Abstract
The transcriptional coactivator PGC-1α is a key regulator of energy metabolism, yet little is known about its role in control of substrate selection. We found that physiological stimuli known to induce PGC-1α expression in skeletal muscle coordinately upregulate the expression of pyruvate dehydrogenase kinase 4 (PDK4), a negative regulator of glucose oxidation. Forced expression of PGC-1α in C2C12 myotubes induced PDK4 mRNA and protein expression. PGC-1α-mediated activation of PDK4 expression was shown to occur at the transcriptional level and was mapped to a putative nuclear receptor binding site. Gel shift assays demonstrated that the PGC-1α-responsive element bound the estrogen-related receptor α (ERRα), a recently identified component of the PGC-1α signaling pathway. In addition, PGC-1α was shown to activate ERRα expression. Chromatin immunoprecipitation assays confirmed that PGC-1α and ERRα occupied the mPDK4 promoter in C2C12 myotubes. Additionally, transfection studies using ERRα-null primary fibroblasts demonstrated that ERRα is required for PGC-1α-mediated activation of the mPDK4 promoter. As predicted by the effects of PGC-1α on PDK4 gene transcription, overexpression of PGC-1α in C2C12 myotubes decreased glucose oxidation rates. These results identify the PDK4 gene as a new PGC-1α/ERRα target and suggest a mechanism whereby PGC-1α exerts reciprocal inhibitory influences on glucose catabolism while increasing alternate mitochondrial oxidative pathways in skeletal muscle.
The transcriptional coactivator peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α) is a key regulator of cellular energy metabolism (40), having known roles in thermogenesis (41), mitochondrial biogenesis (28, 56), fatty acid oxidation (51), and hepatic gluconeogenesis (14, 57). PGC-1α is enriched in metabolically active tissues including brown adipose tissue, the heart, and slow-twitch skeletal muscles (30, 41). In contrast to most transcriptional coactivators, expression of the PGC-1α gene is highly inducible in accordance with tissue-specific energy demands. For example, PGC-1α expression is rapidly increased in brown adipose tissue following cold exposure (41), in the liver and heart following short-term starvation (28, 57), in skeletal muscle with exercise (2, 12, 38), and in the postnatal heart coincident with an increase in mitochondrial fatty acid oxidation (28). Gain-of-function and loss-of-function studies have demonstrated that PGC-1α is necessary and sufficient to increase mitochondrial content and respiratory capacity in response to physiological stimuli in adipocytes (31, 41), skeletal muscle (29, 56), and the heart (1, 29, 44, 54). Skeletal-muscle-specific overexpression of PGC-1α results in a fiber type switch from fast-twitch (type II) to slow-twitch (type I) oxidative fibers (30). Slow-twitch fibers are characterized by increased insulin sensitivity, mitochondrial mass, and oxidative capacity. Recent studies have also demonstrated altered expression of PGC-1α in diabetic skeletal muscles (36) and hearts (8).
A variety of transcription factors are known to be coactivated by PGC-1α, including PPARγ (41), PPARα (51), FOXO1 (39), and estrogen-related receptor α (ERRα) (18, 46). In brown adipose tissue, PGC-1α interacts with PPARγ and other transcription factors to regulate the expression of genes involved in adaptive thermogenesis (41). The PGC-1α/PPARα pathway is involved in the regulation of mitochondrial fatty acid oxidation genes in the heart (28, 51). The PGC-1α/FOXO1 pathway activates expression of gluconeogenic genes in the liver (39). More recently, the PGC-1α/ERRα pathway has been shown to play a role in regulating both fatty acid oxidation and mitochondrial genes in heart and skeletal muscle (20, 35, 45, 53).
In contrast to the role of PGC-1α in the regulation of fatty acid oxidation, mitochondrial respiratory function, and hepatic gluconeogenesis, its function as a potential regulator of glucose utilization pathways has not been well defined. Indeed, the few reports relating PGC-1α to glucose uptake are conflicting. PGC-1α has been reported to induce (33) or repress (34) expression of the glucose transporter GLUT4. Given that regulatory mechanisms exist for reciprocal control of fatty acid and glucose oxidation, it is likely that the PGC-1α regulatory circuit directly or indirectly influences both pathways. Cellular glucose utilization is tightly regulated at multiple levels, including uptake by the glucose transporters (e.g., GLUT4), glycolytic flux, and entry of pyruvate into the citric acid cycle via the pyruvate dehydrogenase complex (PDC). In muscle, the PDC serves a critical, rate-limiting step in the regulation of the glucose oxidation pathway by catalyzing the irreversible decarboxylation of pyruvate to acetyl coenzyme A. A significant body of evidence indicates that multiple regulatory pathways converge on the PDC, including allosteric regulation intermediates of fatty acid oxidation and posttranslational control by a family of pyruvate dehydrogenase kinases (PDK1 to -4) and corresponding phosphatases (reviewed in references 13 and 16). The reversible phosphorylation of the PDC inactivates this complex, sparing glucose and favoring fatty acid oxidation. PDK4 has proven to be particularly important in the muscle and liver response to fasting and exercise (15, 37, 54). PDK4 activity and expression are increased in diabetes concurrent with reduced capacity for muscle and hepatic glucose oxidation. Interestingly, PGC-1α is also activated by fasting and exercise. In addition, recent studies have shown that activators of PPARα, a known PGC-1α target, induce the expression of PDK4 (17, 55). However, the mechanism whereby PPARα regulates PDK4 gene expression has not been delineated. Recent studies have also shown that the forkhead transcription factor FOXO1 and the glucocorticoid receptor (GR), two additional potential PGC-1α partners, directly regulate PDK4 expression through consensus binding sites in the PDK4 gene promoter (9, 26).
The present study was designed to investigate the potential regulatory influence of the inducible transcriptional coactivator PGC-1α on glucose oxidation through effects on PDK4 gene expression. We found that PGC-1α directly activates PDK4 gene transcription in skeletal myotubes, leading to a reduction in glucose oxidation rates. Surprisingly, the PGC-1α-mediated activation of PDK4 transcription was shown to be independent of PPARα and FOXO1; rather, it is dependent on the muscle-enriched orphan nuclear receptor ERRα. Collectively, these results identify a new PGC-1α gene target and suggest a mechanism whereby PGC-1α reciprocally downregulates glucose oxidation while activating mitochondrial fatty acid oxidation in skeletal muscle. Given the importance of PDK4-mediated regulation of muscle glucose utilization, these results provide additional insight into the proposed link between PGC-1α and metabolic disorders such as diabetes.
MATERIALS AND METHODS
Animal studies.
All animal experiments and euthanasia protocols were conducted in strict accordance with the National Institutes of Health guidelines for humane treatment of animals and were reviewed and approved by the Animal Care Committee of Washington University. Unless stated otherwise, 6- to 8-week-old male C57BL/6J mice were used for all animal experiments. A modified treadmill protocol was used for acute exercise experiments (29). In brief, animals were acclimated to the treadmill for 10 min, and on each of the following 2 days, animals exercised for 1 h total, alternating 2 min of running (20 m/min at an 18° incline) with 1 min of rest. Twelve hours following the final bout of exercise, animals were sacrificed, and gastrocnemius muscles were dissected and snap-frozen in liquid nitrogen for RNA extraction. For time course studies, animals underwent a single bout of running following 2 days of 10-min acclimation to the treadmill. Runs were staggered so that all samples would be collected at the same time of day (0900 h). For cold exposure experiments, animals were maintained at 4°C for 6 h as previously described (29); immediately afterwards, they were sacrificed and their tissues dissected for RNA extraction.
Plasmid constructs. (i) Mammalian expression vectors.
pcDNA3.1-myc/ his.PGC-1α, pcDNA3.1-ERRα, CDM-PPARα, and CDM-RXRα have been described previously (18, 51). The expression vector pcDNA3-Flag-FKHR (FOXO1) was provided by W. R. Sellers (Dana-Faber Cancer Institute) and has been described previously (49).
(ii) Reporter constructs.
