Abstract
The multifunctional transcription factor TFII-I is tyrosine phosphorylated in response to extracellular growth signals and transcriptionally activates growth-promoting genes. However, whether activation of TFII-I also directly affects the cell cycle profile is unknown. Here we show that under normal growth conditions, TFII-I is recruited to the cyclin D1 promoter and transcriptionally activates this gene. Most strikingly, upon cell cycle arrest resulting from genotoxic stress and p53 activation, TFII-I is ubiquitinated and targeted for proteasomal degradation in a p53- and ATM (ataxia telangiectasia mutated)-dependent manner. Consistent with a direct role of TFII-I in cell cycle regulation and cellular proliferation, stable and ectopic expression of wild-type TFII-I increases cyclin D1 levels, resulting in accelerated entry to and exit from S phase, and overcomes p53-mediated cell cycle arrest, despite radiation. We further show that the transcriptional regulation of cyclin D1 and cell cycle control by TFII-I are dependent on its tyrosine phosphorylation at positions 248 and 611, sites required for its growth signal-mediated transcriptional activity. Taken together, our data define TFII-I as a growth signal-dependent transcriptional activator that is critical for cell cycle control and proliferation and further reveal that genotoxic stress-induced degradation of TFII-I results in cell cycle arrest.
We have learned a great deal over the last several years about the molecular mechanisms that govern cell growth, cell division, and cell death. Despite the fact that cellular growth and division are mechanistically distinct steps, they are usually coordinately regulated, which is critical for normal cellular development (28). Fibroblasts undergo cell cycle arrest and enter a quiescent program upon serum starvation. However, upon mitogenic signaling, they enter the cell cycle and resume their normal growth program (7). Extracellular growth regulatory signals are ultimately transduced to the nucleus through a series of biochemical steps, resulting in spatial and/or temporal activation of a particular constellation of genes. One way external signals are transmitted to the nucleus is via inducible transcription factors that shuttle between the cytoplasm and nucleus in response to signals. TFII-I is one such multifunctional, inducible transcription factor that is activated via tyrosine phosphorylation (46) in response to growth factor signals and translocates to the nucleus (11, 32, 47). Thus, TFII-I may provide a direct link between mitogen-dependent signaling to changes in nuclear gene expression that govern cellular proliferation and cell division (52).
Although TFII-I was originally discovered as a basal transcription factor that binds and functions through the initiator element (Inr) (12, 42, 53, 54), it also behaves as a signaling protein. In response to mitogenic signaling mediated through growth factor receptors, TFII-I is phosphorylated and engenders transcription of its target genes, including the proproliferative c-fos gene (24, 35). The transcriptional activity of TFII-I is dependent on its tyrosine phosphorylation at defined residues (11). TFII-I is also tyrosine phosphorylated by stress signals, and activated TFII-I up-regulates stress-induced chaperones (49). In B cells, TFII-I is associated constitutively with Bruton's tyrosine kinase. However, upon immunoglobulin receptor cross-linking, TFII-I is tyrosine phosphorylated (47) and activated (64) by Bruton's tyrosine kinase.
A variety of growth-promoting and mitogenic stimuli (e.g., epidermal growth factor, platelet-derived growth factor, serum, and tetradecanoyl phorbol acetate) can enhance tyrosine phosphorylation of TFII-I and subsequent activation of the c-fos promoter (24, 35). Transcriptional activity of TFII-I requires an intact Ras pathway, since a dominant-negative Ras can block TFII-I-dependent transcriptional activation of c-fos (35). It has also been shown that TFII-I physically interacts with mitogen-activated protein kinase through its D-box (36). Additionally, there are several consensus Src-phosphorylation sites that may play critical roles in signal transduction and transcription (52). One of the tyrosine-phosphorylation sites (Y248) has been demonstrated to be required for transcriptional activity of TFII-I at several promoters (11, 46). Importantly, integrity of Y248 is also required for interaction with mitogen-activated protein kinase, suggesting that tyrosine phosphorylation of TFII-I is critical for its downstream function (36).
While it is clear that TFII-I has an important function in mitogenic signal-mediated transcriptional regulation of the c-fos gene, its role in cell cycle control has not yet been addressed. Because of the coordinated regulation of cell growth and division, we investigated whether TFII-I also plays a functional role in the latter process. Here we show that stable and ectopic expression of TFII-I in fibroblasts results in accelerated entry to and exit from S phase due to transcriptional activation of cyclin D1. Genotoxic injury causes activation of p53 tumor suppressor protein with a concomitant arrest in the cell cycle (38). Consistent with its necessary role in the cell cycle, the TFII-I protein is degraded under these conditions. We further show that TFII-I undergoes ubiquitination in vitro and in vivo (upon DNA damage) in a p53-dependent fashion, which results in its proteosome-mediated destruction. The ectopic and stable expression of a wild-type TFII-I leads to enhanced cell cycle entry and exit despite irradiation-induced DNA damage. Conversely, stable expression of a tyrosine phosphorylation deficient mutant TFII-I exacerbates the irradiation induced cell cycle arrest. Thus, TFII-I is an important mediator of cellular proliferation and cell division, destruction of which is necessary for genotoxic stress-mediated cell cycle arrest.
MATERIALS AND METHODS
Plasmids.
The construction of the glutathione transferase (GST) fusion plasmids pEBG vector, pEBG-II-I wild type, pEBG-II-I-YY248/249FF+Y611F, and pEBG-II-I-ΔBR has been detailed elsewhere (13, 14). The cyclin D1 reporter construct used for the reporter assays has been previously described (3). The c-Myc-ubiquitin expression construct was obtained from Daniel Finley, and HA-p53 was obtained from Yoshi Shiloh.
Generation of wild-type p53, p53 null, p53 mutant, ATM null, and ATM null plus p53 mutant Abelson virus-transformed pre-B-cell lines.
Pre-B-cell lines were routinely maintained in supplemented RPMI 1640 medium (50 μM 2-mercaptoethanol, 2 mM l-glutamine, 50 μg of streptomycin per ml, and 50 U of penicillin per ml) containing 10% heat-inactivated fetal calf serum (Atlanta Biologicals) at 37°C in a 6% humidified CO2 atmosphere. The establishment of wt p53 and p53 null pre-B-cell lines has been previously described (60). Bone marrow from Atm−/− or Atm+/+ mice (gift of Phillip Leder, Harvard Medical School) was infected with the Abelson virus, and clonal cell lines were established as previously described (60). Characterization of p53 mutant cell lines was done as previously described (59) with isoform-specific antibodies p53 PAb240 and PAb246 (Oncogene).
Generation of stable NIH 3T3 tetracycline-repressible cell lines.
NIH 3T3 fibroblasts from the American Type Culture Collection and derived cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen), supplemented with 10% heat-inactivated calf serum (Gibco), 50 U/ml penicillin, and 50 μg/ml streptomycin (Invitrogen). Cells were grown at 37°C in 5% CO2 and subcultured two to three times weekly.
To establish retroviral vectors for stable TFII-I expression in NIH 3T3 cells, a NotI cloning site was introduced downstream of the BamHI site of the pSFG tcLuc (39) vector as follows. The luciferase nucleotide sequence between the NcoI and BamHI sites was amplified with the following primers: 5′-CCGACCTGCCATGGAAGA-3′ (forward) and 5′-GGGGATCCGCGGCCGCTCTAGAATTA-3′ (reverse), with a NotI site in the 5′ overhang. The PCR product was digested with NcoI and BamHI and cloned into the pSFG tclLuc+ ITEA vector. The positive clones for the pSFG tclLuc+ ITEA vector were then digested with NcoI and NotI and ligated to a tagless TFII-I sequence obtained from sequential BamHI and NotI followed by a NcoI digest of the pEBB-II-I (13) wild-type vector to obtain the pSFG-II-I construct. For establishment of the pSFG-II-I-YY248/249FF+Y611F, the pSFG tclLuc+ ITEA vector was digested with NcoI and BstBI and ligated to a tagless TFII-I-YY248/249FF+Y611F sequence obtained from the NcoI and BstBI digest of the pEBB-TFII-I-YY248/249FF+Y611F vector to obtain the pSFG-TFII-I-YY248/249FF+Y611F construct. The pEBB-II-I-YY248/249FF+Y611F construct was made by swapping the Acc65I- and BstBI-digested fragment of pEBG-II-I-YY248/249FF+Y611F containing the mutated tyrosines into the pEBB-II-I vector digested with Acc65I and BstBI to yield pSFG-II-I-YY248/249FF+Y611F. The pSFG tclLuc+ ITEA vector was then digested with NcoI and NotI and ligated with a tagless TFII-I-YY248/249FF+Y611F sequence obtained from sequential BamHI and NotI followed by a NcoI digest of the pEBG-II-I wild-type vector. Green fluorescent protein (GFP) was added to both pSFG-II-I and pSFG-II-I-YY248/249FF+Y611F by swapping the BstEBI- and Not1-digested fragment of GFP-II-I from the pEBB-GFP-II-I vector (13) and cloning it into the pSFG-derived vector digested with the same enzymes to yield pSFG-GFP-II-I and pSFG-GFP-II-I-YY248/249FF+Y611F. The pSFG-TFII-I vector was then cotransfected with Psi-Eco envelope vector by calcium phosphate transfection in 293T cells to produce infective virions. Virions were used to infected NIH 3T3 cells which were bulk sorted using a MoFlo instrument (Dako Cytomation) 24 h after infection for GFP expression. Bulk-sorted cells were grown, and single-cell sorting yielded clonal colonies.
