Abstract
The JNKs are components of stress signaling pathways but also regulate morphogenesis and differentiation. Previously, we invoked a role for the JNKs in nerve growth factor (NGF)-stimulated PC12 cell neural differentiation (L. Marek et al., J. Cell. Physiol. 201:459-469, 2004; E. Zentrich et al., J. Biol. Chem. 277:4110-4118, 2002). Herein, the role for JNKs in neural differentiation and transcriptional regulation of the marker gene, NFLC, modeled in mouse embryonic stem (ES) cells was studied. NFLC-luciferase reporters revealed the requirement for NFLC promoter sequences encompassing base pairs −128 to −98 relative to the transcriptional start site as well as a proximal cyclic AMP response element-activating transcription factor binding site at −45 to −38 base pairs for transcriptional induction in NGF-treated PC12 cells and neurally differentiated ES cells. The findings reveal common promoter sequences that integrate conserved signal pathways in both PC12 cell and ES cell systems. To test the requirement for the JNK pathway in ES cell neurogenesis, ES cell lines bearing homozygous disruptions of the jnk1, jnk2, or jnk3 genes were derived and submitted to an embryoid body (EB) differentiation protocol. Neural differentiation was observed in wild-type, JNK2−/−, and JNK3−/− cultures but not in JNK1−/− EBs. Rather, an outgrowth of cells with epithelial morphology and enhanced E-cadherin expression but low NFLC mRNA and protein was observed in JNK1−/− cultures. The expression of wnt-4 and wnt-6, identified inhibitors of ES cell neurogenesis, was significantly elevated in JNK1−/− cultures relative to wild-type, JNK2−/−, and JNK3−/− cultures. Moreover, the Wnt antagonist, sFRP-2, partially rescued neural differentiation in JNK1−/− cultures. Thus, a genetic approach using JNK-deficient ES cells reveals a novel role for JNK1 involving repression of Wnt expression in neural differentiation modeled in murine ES cells.
Members of the mitogen-activated protein (MAP) kinase family, including the extracellular signal-regulated kinases (ERKs), the c-Jun N-terminal kinases (JNKs), and the p38 MAP kinases, have been extensively studied for their role in phosphorylating and activating transcription factors that regulate genes instrumental in cell growth, adaptation, differentiation, and transformation. The JNKs are encoded by three genes (jnk1, jnk2, and jnk3) and participate in cellular decisions following diverse forms of stress (14, 19) as well as the responses to growth factors, oncogenes, cytokines, and morphogens (15, 22, 32). Thus, the JNKs are key components of networks that mediate cell adaptation and apoptosis as well as cell fate decisions and differentiation. Moreover, studies in model organisms in which the jnk genes have been disrupted support a role for the JNKs in embryonic development and morphogenesis. In Drosophila melanogaster, JNK is required for two distinct development processes, epithelial sheet movement leading to dorsal closure and planar cell polarity (36). The JNK pathway also mediates a morphogenetic epithelial sheet movement termed convergent extension in vertebrates (39). Finally, mice lacking both jnk1 and jnk2 undergo mid-gestational embryonic lethality associated with defects in neural tube closure and deregulated neural apoptosis (17, 27). Recently, disruption of the mekk4 gene, encoding a proximal MAP kinase kinase kinase that functions within the JNK signaling pathway, was shown to induce marked neural tube defects manifested by exencephaly and spina bifida in mice (10). Thus, ample evidence supports a broad role for the JNK pathway in neural development and morphogenesis.
The aforementioned studies provide strong support for regulation of neural development programs by the JNK pathway, yet the systems employed are relatively intractable to molecular and biochemical approaches. By contrast, the PC12 pheochromocytoma cell line commits to a neural differentiation program in response to NGF and represents a clonal system that can be readily manipulated for biochemical and molecular studies. Previous studies from our laboratory and others have established the collaborative action of the ERKs and JNKs in NGF-induced neural differentiation and induction of the neural-specific gene neurofilament light chain (NFLC), modeled in PC12 cells (21, 37, 41). Using similar transfection approaches with molecular inhibitors of the JNK pathway, studies using P19 murine embryonal carcinoma (EC) cells provide additional evidence for a role for the JNK pathway in neural differentiation and induction of NFLC (31).
The aforementioned approaches using transfection of inhibitory components of the JNK pathway carry the caveats associated with overexpression techniques and do not provide information regarding the specific JNK family members that may participate in neural differentiation in these cell systems. In this study, we employed murine embryonic stem (ES) cells as an experimental system for molecular genetic analysis of cell differentiation that is generally considered to be highly representative of in vivo development (26, 34). ES cells represent a pluripotent cultured cell system that can be directed through in vitro protocols to differentiate into neurons that exhibit physiologic, morphological, and molecular properties of cultured primary neurons (4, 12, 16) and exhibit physiological functionality when transplanted into animals (7, 20). Moreover, ES cells bearing homozygously disrupted jnk genes can be readily derived from preimplantation murine embryos in which specific jnk genes have been disrupted by homologous recombination, thereby providing a molecular genetic approach to assess the role of the JNK pathway in neural differentiation. In this study, we present findings that unveil a novel requirement for JNK1 in neural differentiation modeled in ES cells.
MATERIALS AND METHODS
Derivation of JNK-deficient ES cells.
Breeder pairs of jnk1−/−, jnk2−/−, and jnk3−/− mice were generously provided by Richard Flavell (Yale University, New Haven, Conn.), and a colony of these mice was subsequently established in our animal facility at University of Colorado Health Sciences Center. ES cell lines bearing targeted disruptions of the jnk1, jnk2, or jnk3 gene were prepared from 3.5- to 4-day blastocysts as described previously (1). In brief, 4 days following the mating of JNK−/− breeder pairs, blastocysts were flushed from the uteri of pregnant females and transferred to tissue culture dishes with prepared feeder layers of mitomycin C-treated primary mouse embryo fibroblasts (PMEFs) in ES cell growth medium (Dulbecco's modified Eagle medium [DMEM] containing 15% fetal bovine serum [HyClone, Logan, UT], 150 μM monothioglycerol, and 1,000 U/ml ESGRO [murine leukemia inhibitory factor; Chemicon, Temecula, CA]). After ∼5 days of culture, inner cell mass outgrowths were mechanically dislodged, trypsinized, and seeded onto new PMEF feeder layers in 24-well plates. Wells with visible ES cell colonies were expanded in ES culture medium containing ESGRO on PMEF feeders. Two independent JNK1−/−, JNK2−/−, and JNK3−/− ES cell lines were established in this manner as well as a JNK1−/+ JNK2−/− ES cell line derived from a blastocyst obtained from the mating of JNK1−/+ JNK2−/− compound mutant mice. A wild-type ES cell line (wtES.1) was also isolated as a companion for the wild-type TC1 ES cells and as another control ES cell line that was isolated in our laboratory. All of the ES cell lines were adapted for feeder-free culture by propagating the cells on gelatin-coated dishes in the presence of ESGRO.
ES cell culture and differentiation.
Wild-type TC1 ES cells were propagated on gelatin-coated plates with ES cell culture medium in a humidified 5% CO2 incubator. To initiate neural differentiation of ES cells, the cells from a confluent 6-cm culture dish were seeded into 10-cm-diameter bacteriological petri dishes in 10 ml DMEM-F12 medium containing 1% horse serum (HyClone, Logan, UT) without ESGRO to induce the formation of embryoid bodies (EBs). After 2 days, the EBs were cultured for another 2 days in the same medium supplemented with 1 μM all-trans retinoic acid. These EBs, referred to as 2−/2+ EBs, were then plated on tissue culture dishes previously coated with polyornithine (15 μg/ml) and mouse laminin (20 μg/ml) in DMEM-F12 containing N2 supplement (Invitrogen, Carlsbad, CA). The medium was replaced every other day, and after 5 to 7 days extensive neurite outgrowth from the attached EBs was observed. Consistent with a recent report (18), we observed more extensive morphological neural differentiation and greater induction of NFLC protein with this method relative to previously reported neurogenesis protocols (4, 12, 20) that employed high concentrations of fetal bovine serum (L. Marek, C. Amura, and L. E. Heasley, unpublished observation).
Real-time quantitative PCR.