The mouse PDK4 (mPDK4) gene promoter deletion series was generated by PCR amplification from C57BL/6J genomic DNA followed by cloning into the pGL3 Basic firefly luciferase reporter plasmid using MluI and BglII sites (Promega, Madison, WI). The following 5′ primers were used: 5′-ACGCGTTCTAGATAAGAATATCATTCTTTGT-3′ (mPDK4.Luc.2281), 5′-ACGCGTACATGATCATTTCCTACATGCTTG-3′ (mPDK4.Luc.2121), 5′-ACGCGTACACAAGTGGACACAACATTAGGC-3′ (mPDK4.Luc.1377), 5′-ACGCGTACTGAGCCTCTGGGAAATGAAAGA-3′ (mPDK4.Luc.874), 5′-ACGCGTGGCTAGGAATGCGTGACATTGAGAT-3′ (mPDK4.Luc.371), 5′-ACGCGTCTGAAGTCCTAGCGACCTGGGATCT-3′ (mPDK4.Luc.308), and 5′-ACGCGTAGGCCATGGAAACCGTGTCGGGATCACTGT-3′ (mPDK4.Luc.190). The same 3′ primer was used with all constructs: 5′-AGATCTGAGCCTGGGTGAAGGGTTGACACTT-3′. Site-directed mutagenesis was performed using the QuikChange kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol using complementary oligonucleotides as follows (with mutated nucleotides shown in lowercase): 5′-GATGGCTCTGGAGTTGTAggggAGGACAAGTCTGGG-3′ (IRSmut), 5′-CTGGAGTTGTAAACAgatctAAGTCTGGGCGG-3′ (NRmut), 5′-GGAGTTGTAAACAAGGACAAGTCTaaGCttGCCTGAAGTCCTAGCG-3′ (Sp1mut), and 5′-CTGGAGTTGTAAACAAcGACAAGTCTGGGCGG-3′ (NRptmut). Primers were designed, and clones were analyzed, using Vector NTI Suite 9 (InforMax, Rockville, MD) and the reported mPDK4 gene sequence (GenBank accession number NT_039340). All clones were verified by ABI automated sequencing through the Protein and Nucleic Acid Chemistry Laboratory (PNACL) at Washington University. Numbering was determined by estimating the transcription start site (+1) by homology to the human PDK4 (hPDK4) gene (43) (GenBank accession number NM_002612). Candidate transcription factor response elements were identified using the Match program (http://www.gene-regulation.com).
(iii) Adenovirus expression vectors.
The adenovirus (Ad)-green fluorescent protein (GFP), Ad-PGC-1α, Ad-ERRα, and Ad-PPARα expression vectors have been described previously (18, 19, 28).
Cell culture, transient transfection, and luciferase reporter studies.
C2C12 myoblasts and CV-1 cells were maintained at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. For experiments requiring myotube formation, the medium of C2C12 myoblasts at >80% confluence was changed to DMEM supplemented with 2% horse serum, and the cells were maintained for an additional 72 h. CV-1 cells and C2C12 myotubes were infected at a multiplicity of infection sufficient to infect >95% of the cells based on GFP fluorescence with minimal cell death. ERRα−/− mice for primary cell culture isolation have been described elsewhere (32). Primary mouse fibroblasts from tail tissue of ERRα+/+ and ERRα−/− mice were isolated and cultured as described elsewhere (20). Transient transfections were performed by the calcium phosphate coprecipitation method (4, 11). Briefly, reporter plasmids (4 μg/ml) were cotransfected with simian virus 40 promoter-driven Renilla luciferase (500 ng/ml), to control for transfection efficiency, and the appropriate mammalian expression vector (500 ng/ml) or its corresponding empty vector. For luciferase reporter assays, CV-1 cells and C2C12 myotubes were harvested and analyzed at 48 and 72 h, respectively, using Dual-Glo (Promega) and were read in a Chameleon plate reader (Hidex, Washington, D.C.) according to the manufacturer's protocols. All transfection data are presented as means ± standard errors (SE) for at least three separate transfection experiments done in triplicate.
RNA and protein analyses.
Total cellular RNA was isolated from cells in culture (48 h following adenovirus infection) or muscle tissue using the RNAzol method (Tel-Test, Friendswood, TX). Northern blot analysis was performed with QuikHyb (Stratagene) using random-primed 32P-labeled cDNA clones for human β-actin, mouse PGC-1α, and PDK4. Band intensities were quantified by phosphorimaging using a GS 525 Molecular Imager system (Bio-Rad) and normalized to the expression of β-actin. Real-time quantitative reverse transcription-PCR (RT-PCR) was performed as previously described (20) and results normalized to the expression of 36B4. The following mouse-specific primer pairs were used: PGC-1α forward, 5′-CGGAAATCATATCCAACCAG-3′; PGC-1α reverse, 5′-TGAGGACCGCTAGCAAGTTTG-3′; PDK4 forward, 5′-CCGCTGTCCATGAAGCA-3′; PDK4 reverse, 5′-GCAGAAAAGCAAAGGACGTT-3′; 36B4 forward, 5′-GCAGACAACGTGGGCTCCAAGCAGAT-3′; 36B4 reverse, 5′-GGTCCTCCTTGGTGAACACGAAGCCC-3′.
Protein extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (4 to 15% Criterion Precast; Bio-Rad) and transferred to nitrocellulose membranes (Midwest Scientific, Valley Park, MO). Western blotting was performed using antibodies against PGC-1α (28), medium-chain acyl coenzyme A dehydrogenase (MCAD) (24), PDK4 (a gift from R. A. Harris, Indiana University School of Medicine), α-tubulin (Molecular Probes, Eugene, OR), PPARα, FKHR/FOXO1, and Sp1 (Santa Cruz Biotechnology, Santa Cruz, CA). An antibody against ERRα was raised in rabbits against an N-terminal peptide corresponding to amino acids 16 to 33 of the mouse ERRα protein conjugated to keyhole limpet hemocyanin (KLH) or bovine serum albumin (BSA). This domain is conserved among species; therefore, the antibody recognizes rodent and human ERRα isoforms. The KLH-conjugated peptide in Freund's adjuvant was used for the initial and second booster immunizations. Subsequent boosts were performed with the BSA-conjugated peptide. Peptide synthesis, conjugation, and purification by high-performance liquid chromatography were performed through PNACL (Washington University). Peptide immunization and serum collection were performed through Biosource (Hopkinton, MA). The specificity of the anti-ERRα antibody was determined by the ability of this antibody to detect recombinant ERRα but not ERRγ and by comparison to the previously described anti-ERRα antibody (47). Detection was performed by measuring the chemiluminescent signal as assayed by SuperSignal Ultra (Pierce, Rockford, IL).
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed as described previously (6). Double-stranded complementary oligonucleotides corresponding to the PGC-1α-responsive element (5′-gatccGGCTAGGAATGCGTGACATTGAGATGGCTCTGGAGTTGTAAACAAGGACAAGTCTGGG CGGGCg-3′) or NRmut (5′-gatccGGCTAGGAATGCGTGACATTGAGATGGCTCTGGAGTTGTAAACAGATCTAAGTCTGGGCGGGCg-3′) (overhangs are shown in lowercase; mutation is underlined) of the mPDK4 promoter were used to generate probes to assay recombinant and endogenous protein binding in vitro. Probes were radiolabeled by a Klenow fill-in reaction. Recombinant proteins for ERRα, PPARα, and FOXO1 were synthesized with the TNT Quick T7-coupled translation kit (Promega). Synthesis of appropriately sized proteins was verified in reactions incorporating [35S]methionine followed by SDS-PAGE analysis and autoradiographic detection. Nuclear extract was isolated as previously described (6). Briefly, C2C12 myotubes were infected with Ad-GFP or Ad-PGC-1α, nuclear extract was isolated 48 h following infection, and samples were aliquoted and stored at −80°C until use.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as previously published (21, 52). In brief, C2C12 myotubes were infected with Ad-GFP or Ad-PGC-1α. Cross-linking was performed 36 to 48 h postinfection byincubating cells with 0.4% formaldehyde (10 min) followed by addition of 0.125M glycine to halt cross-linking. Following cell and nuclear lysis, DNA was sheared by sonication; enrichment for 500-bp fragments was determined by agarose gel electrophoresis. Sheared chromatin complexes were precleared with preimmune rabbit serum, followed by dilution of an aliquot in immunoprecipitation dilution buffer and incubation overnight at 4°C with immunoglobulin G (IgG) or a polyclonal antibody against PGC-1α or ERRα (described above). Antibody-chromatin complexes were bound with ImmunoPure immobilized protein A (Pierce), followed by isolation and purification as described previously (20). Primers were designed to amplify a 164-bp amplicon corresponding to the region of the mPDK4 promoter flanking the nuclear receptor (NR) response element (nucleotides −449 to −285). The following mouse-specific primer pairs were used: PDK4pro forward (5′-TGATTGGCTACTGTAAAAGTCCCGC-3′) and PDK4pro reverse (5′-ATCCCAGGTCGCTAGGACTTCAGG-3′). Primers amplifying the 36B4 gene (see above) were used to control for nonspecific enrichment of DNA in the immunoprecipitation. Samples were analyzed by 2% agarose gel electrophoresis and quantified relative to input using SYBR green RT-PCR (Applied Biosystems, Foster City, CA).
Glucose oxidation assay.
