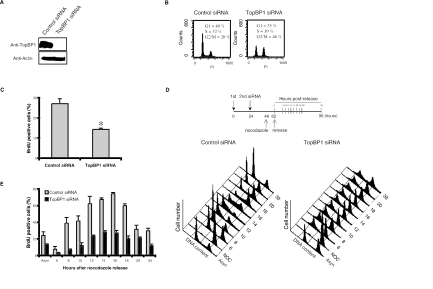
To further explore whether TopBP1 would be required for DNA synthesis, we used siRNA to deplete endogenous TopBP1 expression in HCT116 cells (Fig. 2A). At 72 h after the first siRNA transfection, TopBP1-deficient cells showed a significant accumulation of cells with 4N DNA content, with accompanied decrease of S-phase populations (Fig. 2B). The similar phenotype was also observed with U2OS osteosarcoma, HeLa cervix carcinoma, and A549 lung carcinoma cells (data not shown), suggesting that this phenotype is not cell type specific and does not depend on p53 status. To confirm the decrease of S phase population in the absence of TopBP1, we used BrdU pulse-labeling to determine the percentages of cells with ongoing DNA replication. As shown in Fig. 2C, the depletion of TopBP1 resulted in a modest but significant decrease in the percentage of replicating cells in unsynchronized culture, indicating that TopBP1-deficient cells show an apparent reduction in S-phase population.
FIG. 2.
TopBP1 deficiency results in accumulation of cells with 4N DNA content. (A) At 72 h after siRNA transfection, HCT116 cell lysates were prepared and immunoblotted with anti-TopBP1 or antiactin antibodies. (B) DNA content and cell cycle distributions were determined by flow cytometry. (C) Cells were pulsed with 20 μM BrdU for 30 min and immunostained with anti-TopBP1 and anti-BrdU antibodies. The percentages of cells with BrdU uptake were determined. For each experiment, 500 cells were counted. The results presented here are the average of three independent experiments. *, P < 0.05. (D) Cells transfected with siRNA were arrested at mitosis following nocodazole (NOC) treatment and then released into the next cell cycle. Cell cycle progression was analyzed by flow cytometry. (E) Cells were labeled with 20 μM BrdU for 30 min prior to harvest at each time point, and BrdU-positive cells were determined by immunostaining. An asynchronized (Asyn) cell population was also included as a control.
The apparent reduction of S-phase cells in the absence of TopBP1 could indicate that TopBP1 may be required for normal DNA replication. Alternatively, since we observed an increase of cells with 4N DNA content, it is possible that a large number of cells are arrested with 4N DNA content and cannot reenter the cell cycle. The decrease of S-phase cells in unsynchronized culture could just reflect fewer cycling cells in the absence of TopBP1. To further clarify the cell cycle arrest induced by TopBP1 deficiency, we sought to determine whether there was any defect in overall DNA synthesis in the absence of TopBP1 using synchronized cultures. siRNA-transfected HCT116 cells were synchronized in prometaphase by nocodazole treatment and then released to the cell cycle, following a protocol shown in Fig. 2D. Control siRNA-transfected cells progressed normally into the next S phase. However, many TopBP1-deficient cells were arrested with 4N DNA content throughout the experimental period. Only some TopBP1-deficient cells were able to enter G1 and S phases after release from nocodazole. To carefully monitor the fate of TopBP1-deficient cells, the progression of S phase was determined by pulse-labeling with BrdU. The pattern of S-phase progression appeared to be normal in TopBP1-deficient cells (Fig. 2E). We observed that TopBP1 siRNA-treated cells started to enter S phase 6 to 8 h after being released from nocodazole and S-phase cells peaked at the 14- or 16-h time points. This is very similar to that observed with cells treated with control siRNA (Fig. 2E). There may be a slight delay in S-phase entrance, but largely TopBP1 was not required for the overall DNA replication. Of course, there was an overall decline of BrdU-positive cells in the absence of TopBP1 (Fig. 2E). As shown above, using unsynchronized cultures (Fig. 2B), this decline likely resulted from a blockage of the cell division at 4N DNA status in TopBP1-deficient cells.