Total RNA (3 μg) was reverse transcribed in a volume of 20 μl using random hexamers and Moloney murine leukemia virus reverse transcriptase according to the manufacturer's protocol (Invitrogen). Aliquots (1 μl) of reverse transcription reactions were subjected to PCR in 25-μl reaction mixtures with SYBR green Jumpstart Taq Readymix (Sigma, St. Louis, MO) and the primers listed in Table 1 using an I Cycler (Bio-Rad, Hercules, CA). Initial real-time PCR amplification products were resolved by electrophoresis on 5% polyacrylamide gels to verify that the primer pairs amplified a single product of the predicted (70 to 80 bp) size. GAPDH mRNA levels were measured by quantitative real-time PCR (RT-PCR) as a control housekeeping gene for normalization of the different mRNA expressions and presented as mRNA levels in arbitrary units.
TABLE 1.
Primers for quantitative RT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| oct4 | AAGCTGATTGGCGATGTGAGTGAT | CCGTGTGAGGTGGAGTCTGGAG |
| nanog | GGCAGCCCTGATTCTTCTACCA | CTCCTCAGGGCCCTTGTCAGC |
| brachyury | TGGAAATATGTGAACGGGGAGTGG | ATTGGGCGAGTCTGGGTGGATGTA |
| gata4 | GCCCTCTTTGTCATTCTTCGC | TGCTTTCTGCCTGCTACACACC |
| fgf5 | GATGGCAAAGTCAATGGCTCC | TTCCTACAATCCCCTGAGACACAG |
| nflc | CAAGGACGAGGTGTCGGAAA | AGGCTTCGATCTCCAGGGTC |
| wnt4 | CGCCATCTCTTCAGCAGGTG | GGTCACAGCCACACTTCTCCAG |
| wnt5a | CCAACTGGCAGGACTTTCTCAAG | TGTCTTCGCACCTTCTCCAATG |
| wnt6 | CAGCAGGACATCCGAGAGACAG | TGGAACAGGCTTGAGTGACCG |
| wnt7a | GCTTCGCCAAGGTCTTCGTG | CCCGCCTCGTTATTGTGTAAGTTC |
| bmp4 | AAGCATCACCCACAGCGGTC | CCACAATCCAATCATTCCAGCC |
| gapdh | CGTGGAGTCTACTGGCGTCTTCAC | CGGAGATGATGACCCTTTTGGC |
Immunoblot analysis.
Whole-cell extracts were prepared in MAP kinase lysis buffer (0.5% Triton X-100, 50 mM β-glycerophosphate [pH 7.2], 0.1 mM sodium vanadate, 2 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonylurea, 2 μg/ml leupeptin, and 4 μg/ml aprotinin) supplemented with 300 mM NaCl followed by brief sonication with a microprobe sonicator. The proteins were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and transferred to nitrocellulose. The filters were blocked in Tris-buffered saline (10 mM Tris-Cl, pH 7.4, 140 mM NaCl) containing 0.1% Tween-20 (TTBS) and 3% nonfat dry milk and then incubated with blocking solution containing the indicated antibodies at 1 μg/ml for 12 to 16 h. The filters were extensively washed in TTBS, and bound antibodies were visualized with alkaline phosphatase-coupled secondary antibodies and Lumi-Phos reagent (Pierce, Rockford, IL) according to the manufacturer's directions.
Immunofluorescence analysis.
Neural cultures on gelatin-coated glass Permanox 8-well chamber slides (Nalge Nunc Intl., Naperville, IL) were fixed with 4% paraformaldehyde-3% sucrose for 10 min, washed with phosphate-buffered saline (PBS), permeabilized for 10 min with 0.1% Triton X-100 in PBS, and blocked with 10% goat serum. After three washes with PBS, the slides were incubated with 1 μg/ml anti-NFLC (Chemicon) or isotype control (Jackson Immunoresearch, West Grove, PA) for 1 h, washed five times, and incubated with Alexa Fluor-labeled secondary antibodies (Molecular Probes, Eugene, OR) for an additional 1 h. After extensive washing, slides were mounted with Vectashield (Vector Laboratories, Burlingame, CA) and cells were examined with a Zeiss immunofluorescence microscope.
NFLC promoter activity in NGF-stimulated PC12 cells.
The rat NFLC promoter-firefly luciferase reporter construct containing base pairs −650 and +82 relative to the start site and progressively 5′-truncated reporters (−155, −128, −113, −103, −98, and −39) were generated by PCR as previously described (41). PC12 cells were transfected by electroporation as previously described (41) with 2 μg NFLC-Luc reporter construct, 0.5 μg thymidine kinase (TK)-renilla plasmid, 10 μg of herring sperm DNA and were plated on rat tail collagen-coated dishes in full growth medium (DMEM containing 5% horse serum, 2.5% fetal bovine serum, and 2.5% newborn calf serum). After 24 h, the medium was changed to DMEM containing 1% horse serum and the cells were stimulated with or without 100 ng/ml NGF for 3 days as described previously (41). Cell extracts were prepared in 500 μl of luciferase passive lysis buffer (Promega, Madison, WI), and both firefly and renilla luciferase activities were measured in 80-μl aliquots with a dual luciferase assay according to the manufacturer's instructions using a Luminoskan Ascent (Thermo Electron Corporation, Franklin, MA). The firefly luciferase data were normalized for transfection efficiency with the respective renilla luciferase activities.
Stable transfection of ES cells.
Parental TC1 ES cells were transfected with 2 μg of the NFLC-luciferase (NFLC-luc) reporter constructs (full length, −128, and −98), the full-length NFLC reporter in which the cyclic AMP response element/activating transcription factor (CRE/ATF) site is mutated (41), or empty pA3luc vector as well as pMSCVpuro (Clontech, Palo Alto, CA) as a selectable marker using Lipofectamine 2000 reagent according to the manufacturer's protocol (Promega). ES cells were plated on gelatin-coated dishes in ES cell growth medium for 48 h. The growth medium was subsequently replaced with medium containing 2 μg/ml puromycin for the selection of transfected ES cell clones. After approximately 2 weeks of selection, clones were mechanically isolated with a pipette, trypsinized, and expanded in the same media. Genomic DNA was prepared from individual clones using DNAeasy tissue kits (QIAGEN, Valencia, CA). Transfectants bearing the different NFLC reporters were characterized by PCR with forward primers (−401, 5′-GGTACCGCAGAAAGGGCGAGCCAGGGG-3′; −128, 5′-GGTACCTTTGCTCTTGCGCAGAATCC-3′; −98, 5′-GGTACCGCTGCAGCAGCACGCTGC-3′; −38, 5′-GGTACCGAGTCCCGGCGTATAAAT-3′) and a reverse primer that annealed within the luciferase coding sequences of pA3Luc (5′-TCCAGAGGAATTCATTATCAGTGC-3′). Individual ES cell clones positive for the respective NFLC reporter constructs were pooled and submitted to the neural differentiation protocol described above. Cells were collected with ice-cold PBS and lysed in 500 μl of luciferase passive lysis buffer, and aliquots (80 μl) of the supernatants were assayed for luciferase activity with luciferase assay substrate. The luciferase activity was normalized to cellular protein measured by the Bradford colorimetric assay.
To re-express JNK1 in JNK1−/− ES cells, a cDNA encoding human JNK1α1 was ligated into the murine stem cell viral vector pMSCVpuro, and the resulting vector as well as empty pMSCVpuro were packaged into replication-incompetent retroviruses (9). JNK1−/−.4 ES cells were infected with the resulting retroviruses and selected for resistance to puromycin, and individual clones were screened for expression of the JNK1 polypeptide by immunoblotting with anti-JNK1 antibodies.
RESULTS
We have previously employed NFLC as a prototypic marker of neural differentiation in PC12 pheochromocytoma cells where morphological differentiation and transcriptional induction of NFLC are stimulated by NGF (21, 41). Using this cultured cell system, we have identified critical regulatory regions within the NFLC promoter that integrate stimulatory inputs from the ERK and JNK pathways (41). Moreover, an integrated requirement for the ERK and JNK pathways has been extended to additional genes whose expression is specifically induced by NGF in PC12 cells (21). Because neurogenesis modeled in ES cells is thought to more closely mimic in vivo neurogenesis (16, 34), we assessed the JNK pathway requirement for neural differentiation and NFLC induction in ES cells.
Morphological differentiation and induction of NFLC in neuronally differentiated ES cells.