Cellular glucose oxidation rates were determined using modifications of previously published protocols (5, 22). C2C12 myoblasts were grown in T25 flasks and differentiated into myotubes as described above. Forty-eight hours following infection with Ad-GFP or Ad-PGC-1α, the medium was replaced with MEM supplemented with 0.5% horse serum. Following 18 h of reduced serum incubation, new medium was added containing 0.2 μCi/ml [U-14C]glucose (10 mCi/mmol), 100 nM insulin, and 250 μM oleate. The flasks were immediately capped with rubber stoppers fitted with plastic wells containing a piece of filter paper saturated with 1 M NaOH. The culture flasks were incubated for 30 min at 37°C. Reactions were terminated with perchloric acid, followed by overnight storage at 4°C to trap CO2. The release of 14CO2 from glucose was measured by scintillation counting of the filter paper. Duplicate flasks were plated for each condition. The results were normalized to total protein (determined by the Bradford method).
RESULTS
PGC-1α induces expression of the PDK4 gene in skeletal muscle.
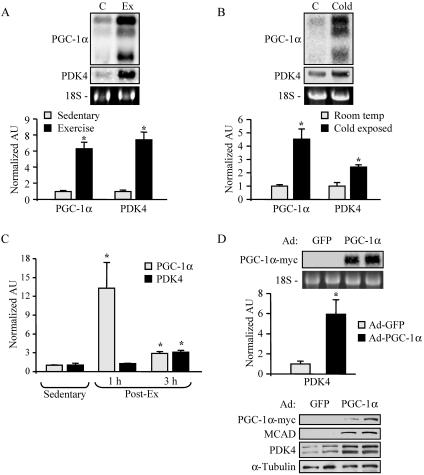
Physiological conditions that increase skeletal muscle energy demands induce the expression of the transcriptional coactivator PGC-1α. As an initial step to investigate a potential role for PGC-1α in the regulation of muscle glucose metabolism via PDK4, we examined levels of mRNAs encoding PGC-1α and PDK4 following acute exercise. For these experiments, a running protocol on a motorized treadmill was employed (29). PGC-1α mRNA levels were markedly induced in mouse gastrocnemius 12 h after two bouts of treadmill running (Fig. 1A). The expression of PDK4 was robustly induced in parallel with PGC-1α expression (Fig. 1A). Similar experiments were conducted following cold exposure, which triggers shivering thermogenesis in skeletal muscle and has been shown to induce PGC-1α expression (41). The expression of PDK4 and PGC-1α genes was coordinately induced (two- to fourfold) in muscle following 6 h of cold exposure (Fig. 1B). To assess the temporal patterns of the physiological induction of PGC-1α and PDK4 gene expression in muscle, a time course experiment was performed following a single bout of exercise. As shown in Fig. 1C, PGC-1α gene expression was markedly induced (at 1 h postexercise) prior to the activation of PDK4 gene expression.
FIG. 1.
Activation of PDK4 gene expression by physiological stimuli and by overexpression of PGC-1α. (A) Northern blot studies performed with 15 μg of total RNA isolated from the gastrocnemii of animals following two bouts of exercise as described in Materials and Methods (Ex) or from sedentary controls (C) (n = 4). Blots were sequentially hybridized with probes specific for the genes encoding PGC-1α and PDK4. Representative autoradiographs are shown at the top. Phosphorimage-based quantification of Northern blot signal intensities is shown in the graph. Data represent mean arbitrary units (±SE) normalized to sedentary control values (taken as 1.0). (B) Results of Northern blot analysis performed as described above on gastrocnemius samples immediately following 6 h of cold exposure (Cold) and from room temperature controls (C) (n = 6). Graph represents mean arbitrary units (±SE) as determined by real-time PCR quantification corrected to the 36B4 transcript and normalized to the values for control littermates (taken as 1.0). (C) RNA analysis of PGC-1α and PDK4 gastrocnemius gene expression 1 h and 3 h after a single bout of exercise (n = 4). The graph represents mean arbitrary units (±SE) as determined by RT-PCR corrected to the 36B4 transcript and normalized to the value for sedentary controls (taken as 1.0). (D) RNA analyses and Western blot analysis were performed with samples isolated from Ad-GFP- or Ad-PGC-1α-infected C2C12 myotubes. (Top) Results of Northern blotting performed as described above. Data represent mean arbitrary units (±SE) as determined by RT-PCR corrected to the 36B4 transcript and normalized to value for control Ad-GFP-infected samples (taken as 1.0). (Bottom) Western blot analysis of whole-cell extracts (25 μg) prepared from C2C12 myotubes infected with adenovirus vectors expressing GFP or PGC-1α. Asterisks indicate significant differences (P < 0.05) from controls.
To determine if PGC-1α plays a direct role in the regulation of PDK4 gene expression, we examined the effect of adenovirus-mediated overexpression of PGC-1α (Ad-PGC-1α) compared to a control vector (Ad-GFP) in C2C12 myotubes. As expected, we observed an increase in levels of MCAD protein, a known PGC-1α target (51), following Ad-PGC-1α infection (Fig. 1D). Overexpression of PGC-1α markedly activated PDK4 mRNA and protein levels (Fig. 1D). Taken together, these results indicate that PGC-1α functions as an upstream activator of PDK4 gene expression.
PGC-1α activates PDK4 gene transcription through a nuclear receptor binding site.
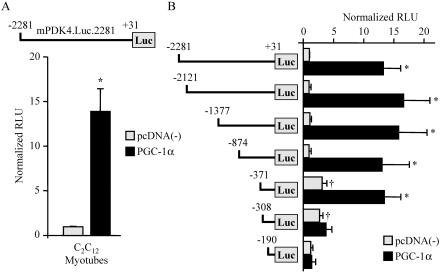
To determine whether PGC-1α induces PDK4 gene expression at the transcriptional level, we performed promoter-reporter transfections in C2C12 myotubes in culture. For these experiments, approximately 2.3 kb of the mPDK4 gene promoter region was cloned into a promoterless luciferase reporter vector (mPDK4.Luc.2281). Cotransfection of mPDK4.Luc.2281 with an expression vector for PGC-1α resulted in robust (14-fold) activation of the reporter (Fig. 2A). To map the cis-acting region conferring the PGC-1α activation, cotransfection experiments were repeated with reporter constructs containing serial 5′ deletions of the mPDK4 promoter (Fig. 2B). The PGC-1α-mediated activation was lost upon deletion of a region of the mPDK4 promoter from −371 to −308 bp upstream of the transcription start site. In addition, the basal promoter activity increased upon deletion of the region between −874 and −371 bp.
FIG. 2.
PGC-1α activates the mPDK4 gene promoter through a cis-acting element within the proximal region. (A) A schematic of the reporter construct mPDK4.Luc.2281, containing the −2281-to-+31 region of the mouse PDK4 gene promoter, is shown at the top. The graph contains results of cotransfection studies with mPDK4.Luc.2281 in the presence (PGC-1α) or absence [pcDNA(−)] of pcDNA-PGC-1α in C2C12 myotubes. The graph represents mean (±SE) relative light units (RLU) corrected for Renilla luciferase activity and normalized to the activity of mPDK4.Luc.2281 (taken as 1.0). (B) Transient transfections performed with a 5′ deletion series of mPDK4.Luc.2281. All values represent at least three independent transfections conducted in triplicate. Asterisks indicate significant differences compared to individual reporters cotransfected with empty vector (P < 0.05). Daggers indicate significant differences in activity compared to mPDK4.Luc.2281 cotransfected with empty vector (P < 0.05).
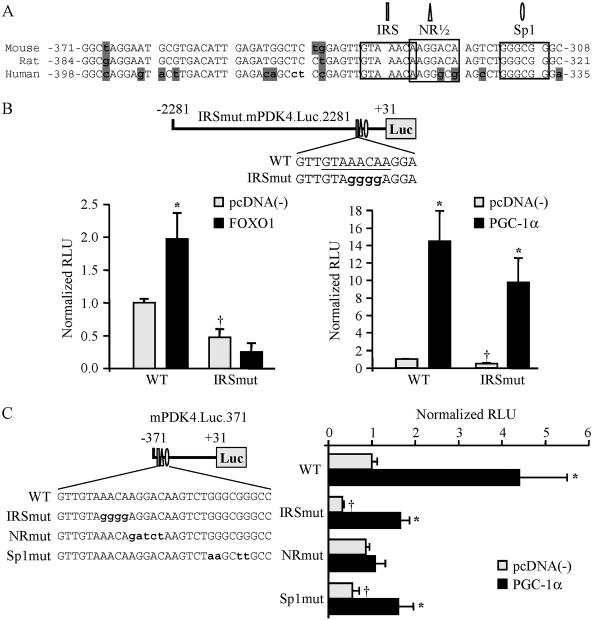
Inspection of the nucleotide sequence of the PGC-1α-responsive region revealed a high degree of nucleotide identity across species (Fig. 3A). A screen of the 63-bp DNA sequence identified several putative transcription factor binding sites that could support PGC-1α coactivation, including an NR half-site, an Sp1 site, and a recently identified FOXO1 binding site (DBE [9], or IRS3 [26]) (Fig. 3A). Interestingly, coactivation of FOXO1 by PGC-1α has been implicated in the regulation of hepatic gluconeogenic gene expression (39). Therefore, we first sought to determine whether the FOXO1 response element was necessary for the PGC-1α-mediated activation. The FOXO1 binding site within the mPDK4.Luc.2281 reporter was abolished using a mutation that has been shown previously to inactivate this element (9). Although the FOXO1 site mutant was incapable of responding to FOXO1, PGC-1α-mediated activation of the mPDK4 promoter was unchanged (Fig. 3B). It is noteworthy that the FOXO1 site mutant resulted in a 50% reduction in basal promoter activity, indicating a PGC-1α-independent role for FOXO1.