TopBP1 deficiency generates spontaneous DNA damage and activates ATM-Chk2 pathway.
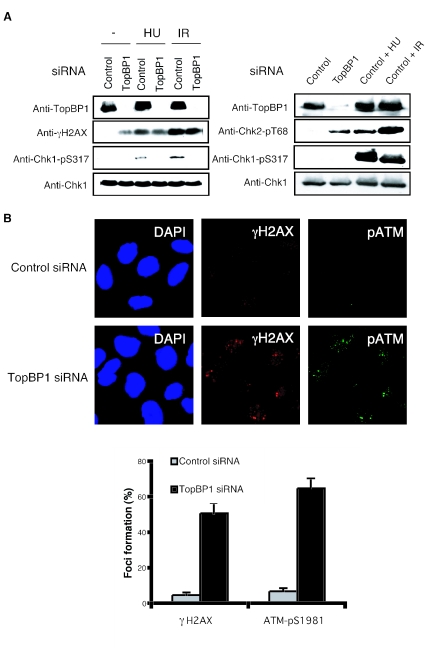
The next question is why TopBP1-deficient cells are arrested with 4N DNA content. The accumulation of cells with 4N DNA content was very similar to the G2/M cell cycle arrest observed with wild-type cells following DNA damage. Given the role of TopBP1 in checkpoint control, it is possible that cells with TopBP1 deficiency bypass the replication checkpoint activated during normal DNA replication. This in turn would lead to the generation of DNA breaks due to the failure to stabilize any stalled replication forks. To test this hypothesis, we examined the phosphorylation of histone H2AX, a marker of double-strand DNA breaks (29). γ H2AX was significantly enhanced in TopBP1-siRNA-transfected U2OS cells compared to control siRNA-transfected cells, either by immunoblotting or by immunostaining (Fig. 3A and B). This indicates that TopBP1 deficiency results in the generation of DNA breaks even in the absence of any external genotoxic stress.
FIG. 3.
TopBP1-deficient cells increase H2AX phosphorylation through the activation of ATM-Chk2 pathway. (A) U2OS (left) or HeLa (right) cells were transfected with control or TopBP1 siRNA. After 72 h, cells were collected and immunoblotted with the indicated antibodies. Cells transfected with indicated siRNA were also treated with 10 mM hydroxyurea (HU) or gamma irradiated (IR; 10 Gy) and then incubated for 1 h before harvest. (B) U2OS cells transfected with the indicated siRNA were immunostained with indicated antibodies. At least 300 cells were counted for each sample.
We next examined whether H2AX phosphorylation induced by TopBP1 siRNA was due to the activation of ATM or ATR kinase. Using an antibody that recognizes the activation-specific ATM autophosphorylation site Ser-1981, we showed a significant activation of ATM colocalized with γ H2AX in TopBP1 siRNA-transfected U2OS cells (Fig. 3B). This phosphorylation of ATM and H2AX was also detected in HeLa cells after TopBP1 depletion (data not shown). Since Chk2 is phosphorylated by active ATM following DNA damage, we would expect to observe Chk2 phosphorylation. We detected a significant Chk2 phosphorylation in TopBP1-deficient HeLa cells (Fig. 3A). Because there is no reliable ATR antibody that can be used as a marker of ATR activation, we did not address directly whether ATR was activated in TopBP1-deficient cells. Instead, we examined Chk1 phosphorylation as an indicator of ATR activation, since ATR is directly required for Chk1 phosphorylation following DNA damage. As shown in Fig. 3A, we failed to observe any Chk1 phosphorylation by immunoblotting. These observations indicate that TopBP1 deficiency causes spontaneous DNA damage, which might resemble some double-strand break-like structures, and thus activate the ATM-Chk2 pathway.