Generation of hairpin loops specific for TFII-I knockdown and lentiviral infection of the NIH 3T3 cell line.
According to previously described methodology (55), the TFII-I cDNA sequence was searched for the specific motif AAG(N18)TT, where the G residue is followed by a string of any 18 nucleotides. The G(N18) sense/antisense sequence was further modified by the addition of a T to the 5′ end for the initiation of transcription at the U6 promoter. A string of six Ts was added to the 3′ end, serving the purpose of a stop codon for the hairpin. Three such sequences were found within the TFII-I cDNA, and sense/antisense oligomers were ordered from IDT. For cloning purposes, HpaI (5′)/XhoI (3′) linkers were added to the ends of the oligonucleotides. In parallel, the Lentilox 3.7 plasmid construct was digested with HpaI/XhoI two times, for 2 h per digestion, and purified using the QIAGEN PCR purification protocol. The three oligonucleotides were ligated overnight at 14°C and transformed into DH5α electrocompetent bacteria. Single colonies were selected, grown overnight in LB broth at 37°C, and purified using a QIAGEN miniprep kit. Subclones were subsequently digested with HpaI/XhoI for 1 h at 37°C and run out on a 2% agarose gel. The hairpins were sequenced and contained no apparent mutations. 293T producer cells were grown to 65% confluence prior to transfection. The lentiviral system consists of three distinct plasmids, the envelope (pVSVG, where VSVG is vesicular stomatitis virus G), the helper (Δ8.9), and the lentilox construct (pLLP3.7). Lentiviral production was done as previously described (40). Briefly, 2 μg of total plasmid was used for the Fugene (Roche) transfection according to the manufacturer's protocol, and production of lentivirus using 0.5 μg of pVSVG, 0.5 μg of Δ8.9, and 0.5 μg of constructs containing TFII-I hairpins 1 and 3. As a control, the exact protocol was followed for the nonsilencing hairpin, hairpin 2, serving the purpose as a control knockdown. Lentivirus was generated for 44 to 48 h in a biosafety level 3 facility, followed by infection. The infections were performed by the addition of 1 ml of lentiviral supernatant containing 8 μg/ml polybrene to 1 × 106 NIH 3T3 cells in log phase growth. Infections were carried out overnight in the biosafety level 3 facility and harvested the following day. Cells were grown out for an additional 24 h and were bulk sorted using a MoFlo instrument (Dako Cytomation) 24 h after infection for GFP expression. Bulk-sorted cells were grown, and single-cell sorting yielded clonal colonies. Cells were grown to log density, harvested, lysed, and analyzed for TFII-I knockdown via Western blot analysis. The clones with the highest level of knockdown were kept and stored in liquid nitrogen.
Transient transfection of COS-7 cells, interaction, and luciferase reporter assays.
COS-7 fibroblasts were cultured in DMEM) (Invitrogen), supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals), 50 U/ml penicillin, and 50 μg/ml streptomycin (Invitrogen). Cells were grown at 37°C in 5% CO2 and subcultured two or three times weekly. For coexpression and interaction studies, 1.5 × 106 COS-7 cells were seeded in 10-cm dishes. After 24 h, cells were transfected using supplemented DMEM with Lipofectamine reagent according to the manufacturer's recommendations (Invitrogen). The following plasmids and amounts were used in our transfections: 4 μg of pEBG-TFII-I, 4 μg of pEBG-TFII-I-248/611, 12 μg of pCWc-Myc-ubiquitin, and increasing hemagglutinin (HA)-p53 at 1, 3, and 9 μg.
For reporter assays, 1 × 105 COS-7 cells were seeded in six-well plates. After 24 h, cells were transfected using supplemented DMEM with Lipofectamine reagent according to the manufacturer's recommendations (Invitrogen). A total of 800 ng of cyclin D1 promoter-luciferase plasmid −1745 CD1LUC (3) or control pFR-Luc (Stratagene) (41) was combined with 35 ng of Renilla luciferase pRL-TK (internal control). Various concentrations of pEBG-II-I wild type, pEBG-II-I phosphorylation mutants, ΔBR mutant, and p53 were added to the mixture. Total DNA concentration was normalized using empty pEBG vector. DNA-Lipofectamine mixtures were added to the cells. At 24 h posttransfection, fresh supplemented DMEM was added to the cells. Cells were harvested 36 h posttransfection according to manufacturer's instructions, using passive lysis (Promega). Relative luciferase activities of firefly and Renilla luminescence were measured using a Dual Luciferase Assay kit (Promega). Transfections were performed in triplicate, and reporter assay experiments were repeated more than three times.
Cell extracts, immunoprecipitation assays, and Western blot analysis.
Cells were washed twice with phosphate-buffered saline (PBS) and lysed for 30 min at 4°C (rotating motion) in lysis buffer (25 mM Tris-HCl, pH 8.0, 100 mM KCl, 5 mM NaF, 2 mM Na3VO4, 1 mM Na2P4O7, 0.4% Triton X-100, and 1% Nonidet P-40), supplemented with EDTA-free 1× antiprotease mixture (Roche Applied Science). For ubiquitination studies, the lysis buffer was supplemented with 1 mM ortho-phenantheoline (Sigma) and 20 μM MG-132 (Calbiochem). Lysates were precleared by centrifugation for 15 min at 14,000 rpm at 4°C. Total protein concentration was measured using the Bradford method (Bio-Rad). A total of 50 μg of lysate was used for Western blot assays and 500 to 1,000 μg of whole-cell lysate was used for GST pull-down or immunoprecipitation assays with anti-TFII-I mouse monoclonal antibody (T81920; Transduction Laboratories).
(i) Coimmunoprecipitations.
For immunoprecipitations, antibodies were incubated with whole-cell lysates for 20 min at 4°C (rotating motion), followed by the addition of protein G-Sepharose “4 Fast Flow” beads (1:1 slurry; 40 μl) (Amersham Biosciences) to the reaction mixture for 2 h at 4°C (rotating motion). Following incubation periods, lysates were washed three times in lysis buffer with 0.6% Nonidet P-40 detergent. For GST pull-down, lysates were incubated with glutathione-agarose beads (1:1 slurry; 40 μl) (Amersham Biosciences) for 2 h at 4°C (rotating motion). Following incubation periods, lysates were washed three times in lysis buffer with 0.6% Nonidet P-40 detergent.
(ii) Western blotting.
The following primary antibodies were used for immunoblotting: anti-GST (1:3,000; Sigma), anti-TFII-I rabbit polyclonal (1:2,500) (46), anti-P-TFII-I rabbit polyclonal (1:2,500) (previously described in reference 11), anti-p53 248 hybridoma supernatant (1:4), anti-cyclin D1 (sc-450; 1:250; Santa Cruz Biotechnology), anti-p21 sc-471 (1:250; Santa Cruz Biotechnology), anti-β-actin (sc-1616; 1:250; Santa Cruz Biotechnology), anti c-Myc (sc-40; 1:500, Santa Cruz Biotechnology), anti-HA (12CA5; 1:2,000; Roche), and antiubiquitin (UG9510; 1:2,000; Biomol International). Secondary horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit antibodies were used at a dilution of 1:10,000 (Zymed Laboratories Inc). Western blots were visualized using enhanced chemiluminescence (PerkinElmer Life Sciences) using standard methods. Autoradiographs were image scanned, and densitometry was performed to quantitate the intensity of Western blot bands using appropriate background correction. Individual experiments were repeated at least three times.