FIG. 3.
PGC-1α activates PDK4 gene transcription through a nuclear receptor binding site. (A) Species comparison of the nucleotide sequence (sense strand is shown) of the PGC-1α-activated region of the PDK4 promoter, demonstrating a high level of sequence identity between mouse, rat, and human genes. The numbers are relative to the transcription start site (+1). Shaded nucleotides indicate sequence that is not conserved. Computer analysis revealed numerous candidate response elements, including a nuclear receptor half-site (NR1/2), an Sp1 site, and the previously identified FOXO1 binding site (IRS) (9, 27), indicated by labels, boxes, and symbols. (B) (Top) Site-directed mutagenesis was used to abolish the FOXO1 response element. Mutated nucleotides are lowercased. (Bottom) The mPDK4.Luc.2281 (wild type [WT]) or IRSmut.mPDK4.Luc.2281 promoter reporter was used in cotransfections in C2C12 myotubes in the presence or absence of FOXO1 (left graph) or PGC-1α (right graph). (C) (Left) Schematic showing the mutations generated by site-directed mutagenesis of the minimal PGC-1α-responsive mPDK4.Luc.371 reporter in FOXO1 (IRSmut), NR (NRmut), or Sp1 (Sp1mut) candidate response elements (mutated nucleotides are lowercased). (Right) Cotransfection studies using WT or mutant mPDK4 promoter reporters were performed with C2C12 myotubes. Data represent mean (±SE) relative light units (RLU) corrected for rLuc and normalized to values for the WT reporter (taken as 1.0). All transient transfection studies represent at least three independent transfections conducted in triplicate. Asterisks indicate significant differences from individual reporters cotransfected with empty vector (P < 0.05). Daggers indicate significant differences from the wild-type reporter cotransfected with empty vector (P < 0.05).
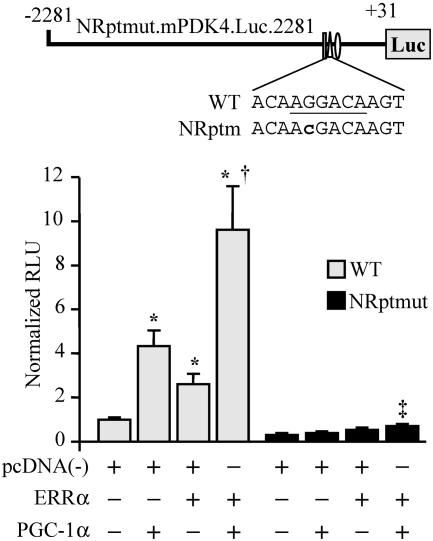
Mutational studies were performed to examine the importance of the putative nuclear receptor and Sp1 binding sites for PGC-1α coactivation. The nuclear receptor site was of particular interest given that several studies have shown that PPARα and its ligands regulate PDK4 gene expression (7, 17, 55). Although mutation of the Sp1 response element significantly reduced the basal promoter activity of the mPDK4.Luc.371 reporter, PGC-1α-mediated activation was maintained (Fig. 3C). In contrast, mutation of the putative NR response element abolished activation by PGC-1α. Together, these results demonstrate that the candidate NR response element is required for the observed PGC-1α-mediated activation of the PDK4 gene promoter.
The orphan nuclear receptor ERRα, but not PPARα, directly confers PGC-1α-mediated coactivation of the mPDK4 promoter.
To determine if PPARα was capable of binding to the PGC-1α-responsive NR site identified within the mPDK4 gene promoter, EMSA were performed using recombinant PPARα and RXRα, its obligate DNA binding partner. Surprisingly, PPARα/RXRα did not bind to this region but did bind to a known PPAR response element from the muscle carnitine palmitoyltransferase I gene promoter (3) (data not shown).
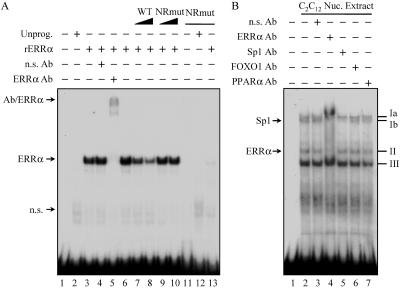
The orphan nuclear receptor ERRα is a recently identified mediator of PGC-1α action (18, 46). ERRα was an attractive candidate for binding the nuclear receptor half-site within the PGC-1α-responsive region of the mPDK4 promoter, because it is capable of binding as a monomer (23) and is enriched in slow-twitch muscle (10, 20, 47). Therefore, EMSA were repeated using recombinant ERRα and the 63-bp region as a probe. As shown in Fig. 4A, ERRα bound the 63-bp PGC-1α-responsive region of the mPDK4 promoter (lane 3). Competition studies with unlabeled wild-type and mutated fragments (Fig. 4A, lanes 6 to 10), as well as ERRα antibody recognition experiments (lanes 3 to 5), confirmed the specificity of the ERRα binding to the NR site. In addition, relative to the wild-type probe, a 32P-labeled mutant probe failed to support ERRα binding (Fig. 4A, lanes 11 to 13).
FIG. 4.
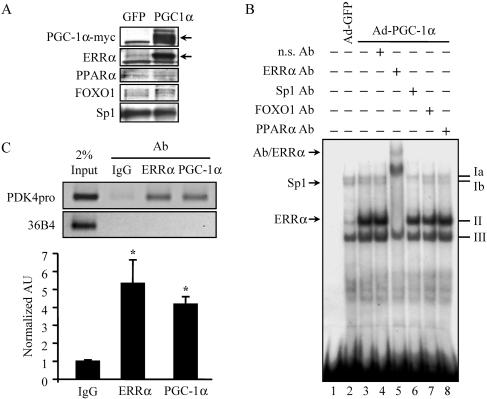
The orphan nuclear receptor ERRα binds to the PGC-1α-responsive region of the mPDK4 promoter. (A) Representative autoradiograph of EMSA conducted with 32P-labeled probes of the 63-bp region of the mPDK4 promoter identified in Fig. 2B. Mutant probe corresponds to the same nucleotide substitutions for NRmut as in Fig. 3C. Probes were incubated with recombinant in vitro-translated ERRα (rERRα) or unprogrammed reticulocyte lysate (Unprog.) as indicated at the top. Antibody (Ab) supershift studies were performed by incubation with an anti-ERRα (lane 5) or nonspecific (n.s.) (lane 4) Ab. Competition analysis was performed by adding a molar excess of the unlabeled wild-type (lanes 7 and 8) or NRmut (lanes 9 and 10) probe in increasing amounts as indicated. For lanes 11 to 13, the 32P-labeled NRmut probe was used. n.s. bands represent protein complexes identified in unprogrammed reticulocyte lysate. (B) Autoradiograph of an EMSA using nuclear (Nuc.) extracts isolated from C2C12 myotubes incubated with the 32P-labeled wild-type probe. Four prominent complexes, labeled Ia, Ib, II, and III, were observed. Complexes were identified by antibody supershift analysis using antibodies specific for the transcription factors indicated at the top.
To determine whether endogenous ERRα bound to the mPDK4 gene promoter, EMSA were repeated with nuclear extracts prepared from C2C12 myotubes. Three complexes formed with the 63-bp mPDK4 probe (Fig. 4B, complexes I to III). Complex I, the slowest-migrating band, exhibited a doublet (Ia and Ib). To identify proteins within the complexes, antibody recognition experiments were performed using antibodies to ERRα, Sp1, FOXO1, and PPARα. Complex Ib was abolished by anti-Sp1 (Fig. 4B, lane 5). Complex II was supershifted with anti-ERRα (Fig. 4B, lane 4). Additionally, the anti-ERRα antibody disrupted complexes Ia and Ib, suggesting a potential requirement for ERRα binding in the formation of these two complexes (Fig. 4B, lane 4). Alternatively, the latter result could be due to the “mass action” of the supershifted complex II. Surprisingly, FOXO1 was not identified within any of the complexes, though the antibody was shown to supershift recombinant FOXO1 (Fig. 4B and data not shown). As expected, none of the complexes were recognized by anti-PPARα. The identities of complexes Ia and III are under investigation. Taken together, the EMSA studies demonstrate that the PGC-1α-mediated region of the mPDK4 gene promoter is capable of binding both exogenous and endogenous ERRα.