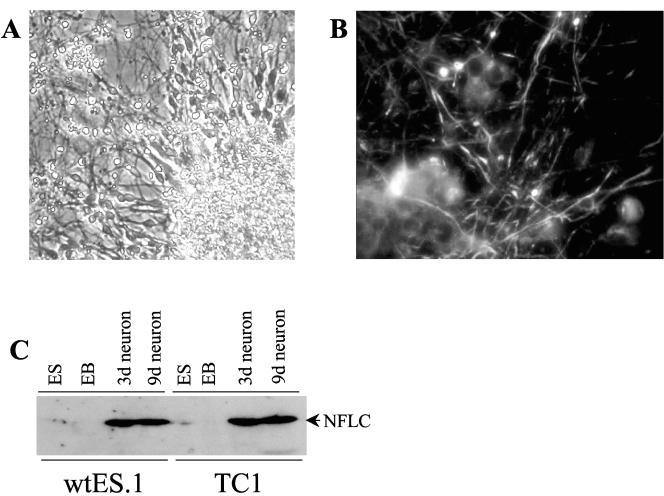
The mouse ES cell line TC1 (11) was submitted to the differentiation protocol described in Materials and Methods where the cells are cultured in suspension in serum-free medium as embryoid bodies (EBs) for 4 days, the final 2 in the presence of 1 μM retinoic acid (referred to as 2−/2+ EBs). When the 2−/2+ EBs are plated on a polyornithine-laminin substrate, the EBs attach and neurite outgrowth is observed within 3 days of culture and is extensive after 6 to 8 days of culture (Fig. 1A). Because the intact EBs were plated and allowed to further differentiate, it was difficult to precisely quantify the percentage of cells undergoing neural differentiation. We estimate that greater than 80% of visible single cells were neurons by morphology. Initial studies were performed with an EB protocol where the ES cells are differentiated for 4 days in DMEM containing 10% fetal bovine serum followed by an additional 4 days in the same medium containing 1 μM retinoic acid. Similar to a recent report (18), we observed greatly enhanced neurogenesis with the serum-depleted 2−/2+ protocol relative to EB differentiation performed in the presence of 10% fetal bovine serum (L. Marek, C. Amura, and L. E. Heasley, unpublished observation). The ES-derived neurons stained positively for the neural specific protein, NFLC (Fig. 1B). In addition, immunoblot analysis demonstrated marked expression of NFLC in ES-derived neurons after 3 or 9 days of culture but not in undifferentiated ES cells or 2×/2+ EBs (Fig. 1C). Thus, neurons expressing NFLC are readily derived by in vitro differentiation of ES cells.
FIG. 1.
Neural differentiation and induction of NFLC protein in ES cells. Undifferentiated TC1 ES cells were submitted to the neural differentiation protocol described in Materials and Methods. (A) Cultures following 6 days of culture on polyornithine-laminin-coated dishes were photographed with a phase-contrast microscope revealing dense outgrowth of neurites from the attached EB in the lower right corner. (B) TC1 ES cell-derived neural cultures on polyornithine-laminin-coated slides were fixed and stained for NFLC as described in Materials and Methods. Staining was visualized with a fluorescence microscope at 400× magnification. (C) TC1 ES cells as well as wtES.1 ES cells derived in our lab were cultured as 2−/2+ EBs and then further cultured for 3 or 9 days on polyornithine-laminin-coated dishes. Whole-cell extracts from the indicated ES cells, 2−/2+ EBs, and neural cultures were prepared and immunoblotted for NFLC with the NR4 monoclonal antibody (Sigma Chemical Co.).
Conserved promoter sequences are required for NFLC regulation in NGF-differentiated PC12 cells and ES cell-derived neurons.
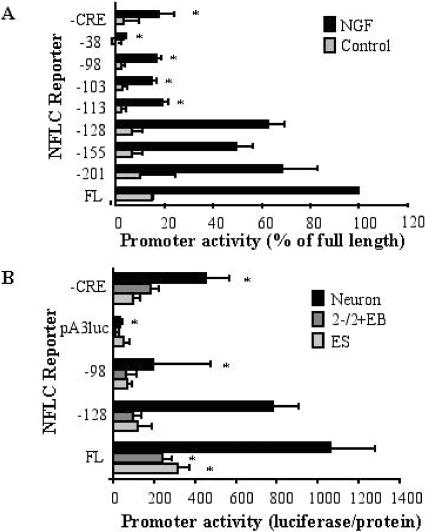
We have previously defined the regulatory regions within the NFLC promoter that mediate induction in PC12 cells by NGF (41). As shown in Fig. 2A, transient transfection experiments using PC12 cells and a panel of NFLC promoter luciferase constructs progressively truncated from the 5′ end indicate that truncation beyond −128 base pairs relative to the transcription start site significantly reduced NFLC-stimulated promoter activity. In the present study, additional reporter constructs where the NFLC promoter was truncated to −113 and −103 base pairs relative to the transcription start site exhibited decreased induction by NGF and indicate the presence of a critical promoter sequence that resides between −128 and −113 for NGF-stimulated promoter activity (Fig. 2A). Deletion of a CRE/ATF-like site residing at −45 to −38 base pairs by truncation to −38 base pairs completely eliminates NGF induction of luciferase activity by the reporter. Also, site-directed mutagenesis of the CRE/ATF-like site within the context of the full-length NFLC promoter reduces, but does not eliminate, promoter activity (Fig. 2A). Thus, NGF induction of the NFLC promoter is accomplished by the action of two independent promoter regions, the CRE/ATF site at −45 to −38 base pairs and a novel site residing between −128 and −113 base pairs relative to the transcription start site. We previously established that NGF-stimulated induction of the NFLC promoter was partially inhibited by the JNK inhibitor SP600125 (21). To define which promoter region is required for JNK-dependent induction of NFLC expression, the degree of inhibition of SP600125 on NGF induction of NFLC-luc reporters containing both promoter regions (−128), only the CRE/ATF-like site (−103) or only the site between −128 and −113 (CRE mutant) was measured. SP600125 (2 μM) reduced the NGF-stimulated induction of the −128, −103, or CRE mutant reporter to 64.1% ± 16.6%, 63.4% ± 14.0%, and 72.4% ± 3.0% of control, respectively. The approximately equal inhibition of the three NFLC promoter constructs by the JNK inhibitor suggests that the JNK pathway may integrate equally at both promoter regions. Alternatively, the JNK pathway may regulate a transcriptional coactivator that interacts with the resident transcription factors that bind to both promoter regions. A recent study (35) provides precedent for the ability of MAP kinases, including JNKs, to phosphorylate coactivators such as SRC-3 as a mechanism for transcriptional regulation.
FIG. 2.
NFLC promoter activity in NGF-stimulated PC12 cells and neuronally differentiated ES cells. (A) PC12 cells were transiently transfected by electroporation with the indicated NFLC-luciferase reporter constructs and a TK-renilla-luciferase vector to measure transfection efficiency. The cells were subsequently incubated with or without NGF in 1% horse serum-DMEM for 3 days as described previously (41). Firefly luciferase activity was normalized to the Renilla luciferase activity measured in replicate aliquots, and the data are presented as the percent of full-length promoter activity in NGF-stimulated cells (means ± standard errors of the means), where an asterisk indicates a statistically significant difference (P < 0.05) from full-length promoter activity. (B) TC1 ES cells stably transfected with the indicated NFLC-luciferase reporters were cultured as ES cells, 2−/2+ EBs, or neural cultures, and luciferase activity was measured. The luciferase activity was normalized to cellular protein in the extracts, and the data are the means and standard errors of the means of at least five independent experiments, where an asterisk indicates a statistically significant difference (P < 0.05) in promoter activity relative to that of the full-length promoter (FL).
To define the NFLC promoter sequences required for induction of this gene during neural differentiation of ES cells, TC1 ES cells were stably transfected with selected NFLC-luciferase reporters (Fig. 2B) as described in Materials and Methods. These NFLC-luciferase transgenic ES cell lines were subsequently cultured as 2−/2+ EBs, plated on polyornithine-laminin substrate, and cultured for 5 days. As shown in Fig. 2B, luciferase activity was significantly induced in neuronal TC1 cells transfected with the full-length promoter (FL) relative to 2−/2+ EBs or undifferentiated ES cells. No neural induction of luciferase activity was observed in TC1 cells stably transfected with empty pA3luc (Fig. 2B). A similar induction of luciferase activity was observed in neuronally differentiated TC1 cells transfected with the NFLC-reporter construct truncated to −128 base pairs relative to the full-length promoter, but induction of luciferase activity directed by the −98 promoter construct or the full-length promoter in which the CRE/ATF-like site was mutated (CRE) was significantly reduced (Fig. 2B). Thus, this experiment indicates that induction of the NFLC promoter during neural differentiation of ES cells requires the same promoter regions necessary for induction of the NFLC promoter in PC12 cells stimulated with NGF.