Semiquantitative RT-PCR.
Total cellular RNA was isolated according to the manufacturer's instructions using Trizol reagent (Invitrogen). Reverse transcription was carried out following the manufacturer's protocol with the SuperScript III First-Strand Synthesis System for reverse-transcription PCR (RT-PCR; Invitrogen). Subsequently, 24 amplification cycles (93°C for 1 min, 55°C for 1 min, and 72°C for 1 min) were carried out with the following primers: for mouse cyclin D1, 5′-CTGACACCAATCTCCTCAACGAC-3′ (forward) and 5′-GCGGCCAGGTTCCACTTGAGC-3′ (reverse). As a control, mouse β-actin cDNA was amplified (93°C for 1 min, 55°C for 1 min, and 72°C for 1 min) with the following primers: 5′-GTGGGAATTCGTCAGAAGGACTCCTATGT-3′ (forward) and 5′-GAAGTCTAGAGCAACATAGCACAGCTTCT-3′ (reverse). To verify the absence of contamination of RNA samples with DNA, we performed the PCR on samples that were processed identically to the target samples but were not reverse transcribed. After RT-PCR, samples were electrophoresed on a 1.5% agarose gel containing 10 μg/ml ethidium bromide.
Real-time ChIP.
3T3 cells were synchronized in G0 by DMEM-0.5% fetal calf serum for serum starvation for 45 h and were serum stimulated or not using DMEM-20% fetal calf serum for 1 h. Cells were fixed directly by adding formaldehyde to a final concentration of 1% for 10 min at room temperature. Chromatin immunoprecipitation (ChIP) assays were performed on chromatin from 1 million cells per condition and sheared by sonication to an average length of 700 kb, according to the protocol of a Chromatin Immunoprecipitation Assay Kit (Upstate Biotechnology Inc., Lake Placid, N.Y.), with minor modifications. Fixation was stopped by incubation with a final concentration of 0.125 M glycine for 5 min, and elution from the beads was performed twice at 65°C for 10 min. Anti-TFII-I directed against the C terminus of TFII-I (BD Biosciences) was used at 2 μl per condition. DNA samples from ChIP assays were analyzed by real-time PCR using a GeneAmp 5700 instrument and Amplitaq PCR master mix (Applied Biosystems, Branchburg, NJ). The primer and probes expressly designed by Applied Biosystems for amplification of region −331 to −101 of the cyclin D1 promoter were used for real-time PCR of rodent glyceraldehyde-3-phosphate dehydrogenase control and of the mouse cyclin D1 promoter: forward, 5′-CTGGCCTCCCTCCTAGCT-3′; reverse, 5′-TCCTCCGAGACCTGTGGA-3′; and probe, 5′-CAGAGCCGCCACTACC-3′. Additional primers and probes were designed by Applied Biosystems to amplify region 2291 to 2491 from the start site of the cyclin D1 coding sequence: forward, 5′-TCACACAGGAGGCTTTTAAACACT-3′; reverse, 5′-TGGTCATGGGCAGCCTTTC-3′; and probe, 5′CTTAAGGCTACAGAAGAGTATTT-3′.
Eukaryotic expression and purification of TFII-I.
For eukaryotic expression of TFII-I, COS-7 cells were transfected with pEBGII-I. Transfections were carried out by the Lipofectamine method according to the manufacturer's protocol (Invitrogen). For transfection, 8 μg of expression plasmid (pEBGII-I) was used per 100-mm-diameter plate. At 36 h posttransfection, cells were harvested, washed twice in PBS, and lysed in ice-cold BC500 buffer (20 mM Tris-HCl [pH 7.9], 500 mM KCl, 20% glycerol) containing 0.1% Nonidet P-40 supplemented with EDTA-free 1× antiprotease mixture (Roche Applied Science). The lysate was clarified by centrifugation for 30 min at 12,000 rpm at 4°C. The GST-six-histidine-tagged TFII-I protein was purified by loading the lysate on a 1-ml cobalt-agarose column (Talon; Pharmacia) at 4°C. The column was washed sequentially with 5 column volumes of BC500 (without protease inhibitors and Nonidet P-40) containing 10 mM imidazole. Finally, the tagged fusion protein was eluted with 3 column volumes of BC500 containing 200 mM imidazole. The TFII-I-containing peak fractions were pooled, dialyzed against buffer B (20 mM Tris-HCl [pH 7.9], 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 10% glycerol) with 100 mM KCl (B100) and concentrated by centrifugation at 3,000 rpm in a Centriprep 50 concentrator (Amicon).
EMSA.
The electrophoretic mobility shift assays (EMSAs) were performed with an Inr element probe based on the cyclin D1 Inr −9 to +16 (5′-CCTCCCGCTCCCATTCTCTGCCGGG-3′) preceded by three guanines at the 5′ ends. The annealed oligonucleotides were labeled with [α-32P]dCTP (3,000 Ci/mmol) and Klenow fragment for 30 min at room temperature. The reaction was terminated by purification of the labeled probes on a G50 nick column according to the manufacturer's protocol (Pharmacia). The specific activity of the labeled probes was approximately 402,657 cpm/μl, and each reaction volume contained 2 μl of probe. Labeled oligonucleotides were incubated alone, with purified TFII-I, with purified TFII-I preincubated with 2 μl of affinity-purified anti-TFII-I antibody raised against the DNA binding domain, or with 30 μg of the mentioned lysates in the presence of a 20-fold molar excess of oligo(dA-dT).
Cell cycle synchronization.
NIH 3T3 and derived cells were synchronized in G0 by serum starvation (0.5% serum) for 45 h as previously described (57). Cells were released with 10% serum and trypsinized at the indicated times, and their cell cycle profiles were determined by propidium iodide staining followed by fluorescence-activated cell sorting (FACS).
Cell cycle analysis.
Briefly, cells were washed with PBS and fixed with ice-cold 70% ethanol for 1 h on ice. Following fixation, cells were washed again and then incubated with 50 μg/ml DNase-free RNase and 50 μg/ml propidium iodide for 1 h at 37°C prior to analysis using a FACScan flow cytometer (Becton Dickenson). ModFit Software was use to interpret the data.
RESULTS
Stable expression of TFII-I enhances cell cycle entry and exit.
To study the potential role of TFII-I in cell cycle regulation and cellular proliferation, we established NIH 3T3 cells stably expressing wild-type human TFII-IΔ isoform (13) under the control of a tetracycline-repressible promoter. Due to the greater than 97% amino acid sequence identity between human and mouse TFII-I orthologs (63), they can be used interchangeably. These cells and the control cells were serum starved for 48 h to synchronize them in G0 and were released with 10% serum-rich medium, and the cell cycle progression was monitored (57). Over a 20-h period, we found that ectopic TFII-IΔ-expressing cells entered, reached their peak DNA content, and exited S phase earlier than control NIH 3T3 cells (Fig. 1A, left panel). We next narrowed our observation period to the G1/S interface and found that ectopic TFII-IΔ-expressing cells entered, reached their peak DNA content, and exited S phase 4 h earlier than control NIH 3T3 cells (Fig. 1A, middle panel). The accelerated S-phase entrance and exit phenotype exhibited by ectopic TFII-IΔ-overexpressing cells could be abolished by prior tetracycline treatment, demonstrating a specific effect of ectopic TFII-I (Fig. 1A, right panel).
FIG. 1.
TFII-I activates cyclin D1 resulting in accelerated cell cycle progression. (A) Control and TFII-IΔ-expressing NIH 3T3 cells were synchronized in G0 by serum starvation (0.5% serum) for 45 h. Cells were released with 10% serum, and every 4 h for 20 h, cells were trypsinized and their cell cycle profiles were determined by propidium iodide staining followed by FACS (left graph). The middle graph shows the results when cells were analyzed every 2 h starting at 10 h and ending at 16 h. The results in the right graph are from cells that were grown in the presence of tetracycline 4 μg/ml and analyzed as described for the middle graph. (B) Control and TFII-IΔ cells were either grown in the presence or absence of 4 μg/ml tetracycline for two days. Whole-cell extracts were prepared and normalized by concentration. The lysates were analyzed by Western blotting using the indicated antibodies. The asterisk denotes a nonspecific band used throughout as an additional loading control.