We further determined how TopBP1 might deal with stalled replication forks induced by external genotoxic stress. It has been reported that TopBP1 homologs are required for the activation of replication checkpoint in yeast and Xenopus systems (26, 33, 42). To confirm that TopBP1 also functions in the DNA replication checkpoint, TopBP1-deficient cells were treated with hydroxyurea, and checkpoint activation was examined by immunoblotting with anti-phospho-Chk1 antibodies. As shown in Fig. 3A, while we readily detected phosphorylated Chk1 in hydroxyurea-treated control U2OS cells, cells transfected with TopBP1 siRNA were defective in Chk1 phosphorylation following hydroxyurea treatment. Similar results were also observed with HeLa cells (data not shown). As shown above in Fig. 1A and B, TopBP1 was also enriched at stalled replication forks. In addition, Chk1 phosphorylation was abolished in TopBP1-deficient cells, following ionizing radiation (Fig. 3A). These data suggest that TopBP1 is involved in sensing and signaling DNA damage and is required for Chk1, following replication stress and double-strand breaks.
TopBP1 partially regulates fragile site expression.
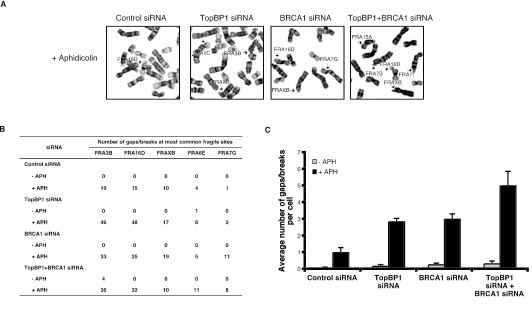
Our findings of spontaneous DNA damage and ATM-Chk2 activation in TopBP1-deficient cells suggest that single-strand and/or double-strand breaks are generated in these cells. One likely source of spontaneous damage is stalled replication forks that accumulate single-strand DNA gaps. In addition, stalled replication forks are also frequently processed to Holliday junction-like “chicken foot” structures. The collapse of such structures leads to DNA double-strand breaks (10, 25, 36). The intra-S-phase checkpoint is required for the stabilization of stalled replication forks and thus maintains genomic stability. One consequence of an inactive or absent replication checkpoint is the generation of fragile sites, characterized by formation of gaps and breaks, which arise upon progression into mitosis (10, 30).
To determine whether TopBP1-deficient cells express common fragile sites, low doses of aphidicolin was used, and the gaps and breaks at fragile sites on metaphase chromosomes were evaluated with trypsin-Giemsa banding (Fig. 4A). Since another checkpoint protein, BRCA1, is required for the fragile-site stability (3), we included BRCA1 siRNA as a control in our experiments. HeLa cells transfected with either TopBP1 siRNA or BRCA1 siRNA showed about a threefold increase in fragile-site expression compared to control cells, indicating that both TopBP1 and BRCA1 partially regulate fragile-site stability (Fig. 4B). As shown in Fig. 4C, the concurrent down-regulation of TopBP1 and BRCA1 resulted in a 1.5-fold increase in fragile-site expression compared to single siRNA-transfected cells, suggesting that TopBP1 and BRCA1 may have overlapping functions in regulating the fragile site. This result is similar to an early study showing the overlapping functions of TopBP1 and BRCA1 in G2/M checkpoint control (46), raising the possibility that the checkpoint function and fragile-site regulation are mechanistically linked.
FIG. 4.
TopBP1 deficiency increases chromosomal breakage. (A) At 48 h after siRNA transfection in HeLa cells, fragile sites were induced by the addition of aphidicolin (APH) 24 h before harvest. Discrimination of fragile sites on metaphase spread was facilitated by trypsin-Giemsa banding. (B) Metaphase spreads (500 cells total) were counted in three independent experiments. The table shows the number of gaps and/or breaks of the most common fragile sites in siRNA-transfected cells. (C) The average number of gaps/breaks per cell transfected with control, TopBP1, or BRCA1 siRNAs was quantified. This number includes all gaps and breaks at common fragile sites, as well as at other random genomic sites.