Cotransfection studies were performed to examine the functional correlate of the binding studies. For these studies, CV-1 cells were used because they are relatively deficient in ERRα and many other nuclear receptors, presenting the opportunity to reconstitute the system with factors required for maximal PGC-1α activation. Cotransfection of a PGC-1α expression vector resulted in fourfold activation of mPDK4.Luc.2281, while ERRα alone resulted in a modest twofold activation (Fig. 5). However, cotransfection of both PGC-1α and ERRα increased reporter activity nearly 10-fold, comparable to that observed in C2C12 myotubes (compare Fig. 5 and 2A). The PGC-1α/ERRα response was dependent on the NR response element, because the mutant construct did not exhibit activation by either factor alone, showing only a very modest increase with the combination (Fig. 5). These data indicate that ERRα is sufficient to confer the PGC-1α-mediated transcriptional activation.
FIG. 5.
ERRα and PGC-1α regulate the mPDK4 promoter through the NR response element. (Top) Schematic showing the mutation (lowercase) of the NR response element in mPDK4.Luc.2281 (NRptmut.mPDK4.Luc.2281). (Bottom) Graph depicting the results of transient cotransfections in CV-1 cells performed with either the mPDK4.Luc.2281 reporter (wild type [WT]) or NRptmut.mPDK4.Luc.2281 (NRptmut). Asterisks indicate significant differences from WT mPDK4.Luc.2281 cotransfected with empty vector (P < 0.05). Dagger indicates a significant difference from mPDK4.Luc.2281 with ERRα (P < 0.05). Double dagger indicates a significant difference from the value obtained using NRptmut.mPDK4.Luc.2281 with empty vector (P < 0.05).
PGC-1α increases ERRα expression and PDK4 promoter binding in skeletal myotubes.
PGC-1α has been shown to directly coactivate ERRα (18). In addition, in certain cell contexts, PGC-1α activates ERRα gene expression (27, 46). To determine whether the activation of PDK4 expression by PGC-1α involved simultaneous activation of ERRα expression, endogenous ERRα protein expression levels were assessed following adenovirus-mediated overexpression of PGC-1α in C2C12 myotubes. ERRα protein levels were markedly increased in Ad-PGC-1α-infected C2C12 myotubes, while the expression of PPARα, FOXO1, and Sp1 was unchanged (Fig. 6A). Consistent with upregulation of ERRα expression, EMSA performed with nuclear extracts prepared from C2C12 myotubes following Ad-PGC-1α infection revealed that the intensity of complex II on the PGC-1α-responsive region of the PDK4 promoter was markedly increased, accompanied by a modest increase in complex III (Fig. 6B, lanes 2 and 3). Addition of an ERRα-specific antibody again confirmed the identity of complex II (Fig. 6B, lane 5). As a separate control, EMSA using extracts from Ad-PPARα-infected cells did not show any changes in complex intensity compared to the control (data not shown).
FIG. 6.
PGC-1α induces expression of the endogenous ERRα gene in skeletal muscle cells.(A) Representative autoradiograph of Western blot analysis performed on nuclear extracts isolated from C2C12 myotubes following infection with Ad-GFP or Ad-PGC-1α as indicated at the top. The antibodies used are given on the left. Arrows on the right indicate specific (PGC-1α and ERRα) bands. (B) EMSA study performed with nuclear extracts isolated from C2C12 myotubes following infection with Ad-GFP (lane 2) or Ad-PGC-1α (lanes 3 to 8) and the wild-type probe described in the legend to Fig. 4. Complexes II and Ib were identified by antibody (Ab) supershift analysis using Abs specific for ERRα and Sp1, respectively (lanes 5 and 6). n.s., nonspecific. (C) Chromatin immunoprecipitation assays performed on C2C12 myotubes following infection by Ad-PGC-1α. Formaldehyde cross-linked protein-DNA complexes were isolated. Sheared protein-DNA complexes were immunoprecipitated with a nonspecific antibody (IgG), anti-ERRα, or anti-PGC-1α. Isolated fragments were amplified by PCR to detect the enrichment of amplicons corresponding to a 190-bp region of the 36B4 gene (negative control) or a 164-bp region encompassing the NR response element of the mPDK4 gene promoter. (Top) Results of agarose gel analysis of a representative trial showing relative band intensities. Input represents 2% of the total chromatin used in the immunoprecipitation reactions. (Bottom) Graphs contain mean values as determined by SYBR green quantification of three independent chromatin isolations and immunoprecipitations (arbitrary units ± SE) normalized to the value for the IgG control (taken as 1.0). Asterisks indicate significant differences from the IgG control (P < 0.05).
The transfection and gel shift studies strongly suggested that a cooperative interaction between ERRα and PGC-1α at the NR half-site leads to activation of PDK4 gene transcription. To determine whether these factors simultaneously occupy the endogenous promoter site in cells, ChIP assays were performed on chromatin isolated from Ad-PGC-1α-infected C2C12 myotubes. Neither the anti-ERRα nor the anti-PGC-1α antibody enriched for the promoter region over a nonspecific IgG control following infection with the Ad-GFP backbone (data not shown). However, following Ad-PGC-1α infection, antibodies specific to ERRα or PGC-1α significantly enriched for the endogenous PDK4 promoter region, four- to fivefold over the IgG control (Fig. 6C). These results confirm that both PGC-1α and ERRα occupy the PGC-1α-responsive region of the mPDK4 promoter in skeletal myotubes and that PGC-1α enriches the level of binding, likely via increasing ERRα gene expression.
ERRα is required for PGC-1α-mediated activation of the PDK4 gene promoter.
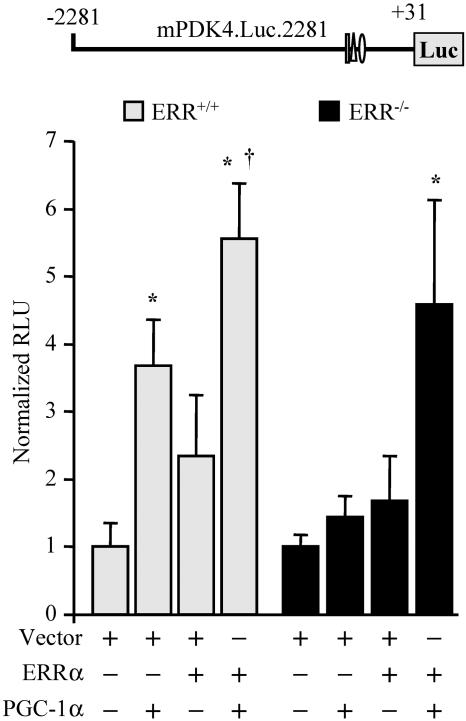
The data shown above support the conclusion that both ERRα and PGC-1α are capable of activating PDK4 gene transcription. To determine if ERRα is actually required for PGC-1α-mediated activation, we assessed the ability of PGC-1α to activate the PDK4 promoter in ERRα-null cells. For these experiments, fibroblasts were isolated from ERRα-null (ERRα−/−) (32) and wild-type (ERRα+/+) mice. Cotransfection of a PGC-1α or ERRα expression vector with the mPDK.Luc.2281 reporter in wild-type cultured primary fibroblasts led to 3.5-fold and 2.5-fold increases in reporter activity, respectively; while addition of both PGC-1α and ERRα increased reporter activity 6-fold (Fig. 7). In ERRα−/− fibroblasts, cotransfection of either a PGC-1α or an ERRα expression vector alone failed to induce mPDK4.Luc.2281 reporter activity (Fig. 7). However, cotransfection of both ERRα and PGC-1α rescued PGC-1α-mediated activation in ERRα−/−fibroblasts, resulting in a nearly fivefold induction (Fig. 7). These results demonstrate that ERRα is required for PGC-1α-mediated activation of the mPDK4 gene promoter.
FIG. 7.
ERRα is required for PGC-1α-mediated activation of PDK4 gene transcription. Primary fibroblasts isolated from wild-type or ERR−/− mice were cotransfected with mPDK4.Luc.2281 and an expression vector for PGC-1α or ERRα, or both. Asterisks indicate significant differences from wild-type mPDK4.Luc.2281 cotransfected with empty vector (P < 0.05). Dagger indicates a significant difference from mPDK4.Luc.2281 cotransfected with ERRα (P < 0.05).