Stable TFII-I overexpression leads to elevated cyclin D1.
It has been shown that cyclin D1 is important for cells to exit from G1 and enter S phase because destruction of cyclin D1 leads to G1 arrest (1, 20). The promoter region of cyclin D1 does not have a TATA box but contains a functional Inr element that could be potentially regulated by TFII-I (19, 50). Hence, we directly tested the endogenous levels of cyclin D1 protein in cells that stably express wild-type TFII-I in the absence and presence of tetracycline. We found that cyclin D1 protein levels were significantly elevated in cells expressing TFII-IΔ. However, upon tetracycline treatment, cyclin D1 levels were reduced, thus directly implicating TFII-I in altered cyclin D1 levels (Fig. 1B). While the cyclin D1 protein expression was reduced, it remained slightly above the wild-type levels when normalized to the β-actin loading control (Fig. 1B). This is likely due to incomplete tetracycline shutdown of ectopically expressed TFII-I or possible cyclin D1 stabilization through other mechanisms. Regardless, it was clear that ectopic expression of TFII-I could enhance cyclin D1 expression.
TFII-I knockdown leads to cell cycle block and reduced cyclin D1.
If our hypothesis that TFII-I promotes cell cycle advancement via cyclin D1 is correct, then one can predict that interfering with TFII-I will result in reduced cyclin D1 levels and cell cycle block. We tested this using stable RNA interference knockdown of TFII-I. When analyzed, cells stably expressing TFII-I knockdown hairpin loops and exhibiting reduction of TFII-I protein by Western blotting not only showed a significant reduction of cyclin D1 expression (Fig. 2A) but also exhibited a marked arrest in the G0-G1 phase of the cell cycle. Indeed, 27% more cells were found in G0-G1 upon TFII-I knockdown (Fig. 2B).
FIG. 2.
TFII-I stable knockdown leads to cell cycle block and decreased cyclin D1. (A) Whole-cell extracts were prepared from control and TFII-I stable knockdown NIH 3T3 cells and normalized by protein concentration. The lysates were analyzed by Western blotting. (B) Control and TFII-I knockdown cells were grown, cells were trypsinized, and their cell cycle profiles were determined by propidium iodide staining followed by FACS analysis using ModFit software.
TFII-I is recruited to the cyclin D1 promoter in vivo, binds to the Inr element, and mediates activation of the promoter.
We directly tested the endogenous levels of cyclin D1 RNA in cells that stably express wild-type TFII-I in the absence and presence of tetracycline. We found that cyclin D1 RNA levels were elevated in cells expressing TFII-IΔ. However, upon tetracycline treatment, cyclin D1 RNA was reduced to the background levels, thus directly implicating TFII-I in altered cyclin D1 levels (Fig. 2A). These results revealed that TFII-I might transcriptionally activate the cyclin D1 promoter. To investigate the transcriptional effect of TFII-I on the cyclin D1 promoter, we performed a cyclin D1-luciferase reporter assay. While TFII-I activated the cyclin D1 reporter −1745 CD1LUC (3) in a dose-dependent manner, it failed to activate a control (pFR-Luc) promoter under the same conditions (Fig. 3B). Importantly, TFII-I-mediated activation of the cyclin D1 promoter was dependent on intact tyrosine residues at positions 248 and 611 (Fig. 3C). In addition, the transcriptional activation of cyclin D1 by TFII-I requires its DNA binding ability. Mutation of either the critical tyrosine residues (Y-F) or deletion of the basic region/DNA binding domain resulted in repressed promoter activity, suggesting that these mutants act as dominant negatives (Fig. 3C). Furthermore, purified recombinant TFII-I directly bound the cyclin D1 Inr in EMSA, and the binding could be abrogated by an antibody raised against the TFII-I DNA binding domain (Fig. 3D). To demonstrate in vivo recruitment of TFII-I to the endogenous cyclin D1 promoter, we then performed a ChIP assay under conditions when quiescent cells are stimulated to reenter the cell cycle. We found that there was a significant enrichment of the cyclin D1 promoter with anti-TFII-I antibodies only upon serum stimulation (Fig. 3E), indicating that TFII-I is recruited to the cyclin D1 promoter in vivo. However, significant recruitment of TFII-I to a control region downstream of the Inr region was not observed in either the absence or the presence of serum (Fig. 3E). Because the transcriptionally active form of TFII-I is tyrosine phosphorylated and because we have shown activated (serum stimulated) TFII-I to be recruited to the cyclin D1 promoter, it is likely that the recruited active TFII-I is tyrosine phosphorylated. Taken together, these results strongly suggest that TFII-I plays a role in cell cycle progression from the G0-G1 to S phase boundary via transcriptional regulation of cyclin D1.
FIG. 3.
TFII-I is recruited to and activates the cyclin D1 promoter. (A) Control and TFII-IΔ-expressing NIH 3T3 cells were either grown in the presence or absence of 4 μg/ml tetracycline for 2 days. RNA was prepared and RT-PCR was performed with mouse cyclin D1 and β-actin primers. (B) COS cells were transiently transfected with a cyclin D1-Luc reporter construct or control pFR-Luc reporter construct as well as increasing amounts of TFII-I. Western blotting of transfected extracts demonstrates ectopic TFII-I expression under these conditions. (C) COS cells were transiently transfected with a cyclin D1-Luc reporter construct as well as mutants of TFII-I lacking the tyrosine at position 248 or lacking the DNA binding domain (D-BR). (D) 32P-labeled oligomers representing the cyclin D1 Inr −9 to +16 region (5′-CCTCCCGCTCCCATTCTCTGCCGGG-3′) were incubated alone or with purified TFII-I alone or were preincubated with antibodies against the TFII-I DNA binding domain and in the presence of a 20-fold excess of oligo(dA-dT). (E) NIH 3T3 cells were synchronized in G0 by 0.5% serum starvation for 45 h and were released or not by 20% serum stimulation for 1 h. Chromatin was prepared, and immunoprecipitation was performed with anti-TFII-I antibody followed by real-time quantitative PCR for the cyclin D1 promoter (filled bars) or coding sequence 2,291 pairs downstream of the start site.
Stable and ectopic expression of TFII-I prevents radiation-induced down-regulation of cyclin D1 and promotes progression of cell cycle despite γ-irradiation.
Consistent with previously published results (2), we found decreased cyclin D1 levels in NIH 3T3 cells following γ-irradiation; however, cyclin D1 protein levels were elevated beyond basal levels despite radiation treatment in NIH 3T3 cells stably expressing TFII-IΔ (Fig. 4A). On the contrary, and consistent with the reporter assays, cyclin D1 levels were constitutively decreased in cells stably expressing the Y-F mutant TFII-I and remained low upon radiation treatment (Fig. 4A).
FIG. 4.
TFII-IΔ overexpression overcomes p53-mediated cell cycle arrest via cyclin D1. (A) NIH 3T3 (control), TFII-IΔ and mutant TFII-I cells lacking tyrosine at positions 248 and 611 [248/611(Y-F)] were either irradiated at 30 Gy or not. Whole-cell extracts were prepared 4 h post-gamma-irradiation treatment and normalized by protein concentration. The lysates were analyzed by Western blotting. (B) TFII-IΔ and mutant TFII-I cells lacking tyrosine at positions 248 and 611 [248/611(Y-F)] were either grown in the presence or absence of 4 μg/ml tetracycline for 2 days. Whole-cell extracts were prepared and normalized by protein concentration. The lysates were analyzed by Western blotting using the indicated antibodies. (C) Control cells (1 × 106) and 1 × 106 TFII-IΔ cells and TFII-I cells lacking tyrosine at positions 248 and 611 [248/611(Y-F)] were either grown in the presence or absence of 4 μg/ml tetracycline for 2 days. Then cells were either irradiated at 30 Gy or not. At 36 h postirradiation treatment, cells were trypsinized and their cell cycle profiles were determined by propidium iodide staining followed by FACS analysis using ModFit software. The asterisk denotes a nonspecific band used throughout as an additional loading control.