TopBP1 deficiency results in abnormal cell cycle progression.
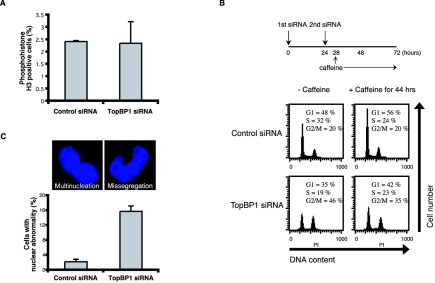
Based on the above observations, it is reasonable to speculate that the inability to correctly process spontaneous replication fork collapse in TopBP1-depleted cells leads to the generation of DNA double-strand breaks, which in turn arrest the cell cycle at G2/M phase. To determine in which cell cycle phase TopBP1-deficient cells are arrested, we examined the percentages of mitotic cells using phosphohistone H3 staining. However, as shown in Fig. 5A, TopBP1-deficient HCT116 cells have approximately the same percentages of cells in mitosis as control siRNA-transfected cells. In addition, the number of cells in late G2 phase, which displays a dotted pattern of phosphohistone H3 staining, also did not increase in TopBP1-deficient cells (data not shown).
FIG. 5.
TopBP1 deficiency leads to aberrant mitosis. (A) HCT116 cells were transfected with control or TopBP1 siRNAs. Percentages of mitotic cells indicated by positive phosphohistone H3 staining were determined 72 h after initial siRNA transfection. (B) HCT116 cells were transfected with control or TopBP1 siRNA in the absence or presence of 5 mM caffeine for the period indicated. Cell cycle distributions were determined by fluorescence-activated cell sorter analyses. (C) HCT116 cells were transfected with control or TopBP1 siRNAs. Cells were fixed and stained with DAPI. The percentages of cells with aberrant nuclear morphologies (indicated by multinucleation or nuclear missegregation) (top) were determined under a microscope. A total of 500 cells per sample were counted, and the results are the average of three independent experiments (bottom).
Because of the lack of proper markers for early G2 phase, we were unable to determine the percentage of TopBP1-deficient cells in early G2 phase. However, if the 4N accumulation observed with TopBP1-deficient cells is the result of G2/M checkpoint activation in response to DNA breaks, it should be overridden by the addition of caffeine, an inhibitor of both ATM and ATR kinases. Earlier studies have indeed shown that the G2/M cell cycle arrest following DNA damage can be bypassed by caffeine (49). Thus, after TopBP1 siRNA transfection, we incubated these cells with caffeine for an additional 44 h (Fig. 5B). To our surprise, the 4N accumulation we observed following TopBP1 depletion largely could not be overcome by caffeine treatment, suggesting that these 4N cells are not likely the normal G2-arrested cells following DNA damage.
To find out the property of these cells with 4N DNA content, we counted the cells with aberrant mitotic morphologies. As shown in Fig. 5C, the number of cells with aberrant mitosis including nuclear missegregation and multinucleation was substantially increased in TopBP1-deficient HCT116 cells. The aberrant phenotypes were also observed with HeLa cells (data not shown). These observations suggest that the TopBP1-deficient cells somehow bypassed the G2/M checkpoint and entered mitosis (see Discussion for details). However, presumably because of the unreplicated DNA regions or remaining DNA breaks, these cells failed to undergo normal mitosis and cytokinesis and subsequently were arrested as tetraploid cells. These cells are unlikely to be able to survive, which is in agreement with our previous report indicating that abolishing TopBP1 expression leads to significant cell death (48).
