Abstract
The U2 snRNP promotes prespliceosome assembly through interactions that minimally involve the branchpoint binding protein, Mud2p, and the pre-mRNA. We previously showed that seven proteins copurify with the yeast (Saccharomyces cerevisiae) SF3b U2 subcomplex that associates with the pre-mRNA branchpoint region: Rse1p, Hsh155p, Hsh49p, Cus1p, and Rds3p and unidentified subunits p10 and p17. Here proteomic and genetic studies identify Rcp10p as p10 and show that it contributes to SF3b stability and is necessary for normal cellular Cus1p accumulation and for U2 snRNP recruitment in splicing. Remarkably, only the final 53 amino acids of Rcp10p are essential. p17 is shown to be composed of two accessory splicing factors, Bud31p and Ist3p, the latter of which independently associates with the RES complex implicated in the nuclear pre-mRNA retention. A directed two-hybrid screen reveals a network of prospective interactions that includes previously unreported intra-SF3b contacts and SF3b interactions with the RES subunit Bud13p, the Prp5p DExD/H-box protein, Mud2p, and the late-acting nineteen complex. These data establish the concordance of yeast and mammalian SF3b complexes, implicate accessory splicing factors in U2 snRNP function, and support SF3b contribution from early pre-mRNP recognition to late steps in splicing.
The eukaryotic spliceosome consists of five small nuclear ribonucleoprotein (snRNP) particles and an unknown number of non-snRNP splicing factors (reviewed in reference 27). In vitro, spliceosome assembly progresses through the sequential recruitment of the U1, U2, and U4/U6.U5 tri-snRNP particles to the pre-mRNP substrate. Subsequent conformational changes within the snRNP complete complex then weaken the association of the U1 and U4 snRNP particles to configure a catalytically active splicing enzyme (reviewed in references 12 and 33). The recovery of native complexes with the predicted snRNA and protein content of mature Saccharomyces cerevisiae (henceforth yeast) spliceosomes provides evidence that spliceosome assembly progresses through a similar activation step in vivo (34, 49).
Early on it was shown that U2 snRNP addition in mammals is stimulated by U2AF65, a protein bound to a pyrimidine-rich sequence near the 3′ end of the intron (40). The UAP56 DExD/H-box protein interacts with U2AF65 to further enhance U2 snRNP recruitment (17). UAP56 likely stimulates a reorganization step within the U1 snRNP-bearing commitment complex that results in the displacement of U2AF65 and the branchpoint binding protein (also called SF1 or BBP) from the targeted site of U2 snRNA/pre-mRNA interaction. The budding yeast homolog of U2AF65 is encoded by the nonessential MUD2 gene (1). Mud2p and yeast BBP (also called Msl5p) are released from (or less stably bound to) the splicing apparatus after U2 snRNP addition, consistent with a conserved mRNP reorganization step during yeast prespliceosome formation (41). The yeast UAP56 homolog, Sub2p, is essential for splicing unless MUD2 is first deleted (28), supporting its involvement in early spliceosome assembly. The ATPase activity of a second DExD/H-box protein, Prp5p, is needed to configure the U2 snRNP into a state competent for prespliceosome assembly (37). Prp5p also serves a separate, ill-defined ATPase-independent function in prespliceosome assembly (36). Recent evidence from the fission yeast system suggests that Prp5p physically tethers the U1 and U2 snRNP particles, although the protein contacts mediating this association are unknown (55).
In 1993, Kramer and coworkers reported the resolution of mammalian splicing factor SF3 into subfractions SF3a and SF3b (11). Binding of SF3a and SF3b to the 12S U2 snRNP core converts this structure into the mature 17S particle recruited during spliceosome assembly (5, 10). Photo-cross-linking experiments show that multiple SF3 subunits, including at least the p14a, SAP49, SAP145, and SAP155 subunits of SF3b, are in close contact with the splicing substrate (22, 23). Intriguingly, SAP155 (22, 23) and possibly the yeast counterpart, Hsh155p (31), straddle the pre-mRNA branchpoint consensus sequence in the splicing complex. The p14a subunit, although buried within a deep cleft of the SF3b particle (20), can bind the bulged branchpoint nucleotide (51) during spliceosome assembly. The contribution of individual SF3b proteins to mammalian SF3b assembly or splicing remains largely untested.
We biochemically purified yeast SF3b and documented the presence of the conserved Hsh49p/SAP49, Hsh155p/SAP155, Cus1p/SAP145, Rse1p/SAP130, and Rds3p/SF3b14b proteins (50). Two small proteins, possibly counterparts to the mammalian p10 and p14a proteins, were observed but could not be identified by mass spectroscopy. No strong match to p10 was reported in the yeast genome (52), and deletion of the gene encoding the proposed p14a homolog, Ist3p (also called Snu17p [21]), reduced but did not eliminate the SF3b p17 band, indicating the presence of a second protein. More recently Dziembowski et al. (16) reported a p10 counterpart in yeast SF3b, although this study did not address its function in splicing nor provide evidence for the presence or identity of p17 in SF3b.
Here we show that the yeast SF3b p10 protein, Rcp10p, is essential for splicing and required for stable SF3b assembly. Yeasts that lack Rcp10p fail to support U2 snRNP recruitment in vitro and have much-reduced levels of the essential SF3b subunit, Cus1p. Genetic and biochemical evidence reveals that two proteins, Ist3p and Bud31p, interact with SF3b as p17. Ist3p independently binds a complex implicated in nuclear pre-mRNA retention called RES (16). Finally a directed yeast two-hybrid analysis reveals an intriguing network of putative interactions suggesting, for instance, that Rcp10p and the Prp5 DExD/H-box factor function in part through contact with the proposed branchpoint straddling protein Hsh155p. These and related observations offer a new perspective on the branchpoint-associated proteins acting from the earliest steps of pre-mRNP assembly through late stages of splicing.
MATERIALS AND METHODS
Gene and yeast strain construction.
To create the rcp10::HIS3 knockout, the YNL138w-a open reading frame (ORF) and flanking sequence were cloned as a PCR product (primers Ynl138wa-1 and Ynl138wa-6) at the SmaI site of YIplac204. The Ynl138w-a ORF was removed by SmaI and PsiI digestion and replaced with HIS3. The disrupted gene was then reamplified by PCR (primers Ynl138wa-9 and Ynl138wa-10), and the resulting fragment was used for gene replacement by homologous recombination in yeast (BY4743; mata/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 met15Δ0/MET15 lys2Δ0/LYS2). The structure of the disrupted allele in this strain (QW201) was confirmed by Southern blotting.
GAL1::RCP10 was prepared by inserting the RCP10 ORF prepared by PCR (primers Ynl138wa-3 and Ynl138wa-4) into the BamHI site of pBM150. Ycplac111-RCP10-TAP was prepared by first inserting the tandem affinity purification (TAP) fragment as previously described (49) into the SmaI site of YCplac111 and then adding RCP10 as a PCR fragment prepared with primers Ynl138wa-1 and Ynl138wa-2. This construct contains approximately 400 bp of RCP10 5′ flanking sequence. The deletions of codons 2 to 19 and 2 to 32 were constructed by inverse PCR on this template using primer RCP10-delta-UP with RCP10Δ19 or RCP10Δ32. The related rcp10fs frameshift allele was fortuitously isolated as a PCR artifact. To compensate for the rcp10fs frameshift (see Results), a 5-bp insertion was placed in the intron by inverse PCR with primers RCP10-intron-1 and RCP10-intron-2 followed by BamHI digestion and ligation. The structures of all gene constructs were confirmed by DNA sequencing and, where appropriate, Western blotting for the TAP fusion. QW201 was transformed with RCP10 derivatives and haploid isolates that uniquely expressed the modified alleles isolated by standard yeast techniques. TAP-tagged versions of all other genes were obtained from Open Biosystems.