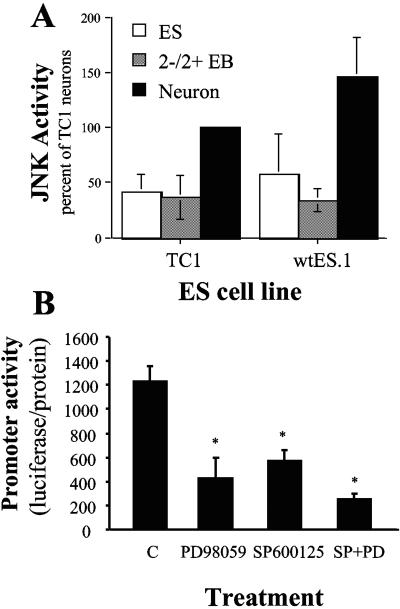
We previously showed that JNK activity is increased in PC12 cells undergoing neural differentiation upon incubation with NGF and that induction of NFLC-luciferase activity in NGF-treated PC12 cells requires the integration of the ERK and JNK pathways (41). Analysis of JNK activity in wild-type TC1 and wtES.1 ES cells, 2−/2+ EBs, and neural cultures revealed elevated JNK activity in neural cultures relative to ES cells or 2−/2+ EBs (Fig. 3A). When TC1 cells transfected with the full-length NFLC-luciferase construct were treated during the final differentiation steps on polyornithine-laminin-coated dishes with 2 μM SP600125, a pharmacological JNK inhibitor, a significant reduction of luciferase activity was observed (Fig. 3B). Likewise, treatment of the EBs with 25 μM PD98059, a MEK inhibitor, also resulted in significantly reduced reporter activity. Treatment with both SP600125 and PD98059 yielded a further reduction in NFLC promoter activity, a finding that is consistent with our previous findings demonstrating the required collaboration of both JNK and ERK MAP kinases in NGF-stimulated NFLC-luciferase induction in PC12 cells (21, 41).
FIG. 3.
Increased JNK activity in ES cell-derived neurons is required for induction of the NFLC promoter. (A) Extracts were prepared from the indicated ES cells, 2−/2+ EBs, and 8-day neural cultures with MAP kinase lysis buffer and assayed for JNK activity with the glutathione S-transferase-c-Jun (1 to 79) adsorption assay as previously reported (8). The data are the means and standard errors of the means of three independent experiments and are presented as the percent of the activity in TC1 ES cell-derived neurons. (B) ES cells stably transfected with full-length NFLC-luc were submitted to the neural differentiation protocol, plated onto polyornithine-laminin-coated plates, and treated with or without 25 μM PD098059 and/or 2 μM SP600125 for 5 days. Luciferase activity and cellular protein were measured, and the data are the means and standard errors of the means of three independent experiments, where an asterisk indicates a statistically significant difference from untreated cells.
Generation of JNK-deficient ES cell lines.
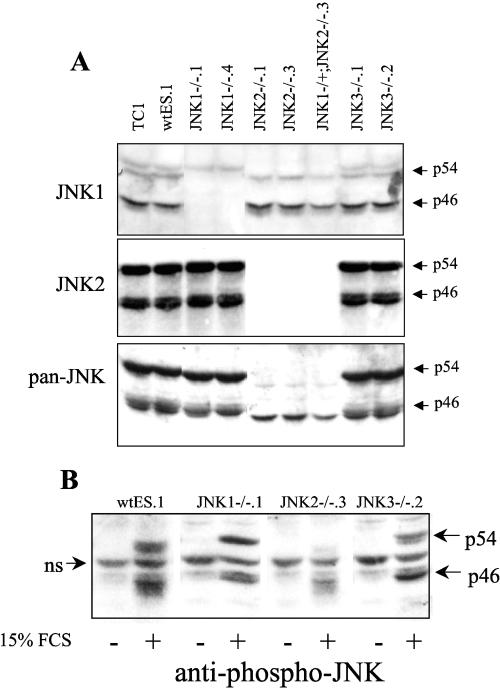
ES cells derived from JNK-deficient embryos would permit a molecular genetic approach for dissection of the role of the JNK signaling pathway in neuronal differentiation. Thus, ES cells in which the jnk1, jnk2, or jnk3 gene was homozygously disrupted were generated from blastocysts (embryonic day 3.5 to 4.0) as described in Materials and Methods. A wild-type ES cell line (wtES.1) was established as a companion for the wild-type TC1 ES cell line as well as two independent isolates of JNK1−/−, JNK2−/−, and JNK3−/− lines and a single JNK1−/+ JNK2−/− ES cell line. The panel of ES cell clones lacking jnk1, jnk2, or jnk3 was characterized by immunoblot analysis with antibodies that selectively detect JNK1 or JNK2 as well as an antibody that recognizes all JNK polypeptides. As shown in Fig. 4A, relative to extracts from wild-type ES cells, extracts from the JNK1−/− ES cells lacked a predominant p46 JNK1 species and a more minor p54 JNK1 species. The JNK1−/+ JNK2−/− ES cell line showed the predicted decrease in JNK1 expression due to the expression of a single jnk1 allele. A p54 JNK species that reacts with the JNK1 antibody was retained in JNK1−/− cell lines and represents weak cross-reactivity of the antibody with p54 JNK2 polypeptides. The JNK2−/− ES cell clones and the JNK1−/+ JNK2−/− ES cell line showed a complete lack of the p46 and p54 JNK2 proteins that were detected in wild-type ES cell extracts with the specific JNK2 antibody (Fig. 4A). The pan-reactive JNK antibody detected a reduction of a p46 species in the JNK1−/− cells and loss of both p46 and p54 JNK polypeptides in JNK2−/− extracts. No detectable loss of a putative JNK polypeptide in the JNK3−/− extracts was observed with the pan-reactive antibody, suggesting that JNK3 proteins are not highly expressed in ES cells. As shown in Fig. 4B, addition of 15% fetal bovine serum to wild-type and JNK3−/− ES cell cultures that had been serum restricted for 4 h strongly increased phosphorylation of multiple p46 and p54 JNK proteins. Similar analysis of extracts from JNK1−/− ES cells revealed increased phosphorylation of the p46 and p54 JNK2 proteins that are retained in the JNK1−/− cells. Likewise, the p46 JNK1 proteins that are retained in the JNK2−/− cells are phosphorylated in a fetal bovine serum-dependent manner. Thus, ES cells bearing disruptions of jnk1 or jnk2 retain serum-stimulated phosphorylation of the remaining JNK proteins.
FIG. 4.
JNK expression and activity in wild-type and JNK−/− ES cells. (A) Extracts were prepared from the indicated ES cell lines with MAP kinase lysis buffer and immunoblotted with monoclonal antibodies against JNK1 (F3; Santa Cruz), JNK2 (D2; Santa Cruz), or a polyclonal antibody that detects all of the known JNK polypeptides (Cell Signaling Biotechnology, Inc., Beverly, MA). (B) The indicated ES cell lines were serum restricted for 4 h and stimulated with 15% fetal bovine serum for 30 min. Extracts were prepared and immunoblotted with anti-phospho-JNK antibodies (Cell Signaling Technology); ns indicates a nonspecifically reacting protein. FCS, fetal calf serum.
The different ES cell lines lacking jnk1, jnk2, or jnk3 exhibited growth properties and morphologies equivalent those of the wild-type ES cell lines (L. Marek, C. Amura and L. E. Heasley, unpublished observation). Moreover, as assessed by quantitative RT-PCR, the stem cell marker Nanog (23) was equally expressed in the wild-type and JNK−/− ES cell lines (Table 2). Expression of a distinct stem cell marker, Oct4 (5), was significantly increased in the JNK1−/− and JNK1−/+ JNK2−/− ES cell lines relative to the wild-type, JNK2−/−, and JNK3−/− ES cells (Table 2). In this regard, Niwa et al. (25) have demonstrated that a 50% increase in Oct4 expression in ES cells can trigger differentiation into primitive endoderm or mesoderm. However, measurement of markers of primitive endoderm (Fgf5 and Gata4) or mesoderm (brachyury) in the panel of ES cell lines did not reveal evidence for statistically significant induction of these marker genes and indicates that the JNK1−/− ES cells have not undergone substantial differentiation towards either of these lineages (Table 2).