DISCUSSION
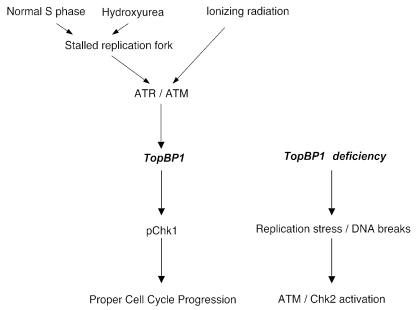
The present study demonstrates that TopBP1 is required for genomic integrity in mammalian cells. Similar to previous studies, we have shown that TopBP1 participates in DNA damage response and is required for Chk1 activation following DNA damage. More importantly, we have shown that TopBP1 is a replication-dependent chromatin binding protein and further accumulates on chromatin following replication stress. TopBP1 is required for faithful DNA synthesis. In the absence of TopBP1, cells accumulate spontaneous DNA damage and activate the ATM-Chk2 pathway, which results in tetraploid cell cycle arrest. Consequently, TopBP1-deficient cells are prone to expression of common fragile sites. These observations allow us to propose a model whereby TopBP1 coordinates cell cycle transitions with the completion of DNA synthesis and DNA checkpoint, thereby safeguarding genomic integrity (Fig. 6).
FIG. 6.
TopBP1 participates in the maintenance of genomic stability during normal S phase, as well as following DNA damage.
S phase is probably one of the most vulnerable cell cycle phases during normal cell proliferation. Either intrinsic or external events can lead to the stalling of replication forks, which if not managed properly will result in the collapse of replication forks and the generation of DNA double-strand breaks. Thus, a guarding system called the replication checkpoint has evolved to deal with these replication stresses and ensure proper DNA duplication (32). Based on our observations, we believe that TopBP1 is one of the key S-phase checkpoint components. Loss of TopBP1 leads to the generation of DNA double-strand breaks and the activation of the ATM-Chk2 pathway, due to the collapse of stalled replication forks in these cells.
Previous studies suggested that components of the replication checkpoint (ATR, BRCA1, BLM/WRN, and Chk1) are all critical for protecting the stability of fragile sites (3, 9, 17, 32, 37). In this study, we have shown that TopBP1 is also required for the prevention of fragile site expression. Interestingly, a recent report shows that in the Xenopus system, the absence of BLM results in chromosomal breaks during S phase (17), a phenomenon similar to that observed here with TopBP1-deficient cells. Chk1 is also proposed to be required for normal S phase to avoid DNA breakage (37). Thus, it implicates that the replication checkpoint is required for faithful DNA replication, suppression of the fragile-site expression, and genomic stability, even in the absence of external DNA damage. Further study of the nature of these stalled replication forks and the molecular mechanisms by which stalled replication forks or replication stress are monitored will yield better understanding of genomic stability in humans.
It remains to be resolved whether or not human TopBP1 is a component of normal DNA replication apparatus. Yeast TopBP1 homologs are required for recruiting Cdc45 or DNA polymerases during replication (2, 42). However, human TopBP1 does not colocalize with PCNA in normal S-phase cells, raising the possibility that TopBP1 is not an essential component of the DNA replication machinery. If TopBP1 is critical for DNA replication, we would expect to detect G1/S-phase or S-phase arrest in TopBP1 siRNA-treated cells. Instead, we observed accumulation of cells with 4N DNA content in the absence of TopBP1. These data suggest that TopBP1 is not necessary for general DNA replication, since cells are able to complete most, if not all, DNA replication without TopBP1. Of course, we cannot rule out that TopBP1 is required for DNA replication at certain regions. DNA replication is not a uniform process, as replication through repetitive sequences or unusual chromatin regions (e.g., telomeres, centromeres, and fragile sites) may need additional auxiliary factors that are not required for the normal DNA replication. Potentially, TopBP1 and/or the replication checkpoint is involved in these special replication events.