The C-terminal V5 epitope fusion alleles were prepared by inserting PCR fragments of BUD31 (primers Bud31-V5-1 and Bud31-V5-2) and IST3 (primers Ist3-3 and Ist3-5) into pYC6/CT (Invitrogen) cleaved at the BamHI site. The resulting constructs were transformed into the corresponding KAN disruption mutants in the BY4742 background (Open Biosystems).
Yeast two-hybrid analysis was conducted by insertion of full-length ORF PCR fragments (primer sequences available on request) into the GAL4 activation domain plasmid pACT and the GAL4 DNA binding plasmid pAS2. Reporter gene transactivation in strain PJ69-4A (26) was scored after 4 days of growth at 30°C on medium lacking adenine and on histidine-deficient medium supplemented with 3-aminotriazole as described previously (26).
DNA oligonucleotides were as shown in Table 1.
TABLE 1.
DNA oligonucleotides
| Name | Sequence |
|---|---|
| Ynl138wa-1 | 5′ CCCGGGATCCTGACTAGGGGAAGCTGGT 3′ |
| Ynl138wa-2 | 5′ CCCGGGATCCTCTCTTCTCGTAAGTAGGC 3′ |
| Ynl138wa-3 | 5′ CGGATCCAAATGGTATGTTACGTTCTATTGC 3′ |
| Ynl138wa-4 | 5′ CGGATCCCTGTTTATGACTTCACGC3′ |
| Ynl138wa-6 | 5′ ATACGAATTCGTAGTATTCGCATCGGC 3′ |
| Ynl138wa-9 | 5′ CCTGACTAGGGGAAGCTGGTGG TA 3′ |
| Ynl138wa-10 | 5′ CGACATACGAGGCGTAATTTTATGTAAAAATTATTTATGGATG TGGCTTCAACGTCGCCTCGTTCAGAATGACACGTATA 3′ |
| RCP10-intron-1 | 5′ ATCGGATCCTAATACTCAAAACAGGCGGA 3′ |
| RCP10-intron-2 | 5′ ATCGGATCCCATTACGTATAAGCCGCACGG 3′ |
| RCP10-delta-UP | 5′ GGATCCCATTTCTTTTCTTAATTTCATGT 3′ |
| RCP10Δ19 | 5′ GGATCCGGACTGGGAGATGAGAGCACTACA 3′ |
| RCP10Δ32 | 5′ GGATTCAGAAATGTGAGAAACGATACAC 3′ |
| Bud31-V5-1 | 5′ AGTTGGATCCTGCTGGTGTCGTTTAAT 3′ |
| Bud31-V5-2 | 5′ AGTTGGATCCGTGCTTGCACATCCACGGCA 3′ |
| Ist3-3 | 5′ GGATCCAAGTTAGCAATCATCTCTTGT 3′ |
| Ist3-5 | 5′ GGATCCTTTTTAGCCAAAATGAGTTTC 3′ |
Cell extract preparation and splicing assays.
Two liters of yeast grown in yeast extract-peptone-dextrose broth to an optical density at 600 nm of 2.0 to 4.0 was collected by centrifugation and washed once with 100 ml of sterile water and once with 50 ml of solution A (10 mM HEPES [pH 7.9], 10 mM KCl, 200 mM NaCl, 10% glycerol, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine, 0.5% NP-40). The cell pellet was dissolved in 8 to 12 ml of solution A, and the yeast cells were broken in liquid nitrogen using a Spex Certiprep 6850 freezer mill (four or five cycles of 2 min of breakage and 2 min of cooling, at a frequency setting of 10). The resultant lysate was then processed for splicing using established protocols (46). Metabolic labeling of yeast with Tran35S-label (ICN) was previously described (49). TAP selection was done according to standard protocols (39) except that the NaCl concentration in the immunoglobulin G (IgG) binding and wash steps varied between 150 mM and 750 mM as indicated. The washed sample was equilibrated with the low-salt cleavage buffer (39) prior to tobacco etch virus (TEV) protease addition. Binding to calmodulin agarose was found to be less consistent at higher salt concentrations, so the binding and wash steps did not exceed 300 mM NaCl (except for the experiment described for Fig. 6).
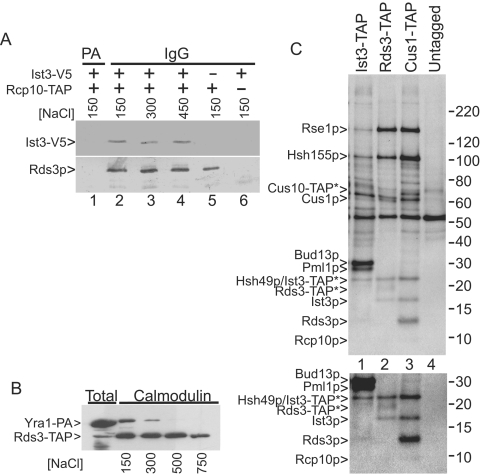
FIG. 6.
Ist3p is recovered with SF3b, and SF3b proteins copurify with Ist3p. A. Western analysis of TAP-selected proteins from yeast strains that express RCP10-TAP (+) or untagged control (−) with or without coexpressed Ist3-V5. The complexes were recovered at the indicated salt concentrations. IgG refers to the initial selection on the initial IgG matrix while PA refers to a protein A agarose control matrix used to score background. Calmodulin agarose selection was subsequently performed in each case. The proteins were detected with appropriate secondary antibodies to a polyclonal Rds3p (SF3b subunit) antibody or the V5 monoclonal antibody. B. Western analysis of calmodulin agarose-selected proteins isolated at the indicated millimolar salt concentrations from yeast strains that simultaneously express a protein A-tagged version of Yra1p (Yra1-PA) and Rds3-TAP. Note that Yra1-PA cannot bind directly to calmodulin agarose. The selected proteins were detected using an antibody against the common protein A domain. C. Metabolically labeled proteins were recovered at 450 mM NaCl from yeast strains that express the IST3-TAP, RDS3-TAP, or CUS1-TAP gene or no epitope tag (Untagged) and resolved by PAGE. The asterisk indicates proteins with a residual (5-kDa) TAP tag. The positions of the SF3b subunits and the RES complex proteins are indicated. The lower panel shows a twofold exposure enhancement of the Rds3p region of this gel. Numbers at right are molecular masses in kilodaltons of unlabeled protein markers run on the same gel.
For mass spectroscopy, samples eluted from calmodulin agarose were trichloroacetic acid precipitated, digested with trypsin, and then resolved by two-dimensional liquid chromatography with strong ion-exchange and C18 packing materials (Whatman) on a Deca mass spectrometer (ThermoFinnigan, San Jose, Calif.) as described previously (49). The tandem mass spectra obtained were converted to mass-intensity lists and searched against the nonredundant Owl database with SEQUEST software and against the nonredundant NCBI database with MASCOT software with a cutoff score of 20. The MASCOT confidence scores obtained for Rcp10-TAP-specific proteins purified at 150 mM NaCl were as follows: Rse1p, 12,050; Hsh155p, 9,163; Cus1p, 4,243; Hsh49p, 1,322; Brr2p, 257; Prp8p, 281; Rds3p, 92; and Bud31, 92. In addition, the Prp19p (158), Smd2p (152), Snu114p (138), Prp9p (130), Prp21p (91), and Prp46p (85) splicing factors were observed. Common high-scoring background proteins included multiple ribosomal proteins, Nam9p (97), and Nop58p (96) and heat shock proteins Ssa1p (145), Ssa2p (145), and Ssb1 (95). Equivalent results were found with multiple Rcp10-TAP and Rds3-TAP (50) preparations.