TABLE 2.
Gene expression in wild-type and JNK−/− ES cell linesa
| ES cell | Oct4 | Nanog | Brachyury | Gata4 | Fgf5 |
|---|---|---|---|---|---|
| TC1 | 0.8 ± 0.1 | 0.7 ± 0.2 | 0.8 ± 0.2 | 0.9 ± 0.1 | 1.2 ± 0.6 |
| wtES.1 | 1.1 ± 0.2 | 1.2 ± 0.2 | 1.0 ± 0.2 | 1.1 ± 0.1 | 0.7 ± 0.3 |
| JNK1−/− .1 | 2.4 ± 0.6* | 1.2 ± 0.3 | 1.2 ± 0.4 | 2.3 ± 1.4 | 1.3 ± 0.4 |
| JNK1−/− .4 | 2.0 ± 0.4* | 1.2 ± 0.2 | 1.2 ± 0.5 | 0.9 ± 0.2 | 2.4 ± 1.3 |
| JNK2−/− .1 | 1.2 ± 0.2 | 1.6 ± 0.5 | 0.8 ± 0.3 | 1.4 ± 0.6 | 0.9 ± 0.3 |
| JNK2−/− .3 | 1.1 ± 0.3 | 2.7 ± 0.4* | 1.6 ± 0.6 | 1.2 ± 0.7 | 1.5 ± 0.9 |
| JNK1−/+ JNK2−/− | 1.6 ± 0.3* | 1.8 ± 0.5 | 3.3 ± 1.9 | 1.9 ± 0.7 | 1.9 ± 0.5 |
| JNK3−/− .1 | 1.3 ± 0.2 | 0.8 ± 0.2 | 1.3 ± 0.3 | 3.1 ± 1.1 | 1.7 ± 0.4 |
| JNK3−/− .2 | 1.2 ± 0.5 | 2.0 ± 0.9 | 1.6 ± 0.4 | 1.1 ± 0.4 | 1.2 ± 0.7 |
Total RNA was prepared from the indicated ES cell lines, and expression of the genes was measured by quantitative RT-PCR as described in Materials and Methods. The values were corrected for GAPDH mRNA expression in replicate samples and normalized to the expression in the wtES.1 and TC1 ES cell lines. The data are presented as mean relative expressions and standard error of the mean of three independent RNA preparations, where an asterisk indicates a significant difference from the TC1 value (P < 0.05) by Student's t test.
JNK1-deficient ES cells exhibit inhibited neurogenesis associated with increased Wnt-4/Wnt-6 expression.
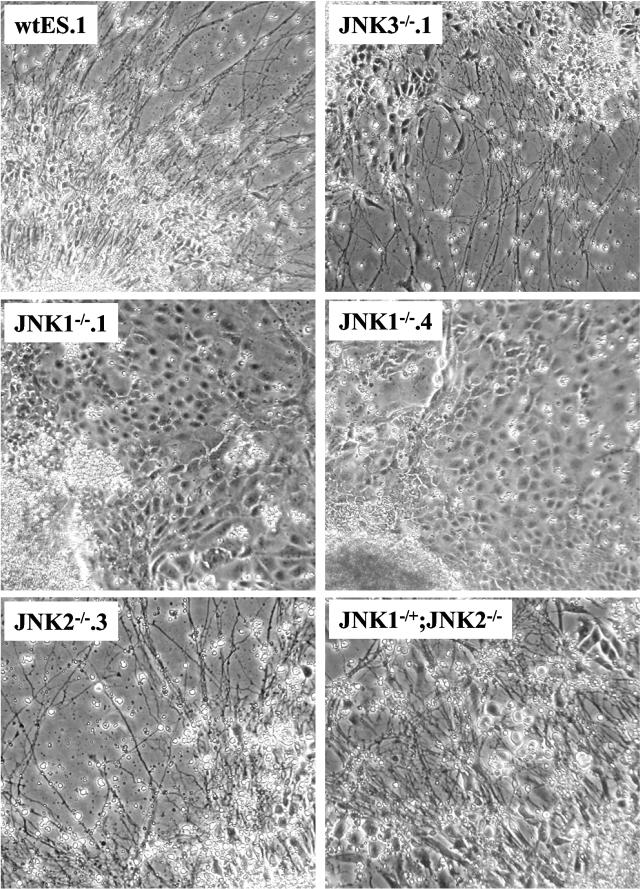
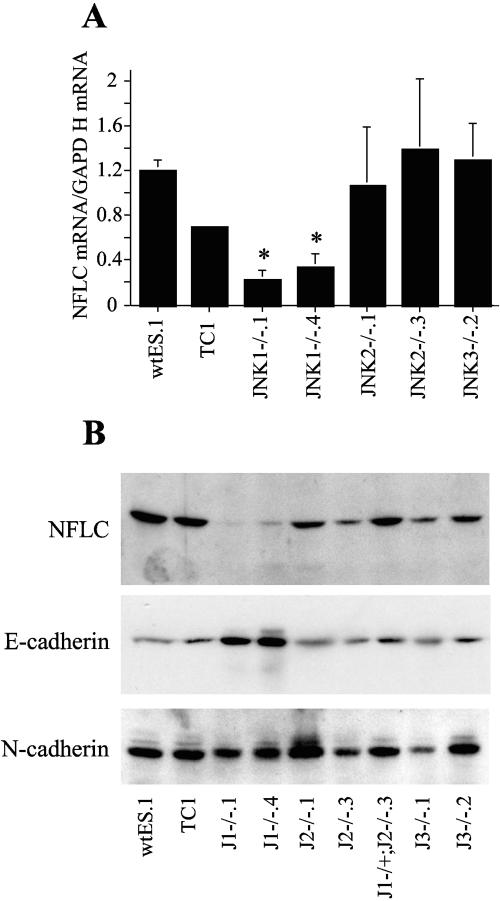
Participation of the JNK pathway in neural differentiation was investigated in the wild-type and JNK-deficient ES cell lines using the in vitro neural differentiation protocol. Compared to 2−/2+ EBs derived from the wild-type, JNK2−/−, and JNK3−/− ES cell lines which exhibited pronounced morphological neural differentiation following culture on polyornithine-laminin substrate (Fig. 5), little or no neurite outgrowth was observed from the plated 2−/2+ EBs derived from JNK1−/−.1 or JNK1−/−.4 ES cells. Rather, an extensive outgrowth of cells with an epithelial morphology was noted. As a molecular measure of neural differentiation distinct from morphological criteria, the levels of NFLC mRNA were measured by quantitative RT-PCR. Figure 6A demonstrates a statistically significant decrease in NFLC mRNA in JNK1−/− cultures, while JNK2−/− and JNK3−/− cultures expressed NFLC mRNA to a level equal to that of the wild-type ES cell-derived cultures. In addition, cell extracts were prepared from the different ES cell-derived neural cultures and immunoblotted for NFLC protein (Fig. 6B). Relative to the expression level of NFLC protein observed in neural cultures derived from wild-type ES, JNK2−/−, JNK1−/+ JNK2−/−, and JNK3−/− cells, significantly reduced levels of NFLC protein were observed in the cultures derived from the replicate JNK1−/− ES cell clones. The expression of N-cadherin protein was approximately equal among the different samples and served as a loading control for the experiment. Notably, increased expression of E-cadherin protein was detected in extracts from JNK1−/− ES cell-derived cultures, consistent with the epithelial morphology manifested by the cellular outgrowths. Thus, JNK1−/− ES cells exhibit an inhibited capacity to undergo neural differentiation relative to wild-type, JNK2−/−, and JNK3−/− ES cells accompanied by increased morphological and molecular evidence of epithelial differentiation.
FIG. 5.
Inhibited neural differentiation by JNK1−/− ES cells. The indicated wild-type and JNK−/− ES cell lines were submitted to the neural differentiation protocol described in Materials and Methods, and 7 days after plating the cultures were photographed under a phase-contrast microscope.
FIG. 6.
NFLC expression is reduced and E-cadherin expression is increased in cell cultures derived from JNK1−/− ES cells. (A) Total RNA was purified from the indicated ES cell-derived cultures, and mRNAs for NFLC and GAPDH were measured by quantitative RT-PCR. The data were normalized for expression of the housekeeping gene GAPDH and are the means and standard errors of the means of three to five independent experiments. (B) Whole-cell extracts prepared from the indicated ES cell-derived neural cultures were submitted to SDS-PAGE and immunoblot analysis using a mouse anti-NFLC antibody (NR4), a monoclonal E-cadherin antibody (BD Biosciences, Palo Alto, CA), or a monoclonal N-cadherin antibody (BD Biosciences).