We observed the accumulation of cells with 4N DNA content in the absence of TopBP1. Since we also detected ATM and Chk2 activation in these cells, we initially suspected that these cells were arrested at G2/M phase, similar to wild-type cells following DNA damage. However, unlike the G2/M checkpoint activation following DNA damage, caffeine treatment failed to rescue the accumulation of 4N cells in the absence of TopBP1. Detailed analyses revealed that these cells are likely to be arrested in tetraploid stage, due to a failure of nuclear segregation or cytokinesis. Then if DNA damage exists in TopBP1-deficient cells (which is clearly indicated by the ATM-Chk2 activation and phosphorylation of H2AX), why these cells are not arrested at G2/M phase? We believe that the absence of cell cycle arrest at G2/M phase is probably due to the failure of Chk1 activation in these cells. Early studies suggest that G2/M cell cycle arrest is mediated by the Chk1-dependent pathway (51). However, in TopBP1-deficient cells, we failed to observe any Chk1 activation in response to either replication stress or ionizing radiation (Fig. 3A). On one hand, these data suggest that TopBP1 is required for Chk1 activation in response to both DNA replication stress and DNA double-strand breaks. On the other hand, the absence of Chk1 activation explains why TopBP1-deficient cells cannot be arrested at G2/M phase, even in the presence of DNA damage and ATM activation.
TopBP1 clearly plays a role in checkpoint control following genotoxic stress. What we emphasize here is that this stress occurs normally during DNA replication. The S phase or replication checkpoint has evolved to specifically deal with the stress or the unusual chromatin structures arising during normal S-phase progression and thus to ensure the faithful replication of genetic material. While TopBP1 does not participate directly in DNA replication, it monitors faithful DNA replication through the ATR pathway. We propose that TopBP1 is recruited to fragile sites and other specific chromosomal regions during normal DNA replication to ensure the successful replication progression through these regions. In the absence of TopBP1, replication forks will stall and likely collapse at these regions, which lead to the generation of DNA strand breaks, the activation of the ATM-Chk2 pathway, and genomic instability. Thus, the normal function of TopBP1 is to maintain genomic integrity by minimizing naturally occurring stalled replication forks and double-strand breaks. Given the importance of deregulated DNA replication and the DNA damage checkpoint in tumorigenesis (6, 13), it will be interesting to explore whether TopBP1 would be deregulated during tumorigenesis and contributes to tumor development in humans.
Acknowledgments
We thank members of the Chen laboratory for discussion, especially Zheunkun Lou, Irene M. Ward, Katherine Minter-Dykhouse, and Jamie Wood.
This work was supported in part by grants from the National Institutes of Health (CA89239, CA92312, and CA100109), the Prospect Creek Foundation, and the Breast Cancer Research Foundation. J.C. is a recipient of the DOD Breast Cancer Career Development Award (DAMD 17-02-1-0472).