Metabolic depletion of Rcp10fs was done by harvesting the GAL1::rcp10fs culture grown in yeast extract-peptone broth with 2% galactose, washing the recovered cells with sterile water, and then resuspending the pellet in 20× to 200× original culture volume with yeast extract-peptone broth containing 2% glucose. After 2 to 16 h, the cells were harvested for the preparation of splicing extracts (described above) or for RNA analysis (7). Conditions for the resolution of snRNP complexes by glycerol gradient centrifugation were previously described (8) as were in vitro splicing assays (50) and the conditions for affinity selection of biotin-substituted splicing complexes (54). Northern hybridization and the 35S-labeled protein images were acquired with a Typhoon 9600 phosphoimager (Amersham/Pharmacia). The anti-Rds3p polyclonal antibody was prepared by Proteintech Group Inc., using a glutathione S-transferase fusion protein expressed from pGEX-2T in the BL21 Escherichia coli strain as recommended by the plasmid distributor (Amersham Biosciences). A polyclonal antibody against the TAP segment present after TEV cleavage was obtained from Open Biosystems. The anti-V5 monoclonal antibody and appropriate secondary antibodies were purchased from Invitrogen.
RESULTS
Identification of the yeast SF3b p10 subunit.
While we were not able to get an identifiable signal for Saccharomyces cerevisiae SF3b p10 by mass spectroscopy (50), a related protein of similar size was found in the conceptual yeast proteome using PSI-BLAST (3) (Fig. 1A). The small YNL138W-A ORF is interrupted after its translational initiation codon by a 122-nucleotide intron with conventional splice site consensus elements (42). Phylogenetic conservation among lower eukaryotes suggests that YNL138W-A is an authentic yeast gene (9, 15), which we named RCP10 (for Rds3p complex protein, 10 kDa), according to the Saccharomyces Genome Database guidelines (4).
FIG. 1.
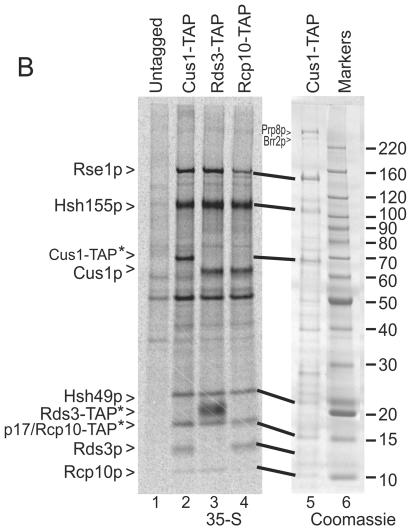
Rcp10p is p10 of SF3b. A. Alignment of the human SF3b p10 protein with the YNL138W-A-encoded protein (Rcp10p) using the BLOSUM 50 scoring matrix. Identical residues are indicated by double dots; similarities are indicated by single dots. B. Yeast strains that express Rcp10-TAP, Cus1-TAP, Rds3-TAP, or no tag were metabolically labeled with [35S]methionine and cysteine. Proteins selected by TAP were resolved on a 15% polyacrylamide gel and visualized on a phosphorimager (35S) or by direct stain (Coomassie blue). The TAP-selected proteins retain approximately 5 kDa of residual tag after TEV cleavage, indicated with an asterisk (e.g., Rcp10-TAP*). Note that Rcp10-TAP* comigrates with p17 in lane 4. Untagged (lane 1) shows background from yeast without a TAP-tagged protein. The positions of the SF3b subunits and the Prp8p and Brr2p splicing factors are indicated. The sizes, in kilodaltons, of the protein molecular mass markers are shown to the right of the Coomassie blue-stained gel.
Since YNL138W-A fell below the arbitrary 100-codon cutoff used for recent systematic analyses of yeast genes (19, 53), we created a null allele by substitution of the yeast HIS3 gene for the RCP10 protein coding sequence (see Materials and Methods). Adiploid strain heterozygous for this mutation was sporulated with and without a functional plasmid-borne copy of RCP10 (GAL1::RCP10, see below, or an equivalent construct expressed from the natural RCP10 promoter). Without this plasmid, no more than two viable offspring germinate and none of the viable haploid progeny contain the disrupted rcp10::HIS3 allele. In contrast, when GAL1::RCP10 is present in the diploid two to four viable meiotic offspring are recovered from each tetrad cultured on galactose-based medium. In addition, all the recovered histidine prototrophs fail to grow on 5-fluoroorotic acid medium, which selects against the URA3-linked plasmid, confirming an essential function for RCP10 in yeast (data not shown).
RCP10 encodes the yeast SF3b p10 protein.
Given the limited sequence similarity of the yeast and human p10 homologs (i.e., roughly 26%), it was critical to show that Rcp10p resides within SF3b. Toward this end, haploid yeast strains that express TAP-tagged (39) versions of Rcp10p, the previously verified SF3b subunits Rds3p and Cus1p, or no TAP tag were metabolically labeled and the tagged proteins were recovered by tandem affinity selection (Fig. 1B). To enhance specificity, the purifications were performed at 0.5 M NaCl rather than at the lower salt concentration originally described (39). Only trace amounts of U2 snRNA copurify under these conditions due to the dissociation of the SF3b structure from the U2 snRNP particle (data not shown but see reference 35). With the exception of the tagged protein, which retains approximately 5 kDa of residual TAP sequence (Fig. 1B, asterisk), equivalent protein banding patterns are observed for the Rds3-TAP, Cus1-TAP, and Rcp10-TAP (lanes 2 to 4) selections, confirming the presence of Rcp10p in the SF3b particle. The identities of five SF3b proteins (i.e., Rse1, Hsh155p, Cus1, Hsh49p, and Rds3p) were verified by mass spectroscopy (see Materials and Methods). While this work was in progress, Dziembowski et al. (16) independently identified Rcp10p (also known as Ysf3) as SF3b associated.
In addition to the proteins mentioned above, an unidentified 17-kDa band is consistently found in the SF3b preparations (Fig. 1B, lanes 2 to 5; note that this protein comigrates with Rcp10-TAP*; also see reference 50). Mass analysis identified a single protein in this size range, the 18-kDa Bud31p. Mass analysis also detected the Prp8p and Brr2p splicing factors as copurifying with SF3b at 150 mM NaCl, and both proteins can be seen in an equivalent preparation stained with Coomassie blue (Fig. 1B, lane 5). Unlike the SF3b subunits, the yields of Prp8p, Brr2p, and the RNA export factor Yra1 (50) (see below) varied from preparation to preparation (for example, compare the region above Rse1p in Fig. 1B, 6C, and 7A). In addition, the Prp9p, Prp21p, Snu114p, and Smd2p splicing factors were occasionally observed by mass spectroscopy (see Materials and Methods). The lower mass analysis confidence scores and the absence of clearly corresponding protein bands when assayed by polyacrylamide gel electrophoresis (PAGE) suggest that these are minor components in the preparation. Contaminants in the preparation include a number of abundant cellular proteins common with TAP selections (see Materials and Methods).
FIG. 7.