Degradation and maintenance of low cyclin D1 levels is an essential component of cell cycle arrest in response to genotoxic stress (2). Therefore, we analyzed cell cycle kinetics following γ-irradiation after ascertaining that p53 was induced normally in all cell lines (data not shown). Control NIH 3T3 cells exhibited a prominent G2 accumulation (72.79%) and arrest, coupled with a nearly threefold reduction in the S-phase fraction from 47.56% to 13.21% (Fig. 4C). However, a smaller percentage (50.44%) of the cells expressing ectopic TFII-IΔ accumulated in G2 and stopped synthesizing DNA in S phase. Indeed, after irradiation the fraction of TFII-IΔ cells in S phase was only reduced from 57.27% to 30.22%. Thus, a significant number of cells which no longer arrested in G1 phase proceeded into S phase despite radiation treatment. These findings demonstrate that, unlike the control NIH 3T3 cells, ectopic TFII-I-expressing cells cycle despite irradiation treatment. In contrast, most of the mutant TFII-I-expressing cells resided in the G0-G1 fraction, regardless of their treatment, accompanied by a reduction in S phase (Fig. 4C). Thus, stable expression of the transcriptionally incompetent mutant TFII-I interfered with cell growth possibly by blocking cell cycle progression at the G1-S phase boundary. We conclude that transcriptional activity of TFII-I is minimally required for progression of cells from G1 to S phase, although it is likely that TFII-I is also necessary for later stages of the cell cycle. The differences we observed result from ectopic wild-type or mutant TFII-I because tetracycline treatment led to a significant reversion of these phenotypes (Fig. 4C). When treated with tetracycline and following γ-irradiation, the wild-type TFII-I-expressing cells exhibited a cell cycle profile closely resembling that of the control cells, while a threefold increase in the fraction of mutant TFII-I-expressing cells residing at G2 phase was observed. However, the incomplete reversion in phenotype in this latter case may result from leaky expression of the mutant TFII-I protein in the presence of tetracycline (Fig. 4B).
TFII-I is degraded in a p53- and ATM-dependent manner following γ-irradiation.
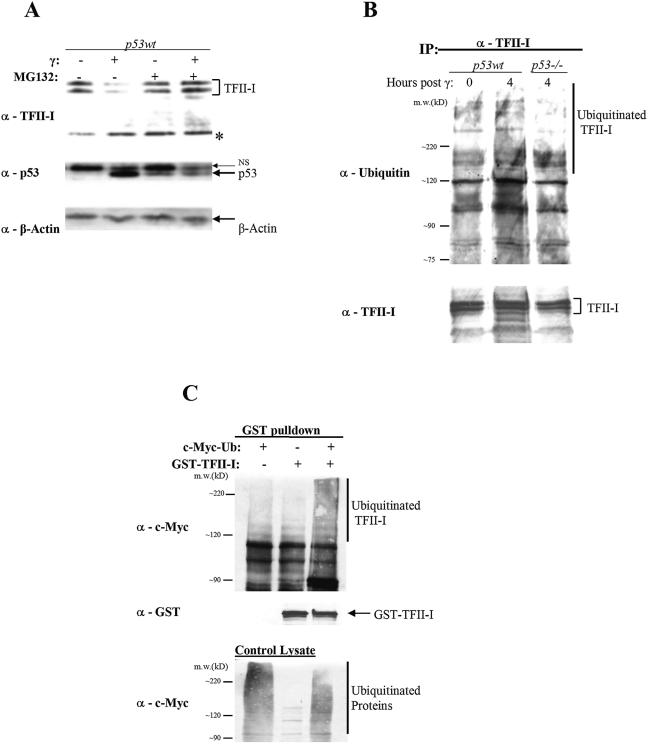
Because we observed that elevated TFII-I levels could enhance cycling of cells, we predicted that either the TFII-I levels may be lowered or its transcriptional activity altered during cell cycle arrest. Hence, we investigated the status of TFII-I upon DNA damage-induced cell cycle arrest. It is well established that γ-irradiation leads to induction of p53, which results in either cell cycle arrest or apoptosis (38). These two outcomes are affected by p53-dependent transcriptional modulation of many proteins implicated in differentiation, proliferation, cell growth, apoptosis, and senescence (30). Gamma-irradiation was used to induce p53 in various cell lines, and TFII-I protein expression was examined via Western blotting. We noted that TFII-I was degraded following γ-irradiation in Abelson virus-transformed pre-B-cell lines. The levels of TFII-I protein decreased gradually, with the reduction being noticeable by 4 h postirradiation (Fig. 5A) and even more striking by 6 h (Fig. 5B). The degradation of TFII-I is p53 dependent because no degradation was observed upon γ-irradiation in p53 null cells, regardless of the time point postirradiation (Fig. 5A and B). Interestingly, TFII-I levels were higher in p53 null cells (Fig. 5A) and in p53 mutant-expressing cells (Fig. 5F). The degradation of TFII-I is not a cell type-specific phenomenon as it is also observed in NIH 3T3 fibroblasts (Fig. 5C) and in primary T cells (D. Banerjee and R. Sen, personal communication). Degradation of TFII-I upon γ-irradiation also required intact ATM as TFII-I levels following radiation treatment were not altered in ATM null cells (Fig. 5F).
FIG. 5.
TFII-I is degraded after γ-irradiation in a p53- and ATM-dependent manner. (A) Abelson virus-transformed murine pre-B cells with either wild-type p53 or lacking p53 were irradiated at 10 Gy. Four hours post-gamma-irradiation treatment, whole-cell extracts were prepared and normalized by protein concentration. Lysates were analyzed by Western blotting. (B) The same procedures were followed as described for panel A except that the lysates were made 6 h after irradiation treatment. (C) NIH 3T3 cells with wild-type p53 were irradiated at 30 Gy. Eight hours post-gamma-irradiation treatment, whole-cell extracts were prepared and normalized by protein concentration. Lysates were analyzed by Western blotting. (D) COS cells were transiently transfected with GST-TFII-I and increasing amounts of HA-p53. Whole-cell extracts were prepared and normalized by protein concentration. Lysates were analyzed by Western blotting. (E) COS cells were transiently transfected with a cyclin D1-Luc reporter construct, p53, and increasing amounts of TFII-I. All concentrations are in nanograms, and experiments were repeated at least three times. (F) Wild-type p53, p53 null, p53 mutant, ATM null, and ATM null plus p53 mutant Abelson virus-transformed murine pre-B cells were irradiated at 10 Gy. Four hours post-gamma-irradiation treatment, whole-cell extracts were prepared and normalized by protein concentration. Lysates were analyzed by Western blotting. The asterisk denotes a nonspecific band used throughout as an additional loading control.
To test a more direct effect of p53 on TFII-I degradation, we coexpressed wild-type GST-TFII-I and HA-p53 in COS cells. The ectopic coexpression of a constant amount of TFII-I with increasing p53 resulted in a dose-dependent reduction of TFII-I, suggesting that this effect is indeed p53 dependent (Fig. 5D). The same dose-dependent reduction of TFII-I protein was also seen when a constant amount of the mutant was coexpressed with increasing p53, implying that phosphorylation at tyrosine 248 or 611 is not necessary for p53-dependent degradation of TFII-I (data not shown). In light of this result and given that TFII-I mediates activation of the cyclin D1 promoter, we examined whether p53 might antagonize this function. It is well established that p53 represses transcription of many genes (66), including cyclin D1 (15). Consistent with this we found that p53 mediated repression of the cyclin D1 promoter (Fig. 5E) and that this repression was relieved by increasing the concentration of TFII-I. In addition, binding of TFII-I to the cyclin D1 Inr was significantly decreased upon γ-irradiation only in cells expressing p53 (data not shown). These results demonstrate that TFII-I is degraded in a p53-dependent manner and suggested that this degradation may contribute to p53-induced cell cycle arrest.
TFII-I is ubiquitinated and targeted for proteasomal degradation in a p53-dependent manner.