PGC-1α decreases glucose oxidation in C2C12 myotubes.
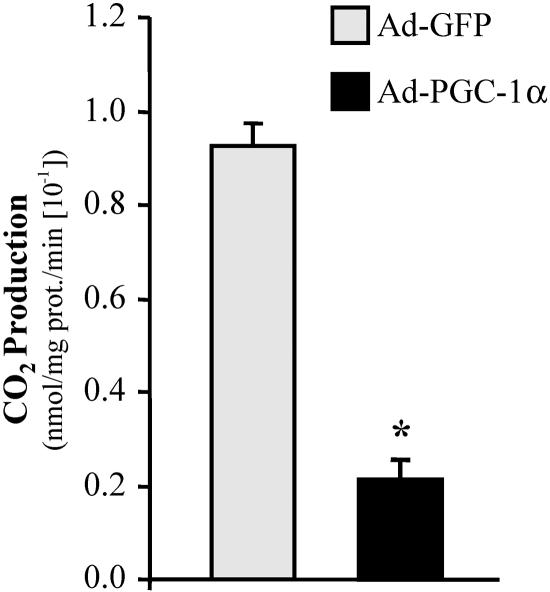
To determine the functional relevance of PGC-1α-mediated activation of PDK4 expression, cellular glucose oxidation rates were determined in the presence and absence of overexpressed PGC-1α. For these experiments, glucose oxidation was measured as the release of 14CO2 from [U-14C]glucose in C2C12 myotubes. Mean glucose oxidation rates were markedly reduced (by approximately 75%) in C2C12 myotubes following infection with Ad-PGC-1α compared to Ad-GFP-infected controls (Fig. 8).
FIG. 8.
PGC-1α decreases glucose oxidation in C2C12 myotubes. The graph represents mean (±SE) [U-14C]glucose oxidation rates in C2C12 myotubes infected with Ad-PGC-1α or Ad-GFP (control). Results are based on three independent experiments performed in triplicate. Asterisk indicates a significant difference from the GFP-infected control (P < 0.001).
DISCUSSION
Skeletal muscle is capable of rapid adjustments in ATP production and fuel preference in accordance with diverse physiological conditions. In order to match energy requirements with activity demands, muscle substrate utilization pathways must be tightly controlled. During intense bouts of exercise, the majority of the muscles' energy demands are met by glucose. Following exercise, muscle glucose is spared through a shift toward mitochondrial fatty acid oxidation so that glycogen levels may be quickly replenished. The molecular basis for the metabolic plasticity of muscle involves rapid changes in the activity of enzymes via allosteric control and posttranslational modifications as well as chronic changes through the regulation of numerous genes controlling mitochondrial content and substrate preference. Delineation of the mechanisms involved in the gene expression changes is critical for the understanding of the molecular pathogenesis of disease states where substrate utilization is chronically deranged, such as heart failure, obesity, and diabetes. In this report, we identify PDK4, an important negative regulator of muscle glucose oxidation, as a new target of PGC-1α-mediated regulation.
The inducible transcriptional coactivator PGC-1α has been identified as a key regulator of multiple ATP-generating pathways in striated muscle through its regulatory influence on genes involved in mitochondrial biogenesis, respiration, oxidative phosphorylation, and fatty acid oxidation (reviewed in references 25 and 40). However, little is known about the potential role of PGC-1α in the control of muscle glucose utilization pathways. The pioneering studies of Randle et al. demonstrated that glucose oxidation is decreased coincident with increased fatty acid oxidation in heart muscle (42). This inhibition of glucose oxidation occurs at the level of the pyruvate dehydrogenase complex, among other enzymes, via allosteric effects of the products of fatty acid oxidation and through changes in the cellular redox state. Our results indicate that PGC-1α may also contribute to this counter-regulatory effect at the level of gene expression in skeletal muscle through modulation of PDK4, a negative regulator of the PDC complex. Recent reports have shown a role for PGC-1α in glucose replenishment in the liver through the regulation of gluconeogenic enzyme genes (14, 39, 57). The regulatory mechanism described by the current study, in which PGC-1α increases muscle PDK4 expression, may serve to replenish glycogen levels in muscle through a different mechanism, by reducing glucose oxidation. Interestingly, PGC-1α has also been shown to activate the expression of the insulin-sensitive glucose transporter GLUT4 in muscle (33), which would also serve to increase glycogen stores in the context of increased PDK4 levels.
Our results identify the orphan nuclear receptor ERRα as the mediator of PGC-1α-induced activation of mPDK4 gene expression. Recently, ERRα was identified as a critical transcriptional regulator of cellular energy metabolism. The activity of ERRα appears to be highly dependent on PGC-1α (18,45). PGC-1α-mediated coactivation of some ERRα targets, such as MCAD (47), ATP synthase β, and cytochrome c (45), is conferred directly by ERRα binding to elements within the promoter regions of these genes. In addition, we recently demonstrated that ERRα is capable of regulating gene expression indirectly by activating the transcription of PPARα (20). ERRα has also been shown to cooperate with the transcription factors NRF-1 and NRF-2 to activate the expression of genes involved in mitochondrial respiratory function (45). The “PGC-1α/ERRα circuit” was further defined when it was shown that PGC-1α could induce ERRα expression through ERRα by an autoregulatory feed-forward mechanism (27). Thus, ERRα serves a central function in the PGC-1α gene regulatory cascade. The current study expands the list of ERRα target genes to include PDK4. Our results show that PGC-1α not only directly coactivates ERRα but also activates its expression through the autoregulatory loop described previously.
Previous work has implicated the nuclear receptor PPARα, a known PGC-1α target factor, in the regulation of PDK4 gene expression. Specifically, the PPARα selective ligand compound WY-14,643 induces PDK4 gene expression (17), and this response is lost in the hearts and kidneys of PPARα-null mice (55). Somewhat surprisingly, our results demonstrate that PGC-1α-mediated activation of PDK4 gene expression occurs independently of PPARα. It should be noted that a direct mechanism for PPARα-mediated activation of PDK4 gene expression has not been shown. However, the current study does not rule out an indirect role for PPARα in regulating PDK4 gene expression or direct PPARα-mediated activation through elements outside of the promoter region examined in our study.
The transcription factor FOXO1 has been shown to function with PGC-1α in the regulation of liver gluconeogenesis (39). Recent studies have shown that the GR in cooperation with FOXO1 activates transcription of the human PDK4 gene. This transcriptional regulatory effect is mediated by a complex mechanism involving the interaction of a single GR response element and three FOXO1 response elements within the hPDK4 gene promoter region (26). This region is not conserved in the mouse PDK4 promoter. As with PPARα, we were also surprised to find that the FOXO1 response element in the proximal mPDK4 promoter was dispensable for the observed PGC-1α effect. To our knowledge, the influence of PGC-1α on the human PDK4 gene promoter has not been studied. However, inspection of the human PDK4 promoter sequence indicates that a PGC-1α/ERRα-responsive region may be conserved in the human promoter. Although the ERR response element we identified in the mPDK4 gene is not spatially conserved in the hPDK4 promoter, a candidate monomeric ERR response element (AAGGACA; bp −823 to −817) does exist, overlapping the previously identified GR response element in the hPDK4 gene (26). Interestingly, a previous report describes the modulation of GR activity by ERR isoforms (50). It is tempting to speculate that ERRα and GR regulatory pathways converge on the human PDK4 gene.
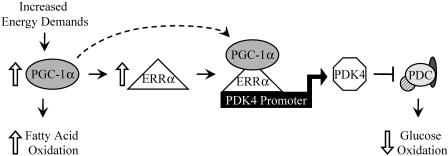
In summary, our findings, together with the known regulatory effects of PGC-1α on cellular energy metabolism, suggest a mechanism by which PGC-1α is able to suppress muscle glucose oxidation while increasing the expression of genes involved in mitochondrial fatty acid oxidation (Fig. 9). Energy demands placed on skeletal muscle result in increased utilization of fatty acids, mediated in part through the activating effects of PGC-1α on the expression of genes involved in cellular fatty acid uptake and oxidation. Induction of PGC-1α activates ERRα expression, leading to occupation of the PGC-1α/ERRα complex on the PDK4 promoter. The resultant increased expression of PDK4 leads to phosphorylation and inactivation of the PDC, which catalyzes the rate-limiting step of glucose oxidation (48). This model is consistent with the response to endurance training, a known stimulus for increasing fatty acid oxidative capacity and mitochondrial biogenesis. Collectively, these findings expand the role of the PGC-1α/ERRα complex to a regulator of muscle substrate selection, leading to a better understanding of transcriptional mechanisms involved in the maintenance of energy homeostasis.