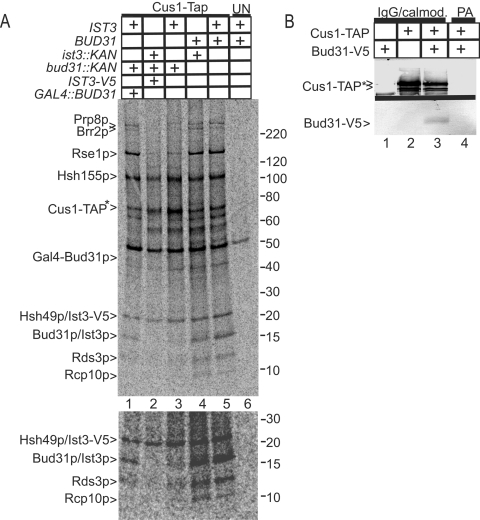
Bud31p and Ist3p contribute to SF3b p17. A. Proteins were recovered by Cus1-TAP selection from yeast metabolic deletion mutants (ist3::KAN bud31::KAN) or wild-type yeast in the presence or absence of epitope-tagged versions of Ist3p (IST3-V5) or Bud31p (GAL4::BUD31). The positions of the relevant proteins are indicated with arrowheads on the left. The similarly sized Bud31-TAP and Hsh49p are resolved poorly on this gel system. The asterisk shows proteins with a residual (5-kDa) TAP tag. The mobility of the molecular weight markers (in thousands) is shown to the right. UN refers to a mock recovery using an untagged yeast strain. The lower panel shows a twofold-enhanced exposure highlighting the low-molecular-weight proteins. B. TAP-selected proteins (IgG/calmod.) recovered at 450 mM NaCl were probed with an antibody against the residual (i.e., after TEV cleavage) TAP tag (Cus1-TAP*, upper panel) and against the V5 epitope (Bud31-V5, lower panel). PA shows the position of mock-selected sample using nonspecific protein A agarose resin.
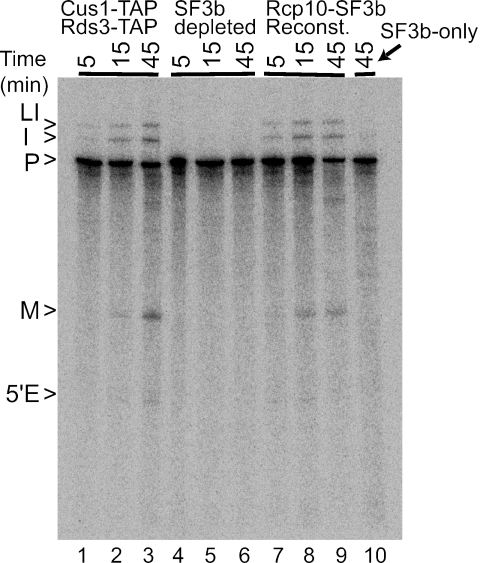
As proof that the Rcp10-TAP recovered complexes are biologically active, we depleted SF3b from an extract that contains TAP-tagged versions of both Cus1p and Rds3p. As expected, passage of this extract through IgG and calmodulin agarose resins blocks the in vitro splicing activity (Fig. 2; compare lanes 1 to 3 with 4 to 6). The affinity-purified Rcp10-TAP complex itself does not splice pre-mRNA (lane 10), but the addition of this preparation to the depleted extract restores splicing (lanes 7 to 9). By this measure, the Rcp10p complex contains all essential SF3b components.
FIG. 2.
The Rcp10p-SF3b complex is biologically active. Shown are results of in vitro splicing of RPS17A pre-mRNA with doubly tagged Cus1-TAP, Rds3-TAP extract before SF3b removal (lanes 1 to 3), after SF3b removal with IgG and calmodulin agarose chromatography (lanes 4 to 6), and after supplementation with Rcp10-TAP-purified SF3b (lanes 7 to 9). In lane 10, the pre-mRNA was incubated only with Rcp10-TAP-purified SF3b. The positions of pre-mRNA (P), lariat intermediate (LI), upstream exon (5′E), excised intron (I), and processed mRNA (M) are noted on the left.
Rcp10p is required for cellular pre-mRNA splicing.
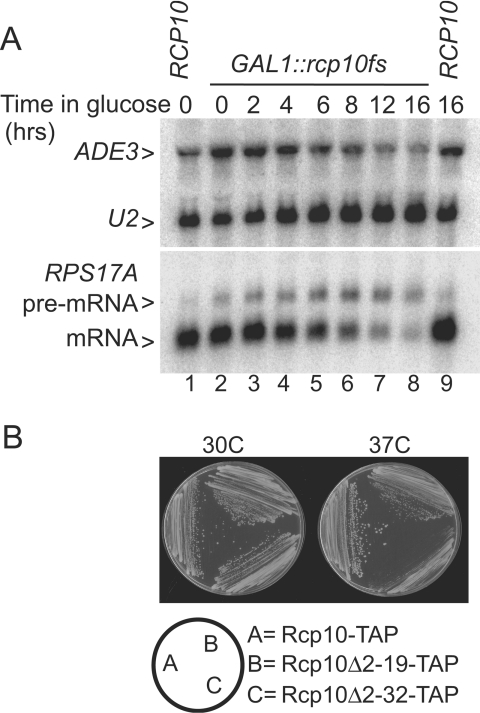
Since RCP10 encodes an essential SF3b subunit, one predicts splicing inhibition in its absence. In support of this, yeast strains that express a GAL1::RCP10 promoter fusion in the rcp10::HIS3 chromosomal background splice well when cultured on galactose-based medium but show impaired splicing on glucose with a maximal pre-mRNA-to-mRNA ratio 9 to 12 h after glucose addition (see Fig. S1 in the supplemental material). Residual GAL1 expression under these conditions is sufficient, however, for continued (albeit inefficient) pre-mRNA processing and cell growth. A fortuitously isolated mutant, rcp10fs, when expressed similarly shows tighter splicing inhibition, resulting at least in part in enhanced protein turnover (Fig. 3A, lower panel; data not shown). Yeast strains that express GAL1::rcp10fs grow well on galactose (equivalent to GAL1::RCP10) but do not form colonies on glucose-based medium (data not shown). No decrease in U2 snRNA is observed even under conditions of severe splicing inhibition, and a modest decrease in the intronless control transcript, ADE3, is seen only after cells are held in a splicing-compromised state for at least 12 h (Fig. 3, upper panel).
FIG. 3.
Rcp10p is essential for cellular pre-mRNA splicing but not for U2 snRNA stability. Shown are results of Northern analysis conducted with RNA from wild-type yeast (RCP10, lanes 1 and 9) and the GAL1::rcp10fs culture before (lane 2) and after (lanes 3 through 8) glucose repression. The upper panel shows the results of hybridization for the intronless ADE3 LSR1 (U2 snRNA) transcripts, and the lower panel shows results for the intron-bearing RPS17A gene. The positions of the unspliced RPS17A pre-mRNA and spliced mRNA are shown. B. Colony formation of wild-type yeast (Rcp10-TAP) and mutant derivatives lacking RCP10 codons 2 to 19 (Rcp10Δ2-19-TAP) and 2 to 32 (Rcp10Δ2-32-TAP) after 2 days of growth on rich medium at 30°C and 37°C.
Curiously, the GAL1::rcp10fs mutation results from a 1-bp deletion within exon II immediately after codon 19. Three closely spaced methionine codons within the intron are in register with the frameshifted rcp10fs exon II. The results of mutational analysis and protein expression studies indicate that one or more of these intronic ATG codons initiate translation, presumably from pre-mRNA that leaks into the cytoplasm when splicing is impaired (see Fig. S2 in the supplemental material). Deletion of codons 2 to 19 or 2 to 32 of RCP10-TAP (including all intron sequence) has a negligible impact on growth when RCP10 is expressed from the natural promoter on a single-copy plasmid at 30°C (Fig. 3B). Both deletion mutants remain viable at 37°C but show more clearly impaired growth defects, with the Δ32 construct being especially temperature sensitive. Based on these observations, we conclude that all essential features of Rcp10p reside 33 to 85 amino acids downstream of the initiating methionine.
Prespliceosome assembly requires Rcp10p.