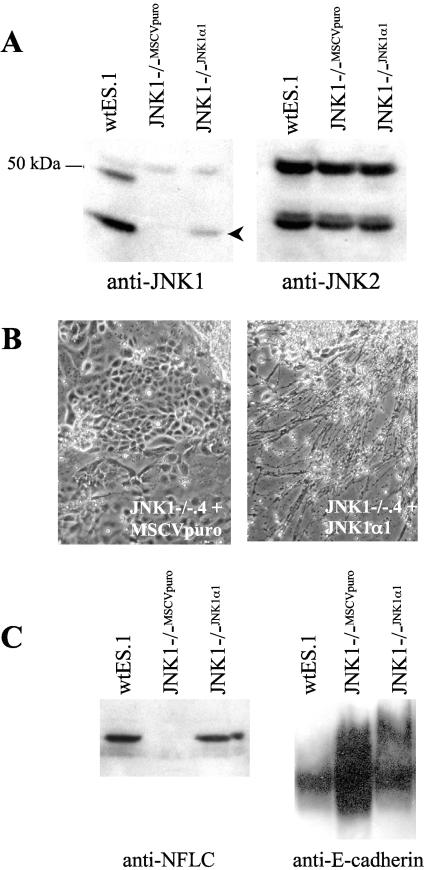
To establish that the phenotype exhibited by the JNK1−/− ES cells is due to the JNK1 deficiency, JNK1 protein was re-expressed in JNK1−/−.4 ES cells using retroviral-mediated gene transfer of a JNK1α1 (p46JNK1α) cDNA (see Materials and Methods). Figure 7A shows that the p46JNK1α protein is present in JNK1−/−.4 ES cells transduced with the pMSCVpuro-JNK1α1 vector relative to JNK1−/−.4 cells transduced with the empty pMSCVpuro vector. The level of JNK1 expression in the JNK1α1 transfectant is less than the endogenous p46 JNK1 expression in wtES.1 cells. When the JNK1−/−.4 cells transfected with JNK1α1 were submitted to the 2−/2+ differentiation protocol and plated on polyornithine-laminin-coated dishes, robust neurite outgrowth (Fig. 7B) and induction of NFLC protein (Fig. 7C) were observed relative to that of the JNK1−/−.4 cells transduced with the empty retroviral vector which exhibited the epithelial phenotype and no neural differentiation. In addition, re-expression of JNK1 significantly reduced the expression of E-cadherin in the differentiated cultures (Fig. 7C), indicating that re-expression of JNK1α1 in JNK1−/−.4 ES cells restored the neural differentiation program to that of wild-type ES cells.
FIG.7.
Re-expression of JNK1 in JNK1−/− ES cells restores neural differentiation and NFLC protein induction. (A) Extracts were prepared from JNK1−/−.4 ES cells transduced with the empty retroviral vector (JNK1−/−MSCVpuro) or with a retroviral vector encoding JNK1α1 (JNK1−/−JNK1α1) as well as wtES.1 cells to define endogenous levels of JNK1 proteins. The extracts were submitted to SDS-PAGE and immunoblotted for JNK1 or JNK2 as a loading control with the antibodies described in the legend to Fig. 4. The arrowhead indicates the migration position of the transduced p46 JNK1 polypeptide. The immunoreactive band of ∼55 kDa present in all three lanes of the JNK1 blot represents cross-reactivity of the JNK1 antibody for p54 JNK2 proteins. (B) The indicated JNK1−/−.4 ES cell transfectants were submitted to the 2−/2+ differentiation protocol, the resulting EBs were plated on polyornithine-laminin-coated dishes, and the cultures were photographed following 7 days of culture. (C) Extracts were prepared from differentiated cultures derived from the JNK1−/−MSCVpuro or JNK1−/−JNK1α1 ES cell transfectants or wtES.1 cells and submitted to immunoblot analysis for NFLC and E-cadherin.
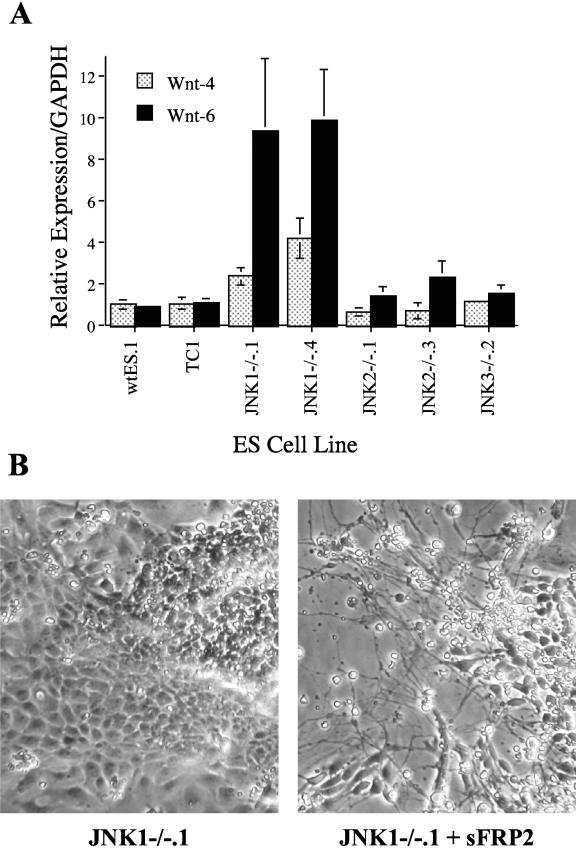
In ES cells, the Wnt pathway has been invoked as a negative regulator of ES-derived neurogenesis (3, 13). Forced expression of Wnt-1 in ES cells inhibited neural differentiation while expression of the Wnt antagonist, sFRP2, enhanced neural differentiation (3). Based on this background, we screened the expression of multiple Wnt genes in cellular outgrowths from wild-type and JNK1−/− EBs to test the hypothesis that the expression of specific Wnt genes was dysregulated in the JNK1−/− cells. Quantitative RT-PCR analysis revealed a statistically significant 10- and ∼3-fold increase in the expression of Wnt-6 and Wnt-4 mRNAs, respectively, in the cellular outgrowths of JNK1−/− EBs relative to the neural cultures derived from wild-type, JNK2−/−, and JNK3−/− ES cells (Fig. 8A). By contrast, the mRNA expression levels of Wnt-5a and Wnt-7a were not different among the various cultures (data not shown). BMP4 has been invoked as a mediator of Wnt inhibition of neurogenesis (24). In addition, Ying et al. (40) have previously demonstrated that addition of BMP4 to wild-type ES cells undergoing neural differentiation suppressed the development of neural precursors and neurons and instead induced the appearance of nonneural differentiated cells with an epithelial morphology similar to that observed in the JNK1−/− cultures. Quantitative RT-PCR analysis revealed a 5.8- ± 1.0- and 3.0- ± 0.8-fold increase in BMP4 mRNA expression in cultures of EBs derived from JNK1−/−.1 and JNK1−/−.4 ES cells, respectively, relative to wtES.1 and TC1 cultures. These findings indicate that JNK1 functions to repress a Wnt-4/Wnt-6 and BMP4 signaling axis in ES cells undergoing neural differentiation.
FIG.8.
Increased expression of Wnt-4 and Wnt-6 is observed in JNK1−/− EB cultures and the Wnt antagonist, sFRP2, increases morphological neural differentiation. (A) The indicated ES cell lines were submitted to the 2−/2+ EB differentiation protocol and subsequently cultured on polyornithine-laminin-coated dishes for 5 to 7 days as described in Materials and Methods. Total RNA was purified, and the expression of Wnt-4 and Wnt-6 mRNAs were measured as described in Materials and Methods by quantitative RT-PCR using the primers listed in Table 1. The results are the average relative expression and standard errors of the means normalized for GAPDH of three to five independent experiments reflecting three independent RNA preparations. (B) JNK1−/−.1 ES cells were submitted to the 2−/2+ EB protocol and subsequently cultured on polyornithine-laminin-coated dishes in medium with or without 1 μg/ml sFRP2 (R&D Systems, Minneapolis, MN) for 6 days and photographed through a phase-contrast microscope.