Ubiquitin-proteasome-mediated proteolysis is established as an important pathway of protein degradation that controls the timed destruction of many cellular regulatory proteins, including cyclins, tumor suppressors, transcription factors, and growth factor receptors (48, 58). Therefore, we tested whether TFII-I degradation was proteasome dependent. We found that treatment of irradiated Abelson virus-transformed pre-B cells with MG-132, a proteasome inhibitor, prevented TFII-I degradation (Fig. 6A), indicating that TFII-I was degraded through the proteasome pathway. Ubiquitin is a ubiquitous 76-amino-acid protein that is highly conserved in all eukaryotes and is covalently linked to lysine residues in target proteins. Polyubiquitination of lysine with more than four moieties targets proteins for degradation via the 26S proteasome (6). Since TFII-I degradation was proteasome dependent, we next examined whether TFII-I was ubiquitinated upon γ-irradiation and whether this is p53 dependent. We found a significant increase in TFII-I ubiquitination following γ-irradiation in p53 wild-type-expressing B cells but not in p53 null cells (Fig. 6B). To demonstrate directly that TFII-I is ubiquitinated, we cotransfected GST-tagged TFII-I and c-Myc-tagged ubiquitin in COS cells. TFII-I was pulled down by glutathione-agarose, and the presence of the ubiquitin moiety was assessed with a c-Myc antibody. Evidence for ubiquitinated TFII-I was seen in the presence but not in the absence of ectopic myc-tagged ubiquitin (Fig. 6C), demonstrating that TFII-I is ubiquitinated in vivo.
FIG. 6.
TFII-I is ubiquitinated and targeted for proteasomal degradation in a p53-dependent manner. (A) Abelson virus-transformed murine pre-B cells containing either wild-type p53 or lacking p53 were pretreated for 1 h with 50 μM of the proteasome inhibitor MG-132 and irradiated at 10 Gy. Four hours post-gamma-irradiation treatment, whole-cell extracts were prepared and normalized by protein concentration. Lysates were analyzed by Western blotting. (B) Abelson virus-transformed murine pre-B cells with wild-type p53 or lacking p53 were irradiated at 10 Gy. Four hours post-gamma-irradiation treatment, whole-cell extracts were prepared and normalized by protein concentration. Proteins were immunoprecipitated with anti-TFII-I-I antibody and analyzed by Western blotting. (C) COS cells were transiently transfected with GST-TFII-I alone or in combination with c-Myc-ubiquitin. Whole-cell extracts were prepared and normalized by protein concentration. Proteins were immunoprecipitated with glutathione-agarose beads and analyzed by Western blotting. The asterisk denotes a nonspecific band used throughout as an additional loading control.
DISCUSSION
Cells must integrate extracellular growth signals prior to making the commitment to replicate their genome (43). Once DNA replication is under way, the cell relies solely on internal signals until the cell ultimately duplicates in mitosis (7). Replication and mitosis are tightly controlled in a manner that maximizes the transmission of accurate copies of their genes. Indeed, failure to do so ultimately leads to pathological conditions such as cancer. Therefore, cells have evolved an elaborate network of mechanisms to sense DNA damage, check cell cycle and attempt repair, when possible, or permanently arrest cell cycle and induce death when the damage is irreparable (5). Over the last decade, p53 has revealed itself as a crucial component of the DNA damage response network (26). Upon genotoxic stress, p53 is stabilized and activated, leading to cell cycle arrest, followed by either DNA repair or apoptosis (38). Despite significant advances, it is still unclear precisely how p53 inhibits the cell cycle. While it is established that p53 leads to delayed and sustained cell cycle arrest upon DNA damage by upregulating a critical cell cycle regulator, p21/WAF1, p21-independent pathways also provide rapid growth arrest at the restriction point (7). Consistent with this latter possibility, we show a pathway by which p53-mediated cell cycle arrest could occur via degradation of the transcription factor TFII-I (Fig. 7).
FIG. 7.
Model on opposing roles of TFII-I and p53 on cell cycle and proliferation. Upon growth factor signaling, TFII-I gets phosphorylated and translocates to the nucleus where it can engender transcription of the cyclin D1 gene and thereby facilitate cell cycle progression. However, upon genotoxic stress p53 is activated and causes an increase in p21. Excess p21 blocks proliferation by inhibiting the cyclin E/cdk2 complex. In addition, p53 directly represses the cyclin D1 promoter. Following gamma-irradiation, TFII-I is ubiquitinated and degraded in a p53-dependent manner, further leading to inactivation of cyclin D1 and cell cycle arrest.
We show that TFII-I is recruited to and mediates transcriptional activation of cyclin D1, thus enhancing the cycling of cells by accelerating the G1 to S phase boundary. Persistent cyclin D1 overexpression is associated with tumorgenesis and increased genetic alterations in transgenic mice (16, 62). It has been shown recently that cyclin D1 upregulation leads to chromosomal abnormalities (45), while reduced levels of cyclin D1 is protective from tumorgenesis (15, 29, 65). Because stable TFII-I overexpression results in sustained and elevated cyclin D1 levels even under basal conditions, it is possible that cells overexpressing TFII-I accumulate genetic damage due to elevated levels of cyclin D1, although the status of DNA in these cells remains to be analyzed. TFII-I undergoes tyrosine phosphorylation in response to growth factor stimulation and, thus, transduces mitogenic signals from the cell surface receptors to the transcriptional machinery. The transcription function of TFII-I also requires an intact tyrosine at position 248, and mutation in this site may behave in a dominant-negative fashion. Accordingly, this mutant suppressed cyclin D1 promoter activity both in stable cell lines and in reporter assays. Furthermore, the transcriptional activation of the cyclin D1 promoter by TFII-I also required its intact DNA binding motif, further suggesting that TFII-I directly activates the cyclin D1 promoter. Growth factor-dependent transcriptional activation of TFII-I may facilitate progression beyond the R-checkpoint, setting in motion transcriptional events that will lead to replication, progression to G2, and ultimately mitosis. Accordingly, cyclin D1 may not be the only TFII-I target gene that is regulated during cell cycle. Indeed, TFII-I regulates c-fos promoter activity in response to various mitogenic stimuli (52). The immediate-early c-fos is a serum-responsive gene that plays a significant role in regulating the transition from G0 to S phase upon cell cycle reentry of quiescent cells. For this reason it is highly plausible that the cell cycle-accelerating effects of TFII-I are also due to upregulation of c-fos (9).
Consistent with its essential role in the cell cycle, radiation-induced DNA damage that activates p53 and leads to cell cycle arrest also results in destruction of TFII-I in various cell lines (Fig. 7). TFII-I is ubiquitinated in vitro and in vivo in p53 wild-type but not in p53 null cells, and p53-dependent TFII-I ubiquitination leads to its proteasomal degradation. Conversely, stable overexpression of TFII-I can bypass p53-mediated cell cycle arrest and lead to cell cycle progression despite irradiation. Upon γ-irradiation, we show that TFII-I is degraded, which likely results in down-regulation of cyclin D1 and cell cycle arrest. The degradation of the ectopic TFII-I is less apparent. While it is possible that the ectopic (and tagged) TFII-I is somehow more stable than the endogenous protein, it appears that a higher dose of radiation might result in degradation of ectopic TFII-I (unpublished data). Interestingly, TFII-I degradation was also ATM dependent as it did not occur in ATM null cells. It will be interesting to determine whether this dependence on ATM reflects the necessity of this protein for p53 activation (34), if TFII-I is directly modified by ATM, or both. While the degradation of TFII-I upon γ-irradiation is clear, how this might be happening is less clear. Sequence analysis of TFII-I protein exhibits three destruction boxes of the type RXXL present at amino acids 186 to 189, 206 to 209, and 887 to 890. This sequence is present in cyclin D1 (2) and a variety of proteins that undergo proteosome-mediated degradation (48). The RXXL destruction box motif is recognized by the anaphase promoting complex (APC) and targets proteins for proteasome-mediated degradation (31). Destruction of cyclins has revealed itself as a key mechanism of cell cycle regulation (44). Indeed, the E3-related ligases of the Skp1/culin/F-box complex and of the APC mediate ubiquitination and destruction of key proteins involved in cell cycle regulation (61). Whereas the APC/C is mostly active during mitosis and G1 and acts as the key antagonist of mitotic Cdks, Skp1/culin/F-box complexes appear to be more versatile, fulfilling a variety of functions at many stages of the cell cycle and beyond (51, 61). Whether the RXXL motif of TFII-I is functionally recognized by the APC and targets TFII-I for destruction remains to be shown.