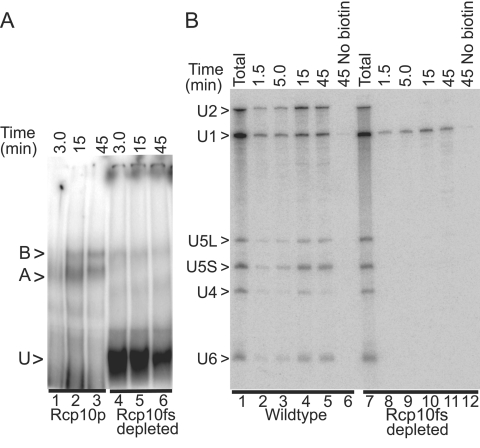
Rcp10p-depleted extracts do not splice pre-mRNA and assemble little or no stable prespliceosome (complex A) or spliceosome (complex B) when assayed by native gel electrophoresis (Fig. 4A). To further investigate the impact of Rcp10p depletion on early events in spliceosome assembly, we used biotin-substituted pre-mRNA to affinity purify in vitro-assembled complexes prepared with or without Rcp10p. Spliceosomes affinity selected from wild-type extract show an accumulation of snRNAs consistent with the assembly profile noted by native gel electrophoresis (Fig. 4B). Background binding of snRNAs to the matrix is negligible when a nonbiotinylated pre-mRNA is used (lanes 6 and 12). As shown previously (38), mature splicing complexes selectively shed U4 snRNA, here indicated by a fivefold reduction in the U4/U6 signal ratio when assembly progresses from 1.5 min (U4/U6, 0.7) to 45 min (U4/U6, 0.15). Although the yeast U1 snRNP association is weakened after prespliceosome formation, causing its dissociation from standard native gels (38), this particle can still be specifically recovered with the splicing apparatus when assayed by gentler selection procedures such as gel filtration or affinity selection (compare lanes 2 to 5 with background in lane 6) (also see references 32 and 54 and review in reference 33).
FIG. 4.
Rcp10p is required for the stable addition of U2 snRNP. A. Native gel electrophoresis of splicing complexes assembled for the indicated times on RPS17A pre-mRNA using extracts from wild-type yeast (Rcp10p) and from yeast depleted of Rcp10p after transcriptional repression of GAL1::rcp10fs (Rcp10fs depleted). U, nonspecific complex; A, prespliceosome; B, spliceosome. B. Time course of spliceosome assembly in Rcp10p-containing (wild-type) or -depleted extracts as described for panel A except using biotin-substituted RPS17A pre-mRNA or a control RNA that lacks biotin (No biotin). Splicing complexes were recovered on streptavidin agarose, and associated snRNAs were compared with that in the starting extract (Total) by Northern blotting. The positions of the snRNAs are noted to the left.
The total (i.e., unfractionated) RNA shows equivalent levels of all spliceosomal snRNAs in the presence or absence of Rcp10p (lanes 1 and 7), yet, unlike the wild type, only U1 snRNA is specifically recovered on splicing complexes assembled in the absence of Rcp10p (compare lanes 8 to 11 with lane 12). The amount of U1 snRNP recovered is roughly equivalent to that observed with the wild-type extract and is well above background (lane 12). Similar to treatments that block U2 snRNP function by targeted snRNA degradation (29) or protein subunit removal (e.g., reference 50), the U1 snRNP-bearing commitment complexes assembled in the Rcp10-depleted extract remain active and can be chased through the splicing pathway with functional extract added together with saturating amounts of cold substrate (data not shown). Thus, Rc10p is required for prespliceosome formation but not for earlier events in assembly.
Rcp10p depletion influences SF3b integrity.
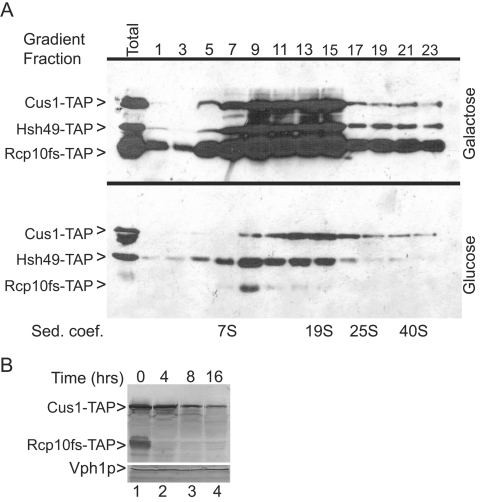
Cellular lysates prepared before and after transcriptional repression of a TAP-tagged GAL1::rcp10fs allele were resolved on a 15 to 40% glycerol gradient and assayed by Western blotting for two SF3b-associated proteins (Fig. 5A). In galactose, the coexpressed Cus1-TAP and Hsh49-TAP proteins colocalize and are most enriched in the SF3b region of the gradient (fractions 9 to 15 between the 7S and 19S markers). Note that, in the galactose-grown culture, the ratio of Cus1-TAP to Hsh49-TAP is constant throughout the first half of the gradient, even where the overall levels are low (e.g., fractions 5 and 7). The overexpressed Rcp10fs-TAP is less tightly focused but is enriched in the same fractions as Cus1-TAP and Hsh49-TAP. Extracts prepared from cells that express Rcp10-TAP from its natural promoter show an equivalent pattern except that the Rcp10-TAP is restricted to the same fractions as Hsh49-TAP and Cus1-TAP (data not shown). After 4 h of GAL1::rcp10fs-TAP repression, the Rcp10fs-TAP levels are reduced approximately 10-fold (compare lanes labeled Total). Gradient fractions 13 and 15 retain equivalent amounts of Cus1-TAP and Hsh49-TAP but lack Rcp10fs-TAP, showing that at least these two proteins and perhaps all of SF3b can assemble in the absence of its smallest subunit. Equivalent results are observed if TAP selection is used to recover these proteins (data not shown). Fraction 11 is similar but shows a 40% reduction in Cus1-TAP compared with the denser fractions. In fractions 7 and 9 the level of Cus1p is further reduced relative to Hsh49p, an indication of incomplete SF3b particles not observed when RCP10 is expressed.
FIG. 5.
Removal of Rcp10p impairs stable SF3b formation. A. Extracts from yeast strains that express TAP-tagged versions of Cus1p and Hsh49p were resolved in parallel on 15 to 40% glycerol gradients before (Galactose) and after (Glucose) transcriptional repression of GAL1::rcp10fs-TAP. Alternate gradient fractions (from the gradient top) were resolved by PAGE and imaged by Western blotting for the TAP tag. Total shows the same extracts prior to gradient fractionation. The sedimentation coefficient (Sed. coef.) markers are the yeast U6 snRNP (7S), thyroglobulin (19S), the U4/U6.U5 tri-snRNP (25S), and the 40S ribosomal subunit. B. Western blot of Cus1-TAP, Rcp10fs-TAP, and Vph1p before (lane 1) and after (lanes 2 to 4) transcriptional repression of GAL1::rcp10fs-TAP.
The change in SF3b sedimentation with Rcp10fs-TAP depletion is due in part to decreased Cus1p levels. As shown in Fig. 5B, Cus1-TAP levels progressively diminish with GAL1::rcp10fsTAP repression while the abundance of an unrelated control protein, Vph1p (or Hsh49p; Fig. 5A and data not shown), does not change. While other explanations are conceivable, the Cus1p reduction most likely reflects enhanced decay of this protein in the absence of Rcp10p. Therefore, while SF3b may be able to assemble without Rcp10p, the integrity of this complex and the stability of its Cus1p subunit appear compromised.
Ist3p is associated with SF3b proteins.