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Araki, H., S. H. Leem, A. Phongdara, and A. Sugino. 1995. Dpb11, which interacts with DNA polymerase II(ε) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. USA 92:11791-11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlt, M. F., B. Xu, S. G. Durkin, A. M. Casper, M. B. Kastan, and T. W. Glover. 2004. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol. Cell. Biol. 24:6701-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao, S., T. Lu, X. Wang, H. Zheng, L. E. Wang, Q. Wei, W. N. Hittelman, and L. Li. 2004. Disruption of the Rad9/Rad1/Hus1 (9-1-1) complex leads to checkpoint signaling and replication defects. Oncogene 23:5586-5593. [DOI] [PubMed] [Google Scholar]
- 5.Bartek, J., J. Falck, and J. Lukas. 2001. CHK2 kinase—a busy messenger. Nat. Rev. Mol. Cell Biol. 2:877-886. [DOI] [PubMed] [Google Scholar]
- 6.Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864-870. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, J. B., M. D. Golino, T. D. Nguyen, and M. M. Green. 1976. Isolation and characterization of X-linked mutants of Drosophila melanogaster which are sensitive to mutagens. Genetics 84:485-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callebaut, I., and J. P. Mornon. 1997. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 400:25-30. [DOI] [PubMed] [Google Scholar]
- 9.Casper, A. M., P. Nghiem, M. F. Arlt, and T. W. Glover. 2002. ATR regulates fragile site stability. Cell 111:779-789. [DOI] [PubMed] [Google Scholar]
- 10.Cimprich, K. A. 2003. Fragile sites: breaking up over a slowdown. Curr. Biol. 13:R231-R233. [DOI] [PubMed] [Google Scholar]
- 11.Dolan, W. P., D. A. Sherman, and S. L. Forsburg. 2004. Schizosaccharomyces pombe replication protein Cdc45/Sna41 requires Hsk1/Cdc7 and Rad4/Cut5 for chromatin binding. Chromosoma 113:145-156. [DOI] [PubMed] [Google Scholar]
- 12.Frei, C., and S. M. Gasser. 2000. RecQ-like helicases: the DNA replication checkpoint connection. J. Cell Sci. 113:2641-2646. [DOI] [PubMed] [Google Scholar]
- 13.Gorgoulis, V. G., L. V. Vassiliou, P. Karakaidos, P. Zacharatos, A. Kotsinas, T. Liloglou, M. Venere, R. A. Ditullio, Jr., N. G. Kastrinakis, B. Levy, D. Kletsas, A. Yoneta, M. Herlyn, C. Kittas, and T. D. Halazonetis. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907-913. [DOI] [PubMed] [Google Scholar]
- 14.Guo, Z., A. Kumagai, S. X. Wang, and W. G. Dunphy. 2000. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14:2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartwell, L. H., and T. A. Weinert. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246:629-634. [DOI] [PubMed] [Google Scholar]
- 16.Hekmat-Nejad, M., Z. You, M. C. Yee, J. W. Newport, and K. A. Cimprich. 2000. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 10:1565-1573. [DOI] [PubMed] [Google Scholar]
- 17.Li, W., S. M. Kim, J. Lee, and W. G. Dunphy. 2004. Absence of BLM leads to accumulation of chromosomal DNA breaks during both unperturbed and disrupted S phases. J. Cell Biol. 165:801-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, Q., S. Guntuku, X. S. Cui, S. Matsuoka, D. Cortez, K. Tamai, G. Luo, S. Carattini-Rivera, F. DeMayo, A. Bradley, L. A. Donehower, and S. J. Elledge. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14:1448-1459. [PMC free article] [PubMed] [Google Scholar]
- 19.Makiniemi, M., T. Hillukkala, J. Tuusa, K. Reini, M. Vaara, D. Huang, H. Pospiech, I. Majuri, T. Westerling, T. P. Makela, and J. E. Syvaoja. 2001. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276:30399-30406. [DOI] [PubMed] [Google Scholar]
- 20.Marchetti, M. A., S. Kumar, E. Hartsuiker, M. Maftahi, A. M. Carr, G. A. Freyer, W. C. Burhans, and J. A. Huberman. 2002. A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. Proc. Natl. Acad. Sci. USA 99:7472-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masumoto, H., A. Sugino, and H. Araki. 2000. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 20:2809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McFarlane, R. J., A. M. Carr, and C. Price. 1997. Characterisation of the Schizosaccharomyces pombe rad4/cut5 mutant phenotypes: dissection of DNA replication and G2 checkpoint control function. Mol. Gen. Genet. 255:332-340. [DOI] [PubMed] [Google Scholar]
- 23.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myung, K., A. Datta, and R. D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104:397-408. [DOI] [PubMed] [Google Scholar]
- 25.Osborn, A. J., S. J. Elledge, and L. Zou. 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12:509-516. [DOI] [PubMed] [Google Scholar]