Although MDM2 is the major E3 ubiquitin ligase of p53 (8), it is has been shown to ubiquitinate other substrates such as β-arrestin, PCAF, and the insulin-like growth factor 1 receptor (23, 33, 56). Additional TFII-I candidate E3 RING ligases include SHIA-1, which is activated in a p53-dependent manner (4, 21), COP1 (constitutively photomorphogenic 1), which has recently been shown to be a p53-inducible gene (17), and members of the Cbl family, which are implicated in negative regulation of activated tyrosine kinases and downregulation of growth factor receptors (18, 37). It will be challenging to identify the E3 ligase that is responsible for radiation-induced ubiquitination of TFII-I. Currently, there are no known conserved consensus ubiquitination sites (10), making it daunting to determine by mutagenesis which lysine residues are ubiquitinated in a large protein like TFII-I. However, TFII-I does contain four lysine residues that are in consensus sumoylation motifs. In some cases sumoylation and ubiquitination may directly compete for modification of target lysines (22, 27). Whether any of these sequences play any significant role in destruction of TFII-I is currently unknown. Interestingly, we found that TFII-I protein is less ubiquitinated and expressed at higher levels in the p53 null cells. Taken together with our data implicating TFII-I in enhanced cell cycle progression via cyclin D1 regulation and the fact that cyclin D1 is elevated in many tumors (25), this observation suggests that deregulated expression of TFII-I may contribute to oncogenesis. In the future, it would be worthwhile to look at TFII-I status in tumors known to express high levels of cyclin D1.
Acknowledgments
We thank Venugopalan Cheriyath and Catarina Sacristan for establishment of stable cell lines. We thank Christine Fillmore for technical assistance. We thank Daniel Finley and members of his laboratory for reagents and advice on how to establish whether a mammalian protein is ubiquitinated in vivo. We thank Yoshi Shiloh for the HA-p53 construct.
Z.P.D. is supported by a Training Grant from the National Institutes of Health. This work is supported in part by grants from the National Institutes of Health (CA33771) to N.R. and (AI45150) to A.L.R.
REFERENCES
- 1.Agami, R., and R. Bernards. 2002. Convergence of mitogenic and DNA damage signaling in the G1 phase of the cell cycle. Cancer Lett. 177:111-118. [DOI] [PubMed] [Google Scholar]
- 2.Agami, R., and R. Bernards. 2000. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 102:55-66. [DOI] [PubMed] [Google Scholar]
- 3.Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270:23589-23597. [DOI] [PubMed] [Google Scholar]
- 4.Amson, R. B., M. Nemani, J. P. Roperch, D. Israeli, L. Bougueleret, I. Le Gall, M. Medhioub, G. Linares-Cruz, F. Lethrosne, P. Pasturaud, L. Piouffre, S. Prieur, L. Susini, V. Alvaro, P. Millasseau, C. Guidicelli, H. Bui, C. Massart, L. Cazes, F. Dufour, H. Bruzzoni-Giovanelli, H. Owadi, C. Hennion, G. Charpak, A. Telerman, and et al. 1996. Isolation of 10 differentially expressed cDNAs in p53-induced apoptosis: activation of the vertebrate homologue of the Drosophila seven in absentia gene. Proc. Natl. Acad. Sci. USA 93:3953-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartek, J., and J. Lukas. 2001. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 13:738-747. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Neriah, Y. 2002. Regulatory functions of ubiquitination in the immune system. Nat. Immunol. 3:20-26. [DOI] [PubMed] [Google Scholar]
- 7.Blagosklonny, M. V., and A. B. Pardee. 2002. The restriction point of the cell cycle. Cell Cycle 1:103-110. [PubMed] [Google Scholar]
- 8.Brooks, C. L., and W. Gu. 2004. Dynamics in the p53-Mdm2 ubiquitination pathway. Cell Cycle 3:895-899. [PubMed] [Google Scholar]
- 9.Brown, J. R., E. Nigh, R. J. Lee, H. Ye, M. A. Thompson, F. Saudou, R. G. Pestell, and M. E. Greenberg. 1998. Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol. 18:5609-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catic, A., C. Collins, G. M. Church, and H. L. Ploegh. 2004. Preferred in vivo ubiquitination sites. Bioinformatics 20:3302-3307. [DOI] [PubMed] [Google Scholar]
- 11.Cheriyath, V., Z. P. Desgranges, and A. L. Roy. 2002. c-Src dependent transcriptional activation of TFII-I. J. Biol. Chem. [DOI] [PubMed]
- 12.Cheriyath, V., C. D. Novina, and A. L. Roy. 1998. TFII-I regulates Vβ promoter activity through an initiator element. Mol. Cell. Biol. 18:4444-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheriyath, V., and A. L. Roy. 2000. Alternatively spliced isoforms of TFII-I. Complex formation, nuclear translocation, and differential gene regulation. J. Biol. Chem. 275:26300-26308. [DOI] [PubMed] [Google Scholar]
- 14.Cheriyath, V., and A. L. Roy. 2001. Structure-function analysis of TFII-I. Roles of the N-terminal end, basic region, and I-repeats. J. Biol. Chem. 276:8377-8383. [DOI] [PubMed] [Google Scholar]
- 15.D'Amico, M., K. Wu, M. Fu, M. Rao, C. Albanese, R. G. Russell, H. Lian, D. Bregman, M. A. White, and R. G. Pestell. 2004. The inhibitor of cyclin-dependent kinase 4a/alternative reading frame (INK4a/ARF) locus encoded proteins p16INK4a and p19ARF repress cyclin D1 transcription through distinct cis elements. Cancer Res. 64:4122-4130. [DOI] [PubMed] [Google Scholar]
- 16.Deane, N. G., M. A. Parker, R. Aramandla, L. Diehl, W. J. Lee, M. K. Washington, L. B. Nanney, Y. Shyr, and R. D. Beauchamp. 2001. Hepatocellular carcinoma results from chronic cyclin D1 overexpression in transgenic mice. Cancer Res. 61:5389-5395. [PubMed] [Google Scholar]
- 17.Dornan, D., I. Wertz, H. Shimizu, D. Arnott, G. D. Frantz, P. Dowd, K. O'Rourke, H. Koeppen, and V. M. Dixit. 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429:86-92. [DOI] [PubMed] [Google Scholar]
- 18.Duan, L., A. L. Reddi, A. Ghosh, M. Dimri, and H. Band. 2004. The Cbl family and other ubiquitin ligases: destructive forces in control of antigen receptor signaling. Immunity 21:7-17. [DOI] [PubMed] [Google Scholar]
- 19.Eto, I. 2000. Molecular cloning and sequence analysis of the promoter region of mouse cyclin D1 gene: implication in phorbol ester-induced tumour promotion. Cell Prolif. 33:167-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, M., C. Wang, Z. Li, T. Sakamaki, and R. G. Pestell. 2004. Minireview: cyclin D1: normal and abnormal functions. Endocrinology 145:5439-5447. [DOI] [PubMed] [Google Scholar]
- 21.Germani, A., A. Prabel, S. Mourah, M. P. Podgorniak, A. Di Carlo, R. Ehrlich, S. Gisselbrecht, N. Varin-Blank, F. Calvo, and H. Bruzzoni-Giovanelli. 2003. SIAH-1 interacts with CtIP and promotes its degradation by the proteasome pathway. Oncogene 22:8845-8851. [DOI] [PubMed] [Google Scholar]
- 22.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18:2046-2059. [DOI] [PubMed] [Google Scholar]
- 23.Girnita, L., A. Girnita, and O. Larsson. 2003. Mdm2-dependent ubiquitination and degradation of the insulin-like growth factor 1 receptor. Proc. Natl. Acad. Sci. USA 100:8247-8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grueneberg, D. A., R. W. Henry, A. Brauer, C. D. Novina, V. Cheriyath, A. L. Roy, and M. Gilman. 1997. A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: identity to TFII-I. Genes Dev. 11:2482-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han, E. K., S. C. Ng, N. Arber, M. Begemann, and I. B. Weinstein. 1999. Roles of cyclin D1 and related genes in growth inhibition, senescence and apoptosis. Apoptosis 4:213-219. [DOI] [PubMed] [Google Scholar]