FIG. 9.
Proposed mechanism for the regulation of muscle glucose metabolism by PGC-1α/ERRα. Increased energy demands related to physiological stress (e.g., exercise) result in the induction of muscle PGC-1α gene expression, which in turn induces ERRα gene expression. ERRα then binds to the PDK4 gene promoter, where PGC-1α directly coactivates ERRα to induce PDK4 gene expression. PDK4 has known roles in the negative regulation of the PDC, resulting in decreased glucose oxidation coincident with increased mitochondrial fatty acid oxidation driven by PGC-1α.
Acknowledgments
Special thanks to Robert A. Harris for providing the PDK4 antibody and to William R. Sellers for providing the FKHR expression vector. We thank Mary Wingate and Teresa Leone for assistance in preparing the manuscript.
This work is supported by NIH grant RO1 DK45416, the Digestive Diseases Research Core Center grant (P30 DK52574), and the Clinical Nutrition Research Unit Core Center (P30 DK56341). A.R.W. is supported by the Washington University School of Medicine Cardiovascular Training grant T32 HL007275.
REFERENCES
- 1.Arany, Z., H. He, J. Lin, K. Hoyer, C. Handschin, O. Toka, F. Ahmad, T. Matsui, S. Chin, P.-H. Wu, I. I. Rybkin, J. M. Shelton, M. Manieri, S. Cinti, F. J. Schoen, R. Bassel-Duby, A. Rosenzweig, J. S. Ingwall, and B. M. Spiegelman. 2005. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 1:259-271. [DOI] [PubMed] [Google Scholar]
- 2.Baar, K., A. R. Wende, T. E. Jones, M. Marison, L. A. Nolte, M. Chen, D. P. Kelly, and J. O. Holloszy. 2002. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1α. FASEB J. 16:1879-1886. [DOI] [PubMed] [Google Scholar]
- 3.Brandt, J., F. Djouadi, and D. P. Kelly. 1998. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor α. J. Biol. Chem. 273:23786-23792. [DOI] [PubMed] [Google Scholar]
- 4.Carter, M. E., T. Gulick, B. D. Raisher, T. Caira, J. A. Ladias, D. D. Moore, and D. P. Kelly. 1993. Hepatocyte nuclear factor-4 activates medium chain acyl-CoA dehydrogenase gene transcription by interacting with a complex regulatory element. J. Biol. Chem. 268:13805-13810. [PubMed] [Google Scholar]
- 5.Caruso, M., C. Miele, P. Formisano, G. Condorelli, G. Bifulco, A. Oliva, R. Auricchio, G. Riccardi, B. Capaaldo, and F. Beguinot. 1997. In skeletal muscle, glucose storage and oxidation are differentially impaired by the IR1152 mutant receptor. J. Biol. Chem. 272:7290-7297. [DOI] [PubMed] [Google Scholar]
- 6.Disch, D. L., T. A. Rader, S. Cresci, T. C. Leone, P. M. Barger, R. Vega, P. A. Wood, and D. P. Kelly. 1996. Transcriptional control of a nuclear gene encoding a mitochondrial fatty acid oxidation enzyme in transgenic mice: role for nuclear receptors in cardiac and brown adipose expression. Mol. Cell. Biol. 16:4043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finck, B. N., C. Bernal-Mizrachi, D. H. Han, T. Coleman, N. Sambandam, L. L. LaRiviere, J. O. Holloszy, C. F. Semenkovich, and D. P. Kelly. 2005. A potential link between muscle peroxisome proliferator-activated receptor α signaling and obesity-related diabetes. Cell Metab. 1:133-144. [DOI] [PubMed] [Google Scholar]
- 8.Finck, B. N., J. J. Lehman, T. C. Leone, M. J. Welch, M. J. Bennett, A. Kovacs, X. Han, R. W. Gross, R. Kozak, G. D. Lopaschuk, and D. P. Kelly. 2002. The cardiac phenotype induced by PPARα overexpression mimics that caused by diabetes mellitus. J. Clin. Investig. 109:121-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuyama, T., K. Kitayama, H. Yamashita, and N. Mori. 2003. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem. J. 375:365-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giguère, V., N. Yang, P. Segui, and R. M. Evans. 1988. Identification of a new class of steroid hormone receptors. Nature 331:91-94. [DOI] [PubMed] [Google Scholar]
- 11.Gorman, C. 1985. High efficiency gene transfer into mammalian cells, p. 143-190. In D. M. Glover (ed.), DNA cloning, a practical approach. IRL Press, Oxford, United Kingdom.
- 12.Goto, M., S. Terada, M. Kato, M. Katoh, T. Yokozeki, I. Tabata, and T. Shimokawa. 2000. cDNA cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochem. Biophys. Res. Commun. 274:350-354. [DOI] [PubMed] [Google Scholar]
- 13.Harris, R. A., M. M. Bowker-Kinley, B. Huang, and P. Wu. 2002. Regulation of the activity of the pyruvate dehydrogenase complex. Adv. Enzyme Regul. 42:249-259. [DOI] [PubMed] [Google Scholar]
- 14.Herzig, S., F. Long, U. S. Jhala, S. Hedrick, R. Quinn, A. Bauer, D. Rudolph, G. Schutz, C. Yoon, P. Puigserver, B. Spiegelman, and M. Montminy. 2001. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413:179-183. [DOI] [PubMed] [Google Scholar]
- 15.Hildebrandt, A. L., H. Pilegaard, and P. D. Neufer. 2003. Differential transcriptional activation of select metabolic genes in response to variations in exercise intensity and duration. Am. J. Physiol. Endocrinol. Metab. 285:E1021-E1027. [DOI] [PubMed] [Google Scholar]
- 16.Holness, M. J., and M. C. Sugden. 2003. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem. Soc. Trans. 31:1143-1151. [DOI] [PubMed] [Google Scholar]
- 17.Huang, B., P. Wu, M. M. Bowker-Kinley, and H. A. Harris. 2002. Regulation of pyruvate dehydrogenase kinase expression by peroxisome proliferator-activated receptor-α ligands, glucocorticoids, and insulin. Diabetes 51:276-283. [DOI] [PubMed] [Google Scholar]
- 18.Huss, J. M., R. P. Kopp, and D. P. Kelly. 2002. PGC-1α coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. J. Biol. Chem. 277:40265-40274. [DOI] [PubMed] [Google Scholar]
- 19.Huss, J. M., F. H. Levy, and D. P. Kelly. 2001. Hypoxia inhibits the PPARα/RXR gene regulatory pathway in cardiac myocytes. J. Biol. Chem. 276:27605-27612. [DOI] [PubMed] [Google Scholar]
- 20.Huss, J. M., I. Pinéda Torra, B. Staels, V. Giguère, and D. P. Kelly. 2004. ERRα directs PPARα signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 24:9079-9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im, H., J. A. Grass, K. D. Johnson, M. E. Boyer, J. Wu, and E. H. Bresnick. 2004. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol. Biol. 284:129-146. [DOI] [PubMed] [Google Scholar]
- 22.Itoh, Y., T. Esaki, K. Shimoji, M. Cook, J. Law, E. Kaufman, and L. Sokoloff. 2003. Dichloroacetate effects on glucose and lactate oxidation by neurons and astroglia in vitro and on glucose oxidation by brain in vivo. Proc. Natl. Acad. Sci. USA 100:4879-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston, S. D., Z. Liu, F. Zuo, T. L. Eisenbraun, S. R. Wiley, R. J. Krauss, and J. E. Mertz. 1997. Estrogen-related receptor α1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen-response elements. Mol. Endocrinol. 11:342-352. [DOI] [PubMed] [Google Scholar]
- 24.Kelly, D. P., J. J. Kim, J. J. Billadello, B. E. Hainline, T. W. Chu, and A. W. Strauss. 1987. Nucleotide sequence of medium-chain acyl-CoA dehydrogenase mRNA and its expression in enzyme-deficient human tissue. Proc. Natl. Acad. Sci. USA 84:4068-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly, D. P., and R. C. Scarpulla. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 18:357-368. [DOI] [PubMed] [Google Scholar]
- 26.Kwon, H. S., B. Huang, G. Unterman, and R. A. Harris. 2004. Protein kinase B-α inhibits human pyruvate dehydrogenase kinase-4 gene induction by dexamethasone through inactivation of FOXO transcription factors. Diabetes 53:899-910. [DOI] [PubMed] [Google Scholar]
- 27.Laganiere, J., G. B. Tremblay, C. R. Dufour, S. Giroux, F. Rousseau, and V. Giguère. 2004. A polymorphic autoregulatory hormone response element in the human estrogen-related receptor α (ERRα) promoter dictates peroxisome proliferator-activated receptor γ coactivator-1α control of ERRα expression. J. Biol. Chem. 279:18504-18510. [DOI] [PubMed] [Google Scholar]