- 26.Parrilla-Castellar, E. R., and L. M. Karnitz. 2003. Cut5 is required for the binding of Atr and DNA polymerase alpha to genotoxin-damaged chromatin. J. Biol. Chem. 278:45507-45511. [DOI] [PubMed] [Google Scholar]
- 27.Post, S. M., A. E. Tomkinson, and E. Y. Lee. 2003. The human checkpoint Rad protein Rad17 is chromatin-associated throughout the cell cycle, localizes to DNA replication sites, and interacts with DNA polymerase epsilon. Nucleic Acids Res. 31:5568-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rappold, I., K. Iwabuchi, T. Date, and J. Chen. 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 153:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 30.Richards, R. I. 2001. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 17:339-345. [DOI] [PubMed] [Google Scholar]
- 31.Ritzi, M., M. Baack, C. Musahl, P. Romanowski, R. A. Laskey, and R. Knippers. 1998. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem. 273:24543-24549. [DOI] [PubMed] [Google Scholar]
- 32.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 33.Saka, Y., P. Fantes, T. Sutani, C. McInerny, J. Creanor, and M. Yanagida. 1994. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 13:5319-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saka, Y., and M. Yanagida. 1993. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+. Cell 74:383-393. [DOI] [PubMed] [Google Scholar]
- 35.Shechter, D., V. Costanzo, and J. Gautier. 2004. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6:648-655. [DOI] [PubMed] [Google Scholar]
- 36.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 37.Syljuasen, R. G., C. S. Sorensen, L. T. Hansen, K. Fugger, C. Lundin, F. Johansson, T. Helleday, M. Sehested, J. Lukas, and J. Bartek. 2005. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 25:3553-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takayama, Y., Y. Kamimura, M. Okawa, S. Muramatsu, A. Sugino, and H. Araki. 2003. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 17:1153-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Bosch, M., R. T. Bree, and N. F. Lowndes. 2003. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 4:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Hatten, R. A., A. V. Tutter, A. H. Holway, A. M. Khederian, J. C. Walter, and W. M. Michael. 2002. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 159:541-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verkade, H. M., and M. J. O'Connell. 1998. Cut5 is a component of the UV-responsive DNA damage checkpoint in fission yeast. Mol. Gen. Genet. 260:426-433. [DOI] [PubMed] [Google Scholar]
- 42.Wang, H., and S. J. Elledge. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, H., J. Guan, A. R. Perrault, Y. Wang, and G. Iliakis. 2001. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer Res. 61:8554-8563. [PubMed] [Google Scholar]
- 44.Ward, I. M., X. Wu, and J. Chen. 2001. Threonine 68 of Chk2 is phosphorylated at sites of DNA strand breaks. J. Biol. Chem. 276:47755-47758. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto, R. R., J. M. Axton, Y. Yamamoto, R. D. Saunders, D. M. Glover, and D. S. Henderson. 2000. The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C terminus domains. Genetics 156:711-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamane, K., J. Chen, and T. J. Kinsella. 2003. Both DNA topoisomerase II-binding protein 1 and BRCA1 regulate the G2-M cell cycle checkpoint. Cancer Res. 63:3049-3053. [PubMed] [Google Scholar]
- 47.Yamane, K., M. Kawabata, and T. Tsuruo. 1997. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur. J. Biochem. 250:794-799. [DOI] [PubMed] [Google Scholar]
- 48.Yamane, K., X. Wu, and J. Chen. 2002. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 22:555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yarden, R. I., S. Pardo-Reoyo, M. Sgagias, K. H. Cowan, and L. C. Brody. 2002. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 30:285-289. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, H., and H. Piwnica-Worms. 2001. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21:4129-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao, H., J. L. Watkins, and H. Piwnica-Worms. 2002. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc. Natl. Acad. Sci. USA 99:14795-14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zou, L., D. Cortez, and S. J. Elledge. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16:198-208. [DOI] [PMC free article] [PubMed] [Google Scholar]