- 26.Haupt, Y., A. I. Robles, C. Prives, and V. Rotter. 2002. Deconstruction of p53 functions and regulation. Oncogene 21:8223-8231. [DOI] [PubMed] [Google Scholar]
- 27.Hay, R. T. 2005. SUMO: a history of modification. Mol. Cell 18:1-12. [DOI] [PubMed] [Google Scholar]
- 28.Hipfner, D. R., and S. M. Cohen. 2004. Connecting proliferation and apoptosis in development and disease. Nat. Rev. Mol. Cell Biol. 5:805-815. [DOI] [PubMed] [Google Scholar]
- 29.Hulit, J., C. Wang, Z. Li, C. Albanese, M. Rao, D. Di Vizio, S. Shah, S. W. Byers, R. Mahmood, L. H. Augenlicht, R. Russell, and R. G. Pestell. 2004. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Mol. Cell. Biol. 24:7598-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iliakis, G., Y. Wang, J. Guan, and H. Wang. 2003. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene 22:5834-5847. [DOI] [PubMed] [Google Scholar]
- 31.Jackson, P. K., A. G. Eldridge, E. Freed, L. Furstenthal, J. Y. Hsu, B. K. Kaiser, and J. D. Reimann. 2000. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10:429-439. [DOI] [PubMed] [Google Scholar]
- 32.Jiang, W., R. Sordella, G. C. Chen, S. Hakre, A. L. Roy, and J. Settleman. 2005. An FF domain-dependent protein interaction mediates a signaling pathway for growth factor-induced gene expression. Mol. Cell 17:23-35. [DOI] [PubMed] [Google Scholar]
- 33.Jin, Y., S. X. Zeng, H. Lee, and H. Lu. 2004. MDM2 mediates p300/CREB-binding protein-associated factor ubiquitination and degradation. J. Biol. Chem. 279:20035-20043. [DOI] [PubMed] [Google Scholar]
- 34.Kastan, M. B., and J. Bartek. 2004. Cell-cycle checkpoints and cancer. Nature 432:316-323. [DOI] [PubMed] [Google Scholar]
- 35.Kim, D. W., V. Cheriyath, A. L. Roy, and B. H. Cochran. 1998. TFII-I enhances activation of the c-fos promoter through interactions with upstream elements. Mol. Cell. Biol. 18:3310-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, D. W., and B. H. Cochran. 2000. Extracellular signal-regulated kinase binds to TFII-I and regulates its activation of the c-fos promoter. Mol. Cell. Biol. 20:1140-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, M., T. Tezuka, K. Tanaka, and T. Yamamoto. 2004. Cbl-c suppresses v-Src-induced transformation through ubiquitin-dependent protein degradation. Oncogene 23:7903-7904. [DOI] [PubMed] [Google Scholar]
- 38.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 39.Lindemann, D., E. Patriquin, S. Feng, and R. C. Mulligan. 1997. Versatile retrovirus vector systems for regulated gene expression in vitro and in vivo. Mol. Med. 3:466-476. [PMC free article] [PubMed] [Google Scholar]
- 40.Lois, C., E. J. Hong, S. Pease, E. J. Brown, and D. Baltimore. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295:868-872. [DOI] [PubMed] [Google Scholar]
- 41.Ma, J., and M. Ptashne. 1987. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847-853. [DOI] [PubMed] [Google Scholar]
- 42.Manzano-Winkler, B., C. D. Novina, and A. L. Roy. 1996. TFII is required for transcription of the naturally TATA-less but initiator-containing Vbeta promoter. J. Biol. Chem. 271:12076-12081. [DOI] [PubMed] [Google Scholar]
- 43.Massague, J. 2004. G1 cell-cycle control and cancer. Nature 432:298-306. [DOI] [PubMed] [Google Scholar]
- 44.Murray, A. W. 2004. Recycling the cell cycle: cyclins revisited. Cell 116:221-234. [DOI] [PubMed] [Google Scholar]
- 45.Nelsen, C. J., R. Kuriyama, B. Hirsch, V. C. Negron, W. L. Lingle, M. M. Goggin, M. W. Stanley, and J. H. Albrecht. 2004. Short-term cyclin D1 overexpression induces centrosome amplification, mitotic spindle abnormalities, and aneuploidy. J. Biol. Chem. [DOI] [PubMed]
- 46.Novina, C. D., V. Cheriyath, and A. L. Roy. 1998. Regulation of TFII-I activity by phosphorylation. J. Biol. Chem. 273:33443-33448. [DOI] [PubMed] [Google Scholar]
- 47.Novina, C. D., S. Kumar, U. Bajpai, V. Cheriyath, K. Zhang, S. Pillai, H. H. Wortis, and A. L. Roy. 1999. Regulation of nuclear localization and transcriptional activity of TFII-I by Bruton's tyrosine kinase. Mol. Cell. Biol. 19:5014-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pagano, M. 1997. Cell cycle regulation by the ubiquitin pathway. FASEB J. 11:1067-1075. [DOI] [PubMed] [Google Scholar]
- 49.Parker, R., T. Phan, P. Baumeister, B. Roy, V. Cheriyath, A. L. Roy, and A. S. Lee. 2001. Identification of TFII-I as the endoplasmic reticulum stress response element binding factor ERSF: its autoregulation by stress and interaction with ATF6. Mol. Cell. Biol. 21:3220-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Philipp, A., A. Schneider, I. Vasrik, K. Finke, Y. Xiong, D. Beach, K. Alitalo, and M. Eilers. 1994. Repression of cyclin D1: a novel function of MYC. Mol. Cell. Biol. 14:4032-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pickart, C. M. 2004. Back to the future with ubiquitin. Cell 116:181-190. [DOI] [PubMed] [Google Scholar]
- 52.Roy, A. L. 2001. Biochemistry and biology of the inducible multifunctional transcription factor TFII-I. Gene 274:1-13. [DOI] [PubMed] [Google Scholar]
- 53.Roy, A. L., H. Du, P. D. Gregor, C. D. Novina, E. Martinez, and R. G. Roeder. 1997. Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 16:7091-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy, A. L., M. Meisterernst, P. Pognonec, and R. G. Roeder. 1991. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature 354:245-248. [DOI] [PubMed] [Google Scholar]
- 55.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, D. L. Rooney, M. M. Ihrig, M. T. McManus, F. B. Gertler, M. L. Scott, and L. Van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 56.Shenoy, S. K., P. H. McDonald, T. A. Kohout, and R. J. Lefkowitz. 2001. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science 294:1307-1313. [DOI] [PubMed] [Google Scholar]
- 57.Slansky, J. E., Y. Li, W. G. Kaelin, and P. J. Farnham. 1993. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol. Cell. Biol. 13:1610-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun, L., and Z. J. Chen. 2004. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 16:119-126. [DOI] [PubMed] [Google Scholar]
- 59.Thome, K. C., A. Radfar, and N. Rosenberg. 1997. Mutation of Tp53 contributes to the malignant phenotype of Abelson virus-transformed lymphoid cells. J. Virol. 71:8149-8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Unnikrishnan, I., A. Radfar, J. Jenab-Wolcott, and N. Rosenberg. 1999. p53 mediates apoptotic crisis in primary Abelson virus-transformed pre-B cells. Mol. Cell. Biol. 19:4825-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vodermaier, H. C. 2004. APC/C and SCF: controlling each other and the cell cycle. Curr. Biol. 14:R787-R796. [DOI] [PubMed] [Google Scholar]
- 62.Wang, T. C., R. D. Cardiff, L. Zukerberg, E. Lees, A. Arnold, and E. V. Schmidt. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369:669-671. [DOI] [PubMed] [Google Scholar]
- 63.Wang, Y. K., L. A. Perez-Jurado, and U. Francke. 1998. A mouse single-copy gene, Gtf2i, the homolog of human GTF2I, that is duplicated in the Williams-Beuren syndrome deletion region. Genomics 48:163-170. [DOI] [PubMed] [Google Scholar]
- 64.Yang, W., and S. Desiderio. 1997. BAP-135, a target for Bruton's tyrosine kinase in response to B cell receptor engagement. Proc. Natl. Acad. Sci. USA 94:604-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 66.Zhao, R., K. Gish, M. Murphy, Y. Yin, D. Notterman, W. H. Hoffman, E. Tom, D. H. Mack, and A. J. Levine. 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14:981-993. [PMC free article] [PubMed] [Google Scholar]