- 28.Lehman, J. J., P. M. Barger, A. Kovacs, J. E. Saffitz, D. Medeiros, and D. P. Kelly. 2000. PPARγ coactivator-1 (PGC-1) promotes cardiac mitochondrial biogenesis. J. Clin. Investig. 106:847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leone, T. C., J. J. Lehman, B. N. Finck, P. J. Schaeffer, A. R. Wende, S. Boudina, M. Courtois, D. F. Wozniak, N. Sambandam, C. Bernal-Mizrachi, Z. Chen, J. O. Holloszy, D. M. Medeiros, R. E. Schmidt, J. E. Saffitz, E. D. Abel, C. F. Semenkovich, and D. P. Kelly. 2005. PGC-1α deficient mice exhibit multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control, and hepatic steatosis. PLoS Biol. 3:672-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, J., H. Wu, P. T. Tarr, C. Y. Zhang, Z. Wu, O. Boss, L. F. Michael, P. Puigserver, E. Isotanni, E. N. Olson, B. B. Lowell, R. Bassel-Duby, and B. M. Spiegelman. 2002. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibers. Nature 418:797-801. [DOI] [PubMed] [Google Scholar]
- 31.Lin, J., P.-H. Wu, P. T. Tarr, K. S. Lindenberg, J. St.-Pierre, C.-Y. Zhang, V. K. Mootha, S. Jãger, C. R. Vianna, R. M. Reznick, L. Cui, M. Manieri, M. X. Donovan, Z. Wu, M. P. Cooper, M. C. Fan, L. M. Rohas, A. M. Zavacki, S. Cinti, G. I. Shulman, B. B. Lowell, D. Krainc, and B. M. Spiegelman. 2004. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell 119:121-135. [DOI] [PubMed] [Google Scholar]
- 32.Luo, J., R. Sladek, J. Carrier, J. A. Bader, D. Richard, and V. Giguère. 2003. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol. Cell. Biol. 23:7947-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michael, L. F., Z. Wu, R. B. Cheatham, P. Puigserver, G. Adelmant, J. J. Lehman, D. P. Kelly, and B. M. Spiegelman. 2001. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. USA 98:3820-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miura, S., Y. Kai, M. Ono, and O. Ezaki. 2003. Overexpression of peroxisome proliferator-activated receptor γ coactivator-1α down-regulates GLUT4 mRNA in skeletal muscles. J. Biol. Chem. 278:31385-31390. [DOI] [PubMed] [Google Scholar]
- 35.Mootha, V. K., C. Handschin, D. Arlow, X. Xie, J. St. Pierre, S. Sihag, W. Yang, D. Altshuler, P. Puigserver, N. Patterson, P. J. Willy, I. G. Schulman, R. A. Heyman, E. S. Lander, and B. M. Spiegelman. 2004. ERRα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA 101:6570-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mootha, V. K., C. M. Lindgren, K.-F. Eriksson, A. Subramanian, S. Sihag, J. Lehar, P. Puigserver, E. Carlsson, M. Ridderstråle, E. Laurila, N. Houstis, M. J. Daly, N. Patterson, J. P. Mesirov, T. R. Golub, P. Tamayo, B. M. Spiegelman, E. S. Lander, J. N. Hirschhorn, D. Altshuler, and L. C. Groop. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267-273. [DOI] [PubMed] [Google Scholar]
- 37.Pilegaard, H., G. A. Ordway, B. Saltin, and P. D. Neufer. 2000. Transcriptional regulation of gene expression in human skeletal muscle during recovery from exercise. Am. J. Physiol. Endocrinol. Metab. 279:E806-E814. [DOI] [PubMed] [Google Scholar]
- 38.Pilegaard, H., B. Saltin, and P. D. Neufer. 2003. Exercise induces transient transcriptional activation of the PGC-1α gene in human skeletal muscle. J. Physiol. 546:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puigserver, P., J. Rhee, J. Donovan, C. J. Walkey, J. C. Yoon, F. Oriente, Y. Kitamura, J. Altomonte, H. Dong, D. Accili, and B. M. Spiegelman. 2003. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423:550-555. [DOI] [PubMed] [Google Scholar]
- 40.Puigserver, P., and B. M. Spiegelman. 2003. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocrine Rev. 24:78-90. [DOI] [PubMed] [Google Scholar]
- 41.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829-839. [DOI] [PubMed] [Google Scholar]
- 42.Randle, P. J., C. N. Hales, and P. B. Garland. 1963. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet i:785-789. [DOI] [PubMed] [Google Scholar]
- 43.Rowles, J., S. W. Scherer, T. Xi, M. Majer, D. C. Nickle, J. M. Rommens, K. M. Popov, R. A. Harris, N. L. Riebow, J. Xia, L. C. Tsui, C. Bogardus, and M. Prochazka. 1996. Cloning and characterization of PDK4 on 7q21.3 encoding a fourth pyruvate dehydrogenase kinase isoenzyme in human. J. Biol. Chem. 271:22376-22382. [DOI] [PubMed] [Google Scholar]
- 44.Russell, L. K., C. M. Mansfield, J. J. Lehman, A. Kovacs, M. Courtois, J. E. Saffitz, D. M. Medeiros, M. L. Valencik, J. A. McDonald, and D. P. Kelly. 2004. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ. Res. 94:525-533. [DOI] [PubMed] [Google Scholar]
- 45.Schreiber, S. N., R. Emter, M. B. Hock, D. Knutti, J. Cardenas, M. Podvinec, E. J. Oakeley, and A. Kralli. 2004. The estrogen-related receptor alpha (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. USA 101:6472-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreiber, S. N., D. Knutti, K. Brogli, T. Uhlmann, and A. Kralli. 2003. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα). J. Biol. Chem. 278:9013-9018. [DOI] [PubMed] [Google Scholar]
- 47.Sladek, R. J., J.-A. Bader, and V. Giguère. 1997. The orphan nuclear receptor estrogen-related receptor α is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol. Cell. Biol. 17:5400-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugden, M. C., and M. J. Holness. 2003. Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am. J. Phys. Endocrinol. Metab. 284:E855-E862. [DOI] [PubMed] [Google Scholar]
- 49.Tang, E. D., G. Nuñez, G. Barr, and K.-L. Guan. 1999. Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 274:16741-16746. [DOI] [PubMed] [Google Scholar]
- 50.Trapp, T., and F. Holsboer. 1996. Nuclear orphan receptor as a repressor of glucocorticoid receptor transcriptional activity. J. Biol. Chem. 271:9879-9882. [DOI] [PubMed] [Google Scholar]
- 51.Vega, R. B., J. M. Huss, and D. P. Kelly. 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 20:1868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinmann, A. S., and P. J. Farnham. 2002. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 26:37-47. [DOI] [PubMed] [Google Scholar]
- 53.Willy, P. J., I. R. Murray, J. Qian, B. B. Busch, W. C. Stevens, R. Martin, R. Mohan, S. Zhou, P. Ordentlich, P. Wei, D. W. Sapp, R. A. Horlick, R. A. Heyman, and I. G. Schulman. 2004. Regulation of PPARγ coactivator 1α (PGC-1α) signaling by an estrogen-related receptor α (ERRα) ligand. Proc. Natl. Acad. Sci. USA 101:8912-8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, P., K. Inskeep, M. M. Bowker-Kinley, K. M. Popov, and R. A. Harris. 1999. Mechanism responsible for inactivation of skeletal muscle pyruvate dehydrogenase complex in starvation and diabetes. Diabetes 48:1593-1599. [DOI] [PubMed] [Google Scholar]
- 55.Wu, P., J. M. Peters, and R. A. Harris. 2001. Adaptive increase in pyruvate dehydrogenase kinase 4 during starvation is mediated by peroxisome proliferator-activated receptor α. Biochem. Biophys. Res. Commun. 287:391-396. [DOI] [PubMed] [Google Scholar]
- 56.Wu, Z., P. Puigserver, U. Andersson, C. Zhang, G. Adelmant, V. Mootha, A. Troy, S. Cinti, B. Lowell, R. C. Scarpulla, and B. M. Spiegelman. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115-124. [DOI] [PubMed] [Google Scholar]
- 57.Yoon, J. C., P. Puigserver, G. Chen, J. Donovan, Z. Wu, J. Rhee, G. Adelmant, J. Stafford, C. R. Kahn, D. K. Granner, C. B. Newgard, and B. M. Spiegelman. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131-138. [DOI] [PubMed] [Google Scholar]