To determine if the increased Wnt-4/Wnt-6 expression mediated inhibition of neurogenesis, the JNK1−/− EBs were cultured in the presence or absence of exogenous sFRP2. As shown in Fig. 8B, inclusion of recombinant sFRP2 in the culture medium resulted in focal regions of morphological neural differentiation of the plated 2−/2+ EBs derived from JNK1−/−.1 ES cells relative to control JNK1−/−.1 cultures which exhibit only the epithelial phenotype. Thus, this result indicates that the increased expression of Wnt-4/Wnt-6 in JNK1−/− cultures is a likely mediator of inhibited neurogenesis as the Wnt antagonist, sFRP2, is able to reverse this phenotype. Interestingly, neither NFLC mRNA nor protein was increased in the sFRP2-treated cultures (data not shown). We interpret this result as evidence for a causative role for the increased Wnt-4/Wnt-6 in inhibition of neurogenesis in JNK1−/− EBs but that JNK1 activity, which is still lacking in sFRP2-treated JNK1−/− cells, is required for induction of NFLC mRNA and protein. Disruption of the NFLC gene in mice reveals that neurogenesis readily proceeds despite the absence of the NFLC protein (42).
DISCUSSION
Previous studies from our laboratory (21, 41) and others (37) have invoked a collaborating role for the JNK pathway with the ERK pathway in neurogenesis modeled in PC12 cells. Using molecular and pharmacological inhibitors, the requirement of the JNKs for full induction of a variety of NGF-stimulated genes including NFLC in PC12 cells has been demonstrated (21). These findings are consistent with other reports demonstrating a requirement for the JNK pathway in neurogenesis and NFLC regulation modeled in the distinct in vitro system, P19 EC cells (31). Using these cells, the investigators demonstrate that a TAK1-MKK4-JNK signaling pathway is activated and participates in the regulation of the neural differentiation of P19 embryonal carcinoma cells (2). Also, ES cells in which the JNK pathway-specific scaffold protein, JSAP1, is homozygously disrupted exhibit reduced in vitro neurogenesis (38). Thus, the findings from our present study with JNK1-deficient ES cells confirm the role previously unveiled for JNK in PC12 cell and P19 EC cell neural differentiation and NFLC gene expression and suggest a general requirement for JNK signaling for neurogenesis. Importantly, our findings highlight JNK1 as a dominant member of the JNK MAP kinase family involved in neurogenesis.
Neural differentiation in murine ES cells is believed to reflect in vivo neurogenesis to a significant degree (26, 34). While the PC12 cell line represents a useful cultured model system for exploring NGF signaling related to neural differentiation, the fact that these cells are derived from a pheochromocytoma demands some caution in extending lessons learned in PC12 cells to the general problem of neurogenesis. The conserved requirement for identical NFLC promoter regulatory regions (Fig. 2B) and a common requirement for integrated signaling through the ERK and JNK pathways for the induction of the NFLC promoter during neuronal differentiation in both PC12 cells (41) and ES cells (Fig. 3B) indicate that common signaling pathways and transcription factors are likely to control NFLC expression in these two distinct model systems. Moreover, the negative influence of the Wnt signaling pathway in both PC12 cells (6, 30) and ES cells (Fig. 7 and references 3 and 13) provides yet another commonality between the two systems. Apparently, observations made in PC12 cells may have a higher degree of relevance to the general question of neural fate specification than previously appreciated.
Our findings reveal significantly increased expression of Wnt-6 and a more modest induction of Wnt-4 in cultured EBs derived from JNK1−/− ES cells (Fig. 8), indicating that JNK1 participates in the repression of the Wnt pathway through inhibition of expression of specific Wnt genes. We therefore extend a novel hypothesis that JNK1-mediated repression of Wnt-4 and Wnt-6 expression regulates neural fate specification in ES cells. In support of this notion, the Wnt pathway has been identified as a negative regulator of in vitro neural differentiation modeled in ES cells (3, 13). Importantly, the Wnt signaling pathway has also been shown to function as an inhibitor of embryonic neurogenesis where repression of Wnt signaling is required in specific embryonic territories during development (28, 29, 33). In contrast to the marked outgrowth of neurons from wild-type, JNK2−/−, and JNK3−/− EBs, a pronounced outgrowth of cells with an epithelial phenotype (Fig. 5) and increased expression of E-cadherin (Fig. 6) was observed in JNK1−/− cultures. With respect to Wnt-6, this Wnt protein has been linked to epithelialization during embryonic development (28, 29). In the chicken embryo, the precise sites of Wnt-6 expression coincide with crucial changes in tissue architecture, namely epithelial remodeling and epithelial-mesenchymal transitions. Moreover, the expression of Wnt-6 is closely associated with regions of BMP signaling. In fact, the “default model” of neurogenesis invokes a Wnt-BMP signaling axis as a dominant repressor of neurogenesis (24). In this regard, Ying et al. (40) have noted that neural differentiation of mouse ES cells in a serum-free system is inhibited by inclusion of BMP4 and causes the appearance of flattened epithelial-like cells similar to the cellular outgrowth we observe from JNK1−/− ES cells (Fig. 5). Similarly, we observe that BMP4 mRNA is significantly increased as well in the differentiated JNK1−/− cultures, suggesting that JNK1 signaling suppresses a Wnt/BMP4 pathway, thereby promoting neural differentiation.
Based on the profound disruption of neural differentiation observed in JNK1−/− ES cells, one would expect mice lacking JNK1 to exhibit a significant disruption of neurogenesis, yet mice bearing homozygous disruption of any single jnk gene develop normally. However, mice in which both jnk1 and jnk2 are disrupted display defective neural tube closure and altered neural apoptosis associated with embryonic lethality (17, 27). One explanation of the discrepancy between our in vitro findings with JNK1−/− ES cells and in vivo neurogenesis is that functional redundancy of JNK1 and JNK2 proteins occurs during in vivo development, preventing a neurogenic defect in JNK1−/− mice. Interestingly, the compound jnk mutant mouse jnk1−/+ jnk2−/− develops normally, while the distinct compound jnk mutant jnk1−/− jnk2−/+ exhibits embryonic neural tube defects, albeit with lower penetrance than that observed in jnk1−/− jnk2−/− embryos (27). These findings support a greater role for JNK1 in neural development relative to JNK2. Recently, disruption of the gene encoding the proximal JNK pathway component, MEKK4, was shown to induce neural tube defects (10) and suggests that a MEKK4-JNK1 signaling axis plays a dominant role in embryonic neurogenesis. Based on our findings that specific Wnt genes are inappropriately expressed in cultures derived from JNK1−/− ES cells and inhibit neurogenesis, we predict that similar events will be observed in the developing neural tubes of compound jnk1/jnk2 mutants and mekk4−/− embryos.
Acknowledgments
The studies were supported by NIH grants GM61718 and DK059756.
We thank Richard Flavell (Yale University, New Haven, Conn.) for generously providing breeder pairs of mice bearing disrupted jnk1, jnk2, and jnk3 alleles.