Ist3p has been alternatively described as a component of SF3b (21, 50) and as an RES complex subunit that does not stably associate with the U2 snRNP (16). To further investigate the proposed Ist3p-SF3b association, we coexpressed a V5-tagged Ist3 protein and Rcp10-TAP and assayed for protein recovery at increasing NaCl concentrations (Fig. 6A). Sequential IgG agarose and calmodulin agarose chromatography was performed, and the eluted samples were probed with a monoclonal anti-V5 antibody and with a polyclonal antibody specific for a second (and untagged) SF3b subunit, Rds3p. The two proteins show similar levels of recovery at 150, 300, and 450 mM NaCl, indicating a stable association. In extracts that lack the epitope-tagged Ist3 protein, only the Rds3p band is observed and only background proteins are recovered if the TAP tag is missing (Fig. 6A). Equivalent Ist3-V5 recovery is observed when Cus1-TAP is used to select SF3b (data not shown). The salt-stable association of Ist3p with SF3b contrasts with the Yra1p mRNA export factor, which we find bound only under low-salt conditions (Fig. 6B) (see reference 50).
To learn what proteins stably associated with Ist3p, we purified the Ist3-TAP complex and compared the recovered protein pattern with that observed with Cus1-TAP and Rds3-TAP under high-salt conditions (Fig. 6C). As previously reported (16) the 30-kDa Bud13p and the 24-kDa Plm1p proteins copurify with Ist3-TAP. Although we do not know the stoichiometry of the associated proteins, the low sulfur content of Ist3p (one methionine and no cysteine residues) likely contributes to its comparatively weak 35S signal. In addition, we consistently observe the Rse1p, Hsh155p, and Cus1p proteins of SF3b in the Ist3-TAP preparation (Fig. 6C). The Rse1p band appears underrepresented relative to Hsh155p and Cus1p, possibly due to the presence of the TAP tag (see Discussion). Bands corresponding to the smaller SF3b subunits are more difficult to evaluate, although a band comigrating with the Rds3 protein can be seen with longer image exposure (lower panel, Fig. 6C). Western analysis confirms this assignment (data not shown). The overlapping migration of Ist3-TAP with Hsh49p makes Hsh49p assignment problematic, and the status of Rcp10p is unknown. Bud13-TAP results in the recovery of a protein set containing SF3b subunits (data not shown). In addition, we note that a minor band the size of Bud13p is observed in the Cus1-TAP preparation in Fig. 6C. A Pml1p-sized band has not been observed, and its association with SF3b is uncertain. This overlapping pattern of recovered proteins is most consistent with Ist3p existing in two states, with and without associated SF3b.
Bud31p contributes to the p17 signal.
Deletion of the nonessential IST3 gene reduces but does not eliminate the 17-kDa signal of SF3b (Fig. 7A, lanes 4 and 5) (see reference 50), showing that at least one other protein exists in this band. Consistent with its detection in this complex by mass spectroscopy, deletion of BUD31 also reduces the p17 signal. Rse1p recovery also drops in the bud31::KAN mutant and can be restored to normal levels by the coexpression of an N-terminal Gal4-Bud31 fusion protein (compare lane 1 with lanes 3 and 5). While weak signals make visualization of the smaller bands difficult, labeled Rds3p and Rcp10p are recovered with the ist3::KAN and bud31::KAN single mutants, although at reduced levels with bud31::KAN (Fig. 7A, lower panel, lanes 3 to 5). Unexpectedly, the ist3::KAN and bud31::KAN double mutant proved to be inviable. To circumvent this synthetic lethality, an Ist3-V5 fusion protein was coexpressed in the double-knockout background. In this case, the p17 band is fully and reproducibly lost and, similar to the original bud31::KAN mutant, reduced recovery of labeled Rse1p, Rds3p, and Rc10p is observed (Fig. 7A, lane 2).
As an alternative means to investigate Bud31p-SF3b association, a Western blot of SF3b recovered at 450 mM NaCl was probed for the presence of an epitope-tagged Bud31-V5 protein. Bud31-V5 copurified with Cus1-TAP (Fig. 7B, lanes 2 and 3) but not from an extract that lacks a TAP-tagged protein (lane 1) or when a nonspecific resin matrix was substituted in TAP selection (lane 4). Together the combined biochemical, immunological, and genetic data make a strong case that Ist3p and Bud31p both contribute to the p17 signal of SF3b.
Two-hybrid interaction map of SF3b.
Although it was first identified as a stable complex over a decade ago (11), surprisingly little is known about the subunit organization of SF3b. Since only a few of the subunits were recovered as interacting partners in the published global two-hybrid screens (18, 25, 45), we constructed full-length (i.e., entire-ORF) two-hybrid clones for each subunit and assayed all pairwise combinations for reporter gene stimulation. Several other genes implicated by genetic or biochemical association with the U2 snRNP function (CLF1, YRA1, BUD13, PRP5, MUD2, and BBP/MSL5) were included as well (Table 2; see Fig. S3 in the supplemental material for photographic images of all positive interactions).
TABLE 2.
Yeast two-hybrid interactionsa
| Construct | BBP | Bud13p | Bud31p | Clf1p | Cus1p | Hsh49p | Hsh155p | Ist3p | Mud2p | Prp5p | Rcp10p | Rds3p | Rse1p | Yra1p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBP | − | − | − | − | − | − | − | − | ++ | − | − | − | − | − |
| Bud13p | − | − | − | − | − | − | − | ++ | − | − | − | − | − | − |
| Bud31p | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Clf1p | ++* | − | − | − | − | − | +* | − | ++ | − | − | − | − | − |
| Cus1p | − | − | − | ++* | − | + | − | − | +* | − | − | − | − | − |
| Hsh49p | − | − | − | − | + | − | − | − | − | − | − | − | − | − |
| Hsh155p | − | ++* | − | ++* | − | − | − | − | +++* | +++* | +* | − | − | − |
| Ist3p | − | +++ | − | − | − | − | − | − | − | − | − | − | − | − |
| Mud2p | +++ | − | − | +++ | +++* | − | +++* | +++* | − | − | +* | − | − | − |
| Prp5p | − | − | − | − | − | − | +* | − | − | − | − | − | − | − |
| Rcp10p | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Rds3p | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Rse1p | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Yra1p | − | − | − | − | − | − | − | − | − | − | − | − | − | ++* |
The two-hybrid scores reflect relative colony sizes after 4 days at 30°C on synthetic medium selective for the HIS3 and ADE3 reporter genes (see data S3 in the supplemental material). Interactions not previously reported are indicated with an asterisk with reciprocal interactions in boldface. x axis, Gal4 DNA binding domain constructs; y axis, Gal4 activation domain constructs. +++ represents growth equivalent to that of a prototroph.
Our observations confirm the previously reported two-hybrid interaction of Mud2p and BBP (2, 18), Cus1p and Hsh49p (18, 24), Bud13p and Ist3p (25, 45), and Clf1p and Mud2p (14). Several previously unpublished interactions were identified (indicated by asterisks in Table 2), including a possible Rcp10p-based interaction with the SF3b protein Hsh155. Intriguingly, both Hsh155p and Rcp10p also appear to associate with Mud2p, although the large number of potential partners for Mud2p (i.e., BBP, Clf1p, Cus1p, Hsh155p, and Ist3P) suggests the possibility of indirect or nonspecific associations. The Ist3p binding partner Bud13p is observed to interact with Hsh155p. Intriguingly, Prp5p, a DExD/H-box protein implicated in U2 snRNP function (see reference 37 and references within), interacts strongly and reciprocally by this assay with Hsh155p. A Yra1p-Yra1p two-hybrid interaction is also observed, consistent with the ability of Ref/Aly-like proteins to self-associate in vitro (48). Finally, the Clf1p subunit of the late-acting nineteen complex (NTC) was observed to interact with SF3b subunits Hsh155p and Cus1p and with BBP (although the BBP interaction is not observed with Clf1p in the DNA binding domain [14]). Most but not all of the interactions were reciprocal; no interactions were found for Rse1p, Rds3p, or Bud31p. Importantly, a number of the novel two-hybrid interactions presented here are supported by additional genetic or biochemical evidence (see Discussion).
DISCUSSION
Here we establish the concordance of the mammalian and yeast SF3b U2 snRNP subunits and demonstrate an essential function for its 10-kDa subunit (50), Rcp10p. This work extends our understanding of RCP10 in several important ways from what was previously reported (9, 15, 16). First, we provide direct proof that Rcp10p is required for cellular pre-mRNA splicing, as its depletion blocks intron removal in vivo. Second, mechanistic insight into this splicing defect is provided by the observation that extracts depleted of Rcp10p assemble the U1-bearing commitment complex but fail to support U2 snRNP addition. Finally, we provide a direct link between Rcp10p function and SF3b integrity by showing that Rcp10 depletion impairs SF3b assembly or stability. The two-hybrid study suggests that Rcp10p may intimately associate with the Hsh155p SF3b protein proposed to span the branchpoint region in splicing complexes. Rcp10p does more than simply bind Hsh155p, however, since Rcp10p is required for maintaining normal cellular levels of the essential Cus1 protein.
Gottschalk and coworkers (21) reported yeast Ist3p/Snu17p as a possible SF3b p14a homolog based on its structural similarity to the mammalian p14a protein and on the specific recovery of U2 snRNA with this RRM-bearing protein (52). Importantly, they also showed splicing inhibition and altered U2 snRNP electrophoretic mobility in an ist3 mutant background, thereby linking Ist3p with U2 snRNP biogenesis and function. More recently, Ist3p was shown to be necessary for Mer1p activation of splicing in strains with mutated branchpoint sequences (43), reinforcing a role for this protein in U2-associated events of intron selection. Here we show that a V5-tagged Ist3p protein specifically copurifies with the SF3b subcomplex of the U2 snRNP. In addition, we show that the purified Ist3-TAP complex(es) contains SF3b subunits. Given the preponderance of evidence for Ist3p-U2 snRNP association, it is surprising that Dziembowski et al. (16) failed to detect Ist3p in SF3b or SF3b subunits with Ist3p. The reason for this discrepancy is unclear but presumably reflects differences in the protein isolation or detection techniques.
The identification of Bud31p as the second p17 protein of SF3b is supported by mass spectroscopy of this complex, Western analysis results showing its salt-stable association with Cus1-TAP, and the reduction in p17 signal in the bud31::KAN background. Bud31p has been found in multiple other splicing-related protein complexes including the endogenous spliceosome (34, 43a, 49). Deletion of this nonessential gene clearly impairs yeast splicing (30), supporting a physiologically relevant basis for these interactions. The partial recovery of Bud31p and other splicing factors such as Brr2p and Prp8p among multiple complexes (e.g., intact snRNP particles, SF3b, and endogenous spliceosome) or intermediates in the extract presumably reflects partitioning based on relative affinity. While little is known about Bud31p function, the synthetic lethality of the bud31::KAN ist3::KAN double knockout suggests a related role in splicing. Conceivably, the lower levels of Rse1p recovered with SF3b from the bud31::KAN mutant reflect impaired SF3b biogenesis or stability. We note with caution, however, that Rse1p interactions appear particularly sensitive to perturbation, as this protein can be released from the endogenous spliceosome with temperature inactivation of the Clf1Δ2p splicing factor (49), from SF3b after Rds3-1p inactivation (50), or seemingly when the TAP tag is introduced into Ist3p (this study). Consequently, the lower level of Rse1p recovered with SF3b from the bud31::KAN mutant may reflect a less direct consequence of splicing inhibition.
Our directed two-hybrid study with full-length bait and prey constructs identified a number of interactions missed in earlier screens of peptide fusion libraries (e.g., references 18, 25, and 45), not a surprising result given the number of unique interactions reported for each of these earlier studies. While not proving direct or functional association, positive two-hybrid results often define interactions worthy of additional consideration, especially when supported by other relevant observations. For instance, the mammalian SAP155 protein interacts with the SF3b p14a subunit that cross-links to the catalytic branchpoint adenosine (21). While we do not observe an Hsh155p-Ist3p interaction, we do detect an interaction between Hsh155p and the Ist3p binding partner, Bud13p (25, 45), that might serve an equivalent role in organizing the pre-mRNP in yeast. Similarly, a reciprocal two-hybrid interaction was found between Hsh155p and the DExD/H-box protein Prp5p. This is particularly interesting since recent results from the fission yeast system suggest that the ATP-independent function of Prp5p may include tethering the U1 and U2 snRNP particles (55); our results suggest that Hsh155p may serve as a U2 snRNP contact. Novel interactions were also observed between the NTC subunit Clf1p and the Cus1p and Hsh155p components of SF3b. Independent support for a Clf1p-U2 snRNP association is provided by the observation that temperature inactivation of Clf1Δ2p in vitro (or in vivo) causes the release of at least two SF3b proteins, Hsh155p and Rse1p, from endogenous spliceosomes (49), suggesting a stabilizing effect of Clf1p on U2 snRNP components of the splicing complex. Indeed, enhanced U2 snRNA expression partially alleviates the growth defect of certain clf1 mutants (6), indicating a mechanism of splicing compensation dependent on the U2 snRNP. Mutations in RDS3 and the RES-associated BUD13 genes were identified in a synthetic lethal screen with clf1Δ2 and shown to independently impair splicing (47). Taken in this context, the Hsh155p-Clf1p-Cus1p two-hybrid interactions reported here suggest a means of physically linking the U2 snRNP with the NTC structure essential for late events in spliceosome assembly and splicing (13, 44). Finally, the previously reported Rse1p-Prp9p and Cus1p-Brr2p two-hybrid interactions (18) are consistent with the SF3b association of Prp9p and Brr2p indicated by our mass analysis of SF3b (see Materials and Methods).
The relationship between the RES and SF3b complexes is an interesting unresolved issue. The results of reporter gene assays suggest that RES components may help retain the unprocessed pre-mRNA in the nucleus independently of U2 snRNP function (16). While Ist3p is structurally similar to mammalian SF3b 14a, an argument has been made that higher eukaryotes possess two 14a-like RNA binding proteins, with the SF3b-specific subunit being absent in budding yeast (16). Since results presented here establish physical association of Ist3p with SF3b, we propose resolution of this conundrum by suggesting that, in yeast, the putative intron retention and splicing functions reside within a single protein. If they are functionally conserved, extrapolation from the yeast system predicts that the second SF3b 14a-like protein in mammals may serve some role in pre-mRNA retention. A speculative model consistent with current data is that RES binds the splicing substrate, possibly near the branchpoint in a pre-mRNA retention capacity prior to prespliceosome formation. With U2 snRNP recruitment, Ist3p and Bud13p (and possibly Pml1p) become SF3b associated and function in splicing. Afterwards, the RES factors may recycle free of U2 snRNP components. Alternatively, the RES proteins may simply serve as accessory splicing factors, intimately associated with the SF3b branchpoint binding subcomplex of the U2 snRNP.
Supplementary Material
Acknowledgments
We thank Bertrand Seraphin and Sakshi Pandit for helpful discussions while this work was in progress. Also, we thank Colleen Stoeppel and Kate Zaytseva for assistance with the two-hybrid constructs and Martha Peterson for comments on the manuscript.