REFERENCES
- 1.Abbondanzo, S. J., I. Gadi, and C. L. Stewart. 1993. Derivation of embryonic stem cell lines. Methods Enzymol. 225:803-823. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, S., T. Yonezawa, T. A. Kudo, M. G. Li, H. Wang, M. Ito, K. Yoshioka, J. Ninomiya-Tsuji, K. Matsumoto, R. Kanamaru, S. Tamura, and T. Kobayashi. 2004. Activation mechanism of c-Jun amino-terminal kinase in the course of neural differentiation of P19 embryonic carcinoma cells. J. Biol. Chem. 279:36616-36620. [DOI] [PubMed] [Google Scholar]
- 3.Aubert, J., H. Dunstan, I. Chambers, and A. Smith. 2002. Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat. Biotechnol. 20:1240-1245. [DOI] [PubMed] [Google Scholar]
- 4.Bain, G., D. Kitchens, M. Yao, J. E. Huettner, and D. I. Gottlieb. 1995. Embryonic stem cells express neuronal properties in vitro. Dev. Biol. 168:342-357. [DOI] [PubMed] [Google Scholar]
- 5.Boiani, M., S. Eckardt, H. R. Scholer, and K. J. McLaughlin. 2002. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 16:1209-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley, R. S., P. Cowin, and A. M. Brown. 1993. Expression of Wnt-1 in PC12 cells results in modulation of plakoglobin and E-cadherin and increased cellular adhesion. J. Cell Biol. 123:1857-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brustle, O., A. C. Spiro, K. Karram, K. Choudhary, S. Okabe, and R. D. G. McKay. 1997. In vitro-generated neural precursors participate in mammalian brain development. Proc. Natl. Acad. Sci. USA 94:14809-14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butterfield, L., B. Storey, L. Maas, and L. E. Heasley. 1997. cJun NH2-terminal kinase-regulation of the apoptotic response of small cell lung cancer cells to ultraviolet radiation. J. Biol. Chem. 272:10110-10116. [DOI] [PubMed] [Google Scholar]
- 9.Butterfield, L., E. Zentrich, A. Beekman, and L. E. Heasley. 1999. Stress and cell type-dependent regulation of transfected cJun N-terminal kinase and mitogen-activated protein kinase kinase isoforms. Biochem. J. 338:681-686. [PMC free article] [PubMed] [Google Scholar]
- 10.Chi, H., M. R. Sarkisian, P. Rakic, and R. A. Flavell. 2005. Loss of mitogen-activated protein kinase kinase kinase 4 (MEKK4) results in enhanced apoptosis and defective neural tube development. Proc. Natl. Acad. Sci. USA 102:3846-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, C., A. Wynshaw-Boris, F. Zhou, A. Kuo, and P. Leder. 1996. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84:911-921. [DOI] [PubMed] [Google Scholar]
- 12.Gottlieb, D. I., and J. E. Huettner. 1999. An in vitro pathway from embryonic stem cells to neurons and glia. Cells Tissue Organs 165:165-172. [DOI] [PubMed] [Google Scholar]
- 13.Haegele, L., B. Ingold, H. Naumann, G. Tabatabai, B. Ledermann, and S. Brandner. 2003. Wnt signalling inhibits neural differentiation of embryonic stem cells by controlling bone morphogenetic protein expression. Mol. Cell Neurosci. 24:696-708. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, G. L., and R. Lapadat. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911-1912. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy, N. J., and R. J. Davis. 2003. Role of JNK in tumor development. Cell Cycle 2:199-201. [PubMed] [Google Scholar]
- 16.Kim, J. H., J. M. Auerbach, J. A. Rodriguez-Gomez, I. Velasco, D. Gavin, N. Lumelsky, S. H. Lee, J. Nguyen, R. Sanchez-Pernaute, K. Bankiewicz, and R. McKay. 2002. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson's disease. Nature 418:50-56. [DOI] [PubMed] [Google Scholar]
- 17.Kuan, C. Y., D. D. Yang, D. R. Samanta Roy, R. J. Davis, P. Rakic, and R. A. Flavell. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron 22:667-676. [DOI] [PubMed] [Google Scholar]
- 18.Kubo, A., K. Shinozaki, J. M. Shannon, V. Kouskoff, M. Kennedy, S. Woo, H. J. Fehling, and G. Keller. 2004. Development of definitive endoderm from embryonic stem cells in culture. Development 131:1651-1662. [DOI] [PubMed] [Google Scholar]
- 19.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807-869. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S.-H., N. Lumelsky, L. Studer, J. M. Auerbach, and R. D. McKay. 2000. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat. Biotechnol. 18:675-678. [DOI] [PubMed] [Google Scholar]
- 21.Marek, L., C. Amura, V. Levresse, E. Zentrich, V. Van Putten, R. A. Nemenoff, and L. E. Heasley. 2004. Multiple signaling conduits regulate global differentiation-specific gene expression in PC12 cells. J. Cell. Physiol. 201:459-469. [DOI] [PubMed] [Google Scholar]
- 22.Martin, P., and S. M. Parkhurst. 2004. Parallels between tissue repair and embryo morphogenesis. Development 131:3021-3034. [DOI] [PubMed] [Google Scholar]
- 23.Mitsui, K., Y. Tokuzawa, H. Itoh, K. Segawa, M. Murakami, K. Takahashi, M. Maruyama, M. Maeda, and S. Yamanaka. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631-642. [DOI] [PubMed] [Google Scholar]
- 24.Munoz-Sanjuan, I., and A. H. Brivanlou. 2002. Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 3:271-280. [DOI] [PubMed] [Google Scholar]
- 25.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 26.O'Shea, K. S. 1999. Embryonic stem cell models of development. Anat. Rec. 257:32-41. [DOI] [PubMed] [Google Scholar]
- 27.Sabapathy, K., W. Jochumm, K. Hochedlinger, L. Chang, M. Karin, and E. F. Wagner. 1999. Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech. Dev. 89:115-124. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt, C., M. Stoeckelhuber, I. McKinnell, R. Putz, B. Christ, and K. Patel. 2004. Wnt 6 regulates the epithelialisation process of the segmental plate mesoderm leading to somite formation. Dev. Biol. 271:198-209. [DOI] [PubMed] [Google Scholar]
- 29.Schubert, F. R., R. C. Mootoosamy, E. H. Walters, A. Graham, L. Tumiotto, A. E. Munsterberg, A. Lumsden, and S. Dietrich. 2002. Wnt6 marks sites of epithelial transformations in the chick embryo. Mech. Dev. 114:143-148. [DOI] [PubMed] [Google Scholar]
- 30.Shackleford, G. M., K. Willert, J. Wang, and H. E. Varmus. 1993. The Wnt-1 proto-oncogene induces changes in morphology, gene expression, and growth factor responsiveness in PC12 cells. Neuron 11:865-875. [DOI] [PubMed] [Google Scholar]
- 31.Wang, H., S. Ikeda, S. Kanno, L. M. Guang, M. Ohnishi, M. Sasaki, T. Kobayashi, and S. Tamura. 2001. Activation of c-Jun amino-terminal kinase is required for retinoic acid-induced neural differentiation of P19 embryonal carcinoma cells. FEBS Lett. 503:91-96. [DOI] [PubMed] [Google Scholar]
- 32.Weston, C. R., and R. J. Davis. 2002. The JNK signal transduction pathway. Curr. Opin. Genet. Dev. 12:14-21. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, S. I., A. Rydstrom, T. Trimborn, K. Willert, R. Nusse, T. M. Jessell, and T. Edlund. 2001. The status of Wnt signalling regulates neural and epidermal fates in the chick embryo. Nature 411:325-330. [DOI] [PubMed] [Google Scholar]
- 34.Wobus, A. M., and K. R. Boheler. 2005. Embryonic stem cells: prospects for developmental biology and cell therapy. Physiol. Rev. 85:635-678. [DOI] [PubMed] [Google Scholar]
- 35.Wu, R. C., J. Qin, P. Yi, J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2004. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol. Cell 15:937-949. [DOI] [PubMed] [Google Scholar]
- 36.Xia, Y., and M. Karin. 2004. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell Biol. 14:94-101. [DOI] [PubMed] [Google Scholar]
- 37.Xiao, J., and Y. Liu. 2003. Differential roles of ERK and JNK in early and late stages of neuritogenesis: a study in a novel PC12 model system. J. Neurochem. 86:1516-1523. [DOI] [PubMed] [Google Scholar]
- 38.Xu, P., K. Yoshioka, D. Yoshimura, Y. Tominaga, T. Nishioka, M. Ito, and Y. Nakabeppu. 2003. In vitro development of mouse embryonic stem cells lacking JNK/stress-activated protein kinase-associated protein 1 (JSAP1) scaffold protein revealed its requirement during early embryonic neurogenesis. J. Biol. Chem. 278:48422-48433. [DOI] [PubMed] [Google Scholar]
- 39.Yamanaka, H., T. Moriguchi, N. Masuyama, M. Kusakabe, H. Hanafusa, R. Takada, S. Takada, and E. Nishida. 2002. JNK functions in the non-canonical Wnt pathway to regulate convergent extension movements in vertebrates. EMBO Rep. 3:69-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ying, Q. L., M. Stavridis, D. Griffiths, M. Li, and A. Smith. 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21:183-186. [DOI] [PubMed] [Google Scholar]
- 41.Zentrich, E., S. Y. Han, L. Pessoa-Brandao, L. Butterfield, and L. E. Heasley. 2002. Collaboration of JNKs and ERKs in nerve growth factor regulation of the neurofilament light chain promoter in PC12 cells. J. Biol. Chem. 277:4110-4118. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, Q., S. Couillard-Despres, and J. P. Julien. 1997. Delayed maturation of regenerating myelinated axons in mice lacking neurofilaments. Exp. Neurol. 148:299-316. [DOI] [PubMed] [Google Scholar]