Support for this work was provided by NIH GM42476 to B.C.R. and infrastructure funds provided by NSF EPS-0132295.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abovich, N., X. C. Liao, and M. Rosbash. 1994. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 8:843-854. [DOI] [PubMed] [Google Scholar]
- 2.Abovich, N., and M. Rosbash. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89:403-412. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balakrishnan, R., K. R. Christie, M. C. Costanzo, K. Dolinski, S. S. Dwight, S. R. Engel, D. G. Fisk, J. E. Hirschman, E. L. Hong, R. Nash, R. Oughtred, M. Skrzypek, C. L. Theesfeld, G. Binkley, C. Lane, M. Schroeder, A. Sethuraman, S. Dong, S. Weng, S. Miyasato, R. Andrada, D. Botstein, and J. M. Cherry. 2004. Saccharomyces Genome Database. [Online.] http://www.yeastgenome.org/gene_guidelines.shtml.
- 5.Behrens, S. E., K. Tyc, B. Kastner, J. Reichelt, and R. Luhrmann. 1993. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol. Cell. Biol. 13:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Yehuda, S., I. Dix, C. S. Russell, M. McGarvey, J. D. Beggs, and M. Kupiec. 2000. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics 156:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanton, S., A. Srinivasan, and B. C. Rymond. 1992. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol. Cell. Biol. 12:3939-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordonne, R., J. Banroques, J. Abelson, and C. Guthrie. 1990. Domains of yeast U4 spliceosomal RNA required for PRP4 protein binding, snRNP-snRNP interactions, and pre-mRNA splicing in vivo. Genes Dev. 4:1185-1196. [DOI] [PubMed] [Google Scholar]
- 9.Brachat, S., F. S. Dietrich, S. Voegeli, Z. Zhang, L. Stuart, A. Lerch, K. Gates, T. Gaffney, and P. Philippsen. 2003. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 4:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosi, R., K. Groning, S. E. Behrens, R. Luhrmann, and A. Kramer. 1993. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science 262:102-105. [DOI] [PubMed] [Google Scholar]
- 11.Brosi, R., H. P. Hauri, and A. Kramer. 1993. Separation of splicing factor SF3 into two components and purification of SF3a activity. J. Biol. Chem. 268:17640-17646. [PubMed] [Google Scholar]
- 12.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333-360. [DOI] [PubMed] [Google Scholar]
- 13.Chen, C. H., W. C. Yu, T. Y. Tsao, L. Y. Wang, H. R. Chen, J. Y. Lin, W. Y. Tsai, and S. C. Cheng. 2002. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 30:1029-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung, S., M. R. McLean, and B. C. Rymond. 1999. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5:1042-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 16.Dziembowski, A., A. P. Ventura, B. Rutz, F. Caspary, C. Faux, F. Halgand, O. Laprevote, and B. Seraphin. 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 23:4847-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleckner, J., M. Zhang, J. Valcarcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11:1864-1872. [DOI] [PubMed] [Google Scholar]
- 18.Fromont-Racine, M., J. C. Rain, and P. Legrain. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16:277-282. [DOI] [PubMed] [Google Scholar]
- 19.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 20.Golas, M. M., B. Sander, C. L. Will, R. Luhrmann, and H. Stark. 2003. Molecular architecture of the multiprotein splicing factor SF3b. Science 300:980-984. [DOI] [PubMed] [Google Scholar]
- 21.Gottschalk, A., C. Bartels, G. Neubauer, R. Luhrmann, and P. Fabrizio. 2001. A novel yeast U2 snRNP protein, Snu17p, is required for the first catalytic step of splicing and for progression of spliceosome assembly. Mol. Cell. Biol. 21:3037-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gozani, O., R. Feld, and R. Reed. 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 23.Gozani, O., J. Potashkin, and R. Reed. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18:4752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igel, H., S. Wells, R. Perriman, and M. Ares, Jr. 1998. Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA 4:1-10. [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 28.Kistler, A. L., and C. Guthrie. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 15:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legrain, P., B. Seraphin, and M. Rosbash. 1988. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol. Cell. Biol. 8:3755-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masciadri, B., L. B. Areces, P. Carpinelli, M. Foiani, G. Draetta, and F. Fiore. 2004. Characterization of the BUD31 gene of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 320:1342-1350. [DOI] [PubMed] [Google Scholar]
- 31.McPheeters, D. S., and P. Muhlenkamp. 2003. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 23:4174-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaud, S., and R. Reed. 1991. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 5:2534-2546. [DOI] [PubMed] [Google Scholar]
- 33.Moore, M., C. C. Query, and P. A. Sharp. 1993. Splicing of precursors to mRNA by the spliceosome, p. 303-358. In R. F. Gesteland and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Ohi, M. D., A. J. Link, L. Ren, J. L. Jennings, W. H. McDonald, and K. L. Gould. 2002. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22:2011-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pauling, M. H., D. S. McPheeters, and M. Ares, Jr. 2000. Functional Cus1p is found with Hsh155p in a multiprotein splicing factor associated with U2 snRNA. Mol. Cell. Biol. 20:2176-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perriman, R., and M. Ares, Jr. 2000. ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 14:97-107. [PMC free article] [PubMed] [Google Scholar]
- 37.Perriman, R., I. Barta, G. K. Voeltz, J. Abelson, and M. Ares, Jr. 2003. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. USA 100:13857-13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pikielny, C. W., B. C. Rymond, and M. Rosbash. 1986. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. Nature 324:341-345. [DOI] [PubMed] [Google Scholar]
- 39.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 40.Ruskin, B., P. D. Zamore, and M. R. Green. 1988. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52:207-219. [DOI] [PubMed] [Google Scholar]
- 41.Rutz, B., and B. Seraphin. 1999. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA 5:819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rymond, B. C., and M. Rosbash. 1992. Yeast pre-mRNA splicing, p. 143-192. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces,vol. 21. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 43.Spingola, M., J. Armisen, and M. Ares, Jr. 2004. Mer1p is a modular splicing factor whose function depends on the conserved U2 snRNP protein Snu17p. Nucleic Acids Res. 32:1242-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Stevens, S. W., D. E. Ryan, H. Y. Ge, R. E. Moore, M. K. Young, T. D. Lee, and J. Abelson. 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9:31-44. [DOI] [PubMed] [Google Scholar]
- 44.Tarn, W. Y., K. R. Lee, and S. C. Cheng. 1993. The yeast PRP19 protein is not tightly associated with small nuclear RNAs but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol. 13:1883-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 46.Umen, J. G., and C. Guthrie. 1995. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 9:855-868. [DOI] [PubMed] [Google Scholar]
- 47.Vincent, K., Q. Wang, S. Jay, K. Hobbs, and B. C. Rymond. 2003. Genetic interactions with CLF1 identify additional pre-mRNA splicing factors and a link between activators of yeast vesicular transport and splicing. Genetics 164:895-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Virbasius, C. M., S. Wagner, and M. R. Green. 1999. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell 4:219-228. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Q., K. Hobbs, B. Lynn, and B. C. Rymond. 2003. The Clf1p splicing factor promotes spliceosome assembly through N-terminal tetratricopeptide repeat contacts. J. Biol. Chem. 278:7875-7883. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Q., and B. C. Rymond. 2003. Rds3p is required for stable U2 snRNP recruitment to the splicing apparatus. Mol. Cell. Biol. 23:7339-7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Will, C. L., C. Schneider, A. M. MacMillan, N. F. Katopodis, G. Neubauer, M. Wilm, R. Luhrmann, and C. C. Query. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 20:4536-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Will, C. L., H. Urlaub, T. Achsel, M. Gentzel, M. Wilm, and R. Luhrmann. 2002. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 21:4978-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 54.Xie, J., K. Beickman, E. Otte, and B. C. Rymond. 1998. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 17:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, Y. Z., C. M. Newnham, S. Kameoka, T. Huang, M. M. Konarska, and C. C. Query. 2004. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 23:376-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.