Abstract
The Forkhead box m1 (Foxm1) gene is critical for G1/S transition and essential for mitotic progression. However, the transcriptional mechanisms downstream of FoxM1 that control these cell cycle events remain to be determined. Here, we show that both early-passage Foxm1−/− mouse embryonic fibroblasts (MEFs) and human osteosarcoma U2OS cells depleted of FoxM1 protein by small interfering RNA fail to grow in culture due to a mitotic block and accumulate nuclear levels of cyclin-dependent kinase inhibitor (CDKI) proteins p21Cip1 and p27Kip1. Using quantitative chromatin immunoprecipitation and expression assays, we show that FoxM1 is essential for transcription of the mitotic regulatory genes Cdc25B, Aurora B kinase, survivin, centromere protein A (CENPA), and CENPB. We also identify the mechanism by which FoxM1 deficiency causes elevated nuclear levels of the CDKI proteins p21Cip1 and p27Kip1. We provide evidence that FoxM1 is essential for transcription of Skp2 and Cks1, which are specificity subunits of the Skp1-Cullin 1-F-box (SCF) ubiquitin ligase complex that targets these CDKI proteins for degradation during the G1/S transition. Moreover, early-passage Foxm1−/− MEFs display premature senescence as evidenced by high expression of the senescence-associated β-galactosidase, p19ARF, and p16INK4A proteins. Taken together, these results demonstrate that FoxM1 regulates transcription of cell cycle genes critical for progression into S-phase and mitosis.
Activation of the Ras-mitogen-activated protein kinase (MAPK) signaling pathway drives cell cycle progression by temporal expression of cyclin regulatory subunits, which activate their corresponding cyclin-dependent kinases (CDKs) through complex formation (45, 46, 61). Progression into DNA replication (S-phase) requires phosphorylation of the retinoblastoma (RB) protein by the Cdk4/Cdk6 proteins in complex with cyclin D (45). Phosphorylated RB dissociates from the E2F transcription factor and alleviates inhibition of E2F to allow transcriptional stimulation of cyclin E and other S-phase-promoting genes (45). Association of cyclin E with Cdk2 leads to hyperphosphorylation of RB, thus stimulating cell cycle progression beyond the G1/S checkpoint. Ras-MAPK signaling is also necessary to induce expression of S-phase-promoting Cdc25A phosphatase, which dephosphorylates inhibitory Cdk2 residues and activates Cdk2-cyclin E activity (45, 52). Cdk2-cyclin E phosphorylation of the CDK inhibitor (CDKI) proteins p27Kip1 and p21Cip1 is required for recognition by the F-Box protein S-phase kinase-associated protein 2 (Skp2) and CDK subunit 1 (Cks1) proteins, which are substrate specificity subunits of the Skp1-Cullin1-F-box protein (SCF) ubiquitin ligase complex (7, 11, 19, 49, 59, 63). During the G1/S transition of the cell cycle, the SCF ubiquitin ligase complex targets these phosphorylated CDKI proteins for ubiquitin modification and subsequent proteasome degradation, thus facilitating activation of the Cdk2-cyclin E complex for progression into S-phase.
Progression through the G2/M transition requires activation of the Cdk1-cyclin B complex through dephosphorylation and activation of Cdk1 by the Cdc25B and Cdc25C phosphatases, the latter of which is activated by Polo-like kinase 1 (PLK1) phosphorylation (4, 52). During mitosis, development of a multiprotein kinetochore complex on centromeres is required for attachment of spindle microtubules to chromosomes that radiate and attach to bipolar centrosomes, thereby mediating segregation of sister chromatids (17). PLK1 protein is critical for centrosome duplication, and both PLK1 and Aurora B kinase are involved in bipolar microtubule spindle attachment to the centromeric kinetochores (4, 48). The Aurora B kinase is also essential for spindle assembly checkpoint, chromosome segregation, and cytokinesis (48). Aurora B kinase deficiency causes endoreduplication and a polyploid genotype due to a failure in the spindle assembly checkpoint, resulting in premature mitotic exit during prophase and reinitiation of S-phase (28, 48, 50, 51). Survivin forms a complex with chromosome passenger proteins Aurora B kinase and inner centromere protein (INCENP), where it plays a critical role in the localization of the Aurora B kinase-INCENP complex to the inner chromosomal region of centromeres at the early stages of mitosis (3). Moreover, replacement of histone H3 in centromeric nucleosomes with the histone variant centromere protein A (CENPA) is a prerequisite for recruitment of CENPB and CENPC proteins, which are necessary for assembly of the kinetochore protein complex on the centromere region of chromosomes (2, 5, 47).
The ARF/INK4A locus encodes two distinct tumor suppressors, the CDKI protein p16INK4A and the mouse 19-kDa alternative reading frame (ARF) protein (14-kDa ARF protein in humans) that are translated from different reading frames on exon 2 (57, 60). The p19ARF protein stabilizes the tumor suppressor p53 protein by interfering with Mdm2-mediated p53 degradation, thereby allowing accumulation of p53 transcription factor and stimulating transcription of the CDKI p21Cip1 gene (39). The p19ARF protein also mediates p53-independent cell cycle arrest, because the mouse ARF protein targets both the E2F1 and c-Myc transcription factors to the nucleolus, thus preventing their transcriptional activation of S-phase-promoting target genes (15, 16, 44, 56). Long-term growth of primary mouse embryonic fibroblasts (MEFs) induces replicative senescence, which is associated with increased expression of senescence-associated β-galactosidase (SA-β-Gal), the CDKIs p21Cip1 and p16INK4A, and the tumor suppressor proteins p19ARF and p53 (1, 20, 53, 54). Moreover, p19ARF−/− MEFs are immortalized and do not undergo growth arrest following long-term passage in culture, suggesting that ARF plays an important role in replicative senescence of rodent cells (29).
The mammalian Forkhead box (Fox) family of transcription factors consists of more than 50 mammalian proteins (9, 26) that share homology in the winged helix DNA binding domain (12, 43). Expression of the Foxm1 transcription factor is induced during the G1 phase of the cell cycle, and its expression continues during S-phase and mitosis (33, 73-75). Foxm1 is expressed in all proliferating mammalian cells and tumor-derived cell lines, and its expression is extinguished in terminally differentiated cells that exit the cell cycle (33, 40, 73-75). Transcription of the Foxm1 locus results in three differentially spliced mRNAs that are almost identical in sequence but differ by the addition of two small exons: the Foxm1b isoform (HFH-11B) contains no additional exons, while the Foxm1c (Trident, WIN, or MPP2) and Foxm1a (HFH-11A) isoforms contain one or two additional exons, respectively (34, 40, 73, 75). Transcriptional activity of the Foxm1 protein is dependent on Ras-MAPK signaling, because the FoxM1 carboxyl-terminal activation domain binds the Cdk-cyclin complexes, which allows efficient phosphorylation of the Cdk site at Thr residue 596 that is essential for recruiting the CREB binding protein (CBP) transcriptional coactivator (42).
We used the albumin promoter/enhancer Cre recombinase transgene (Alb-Cre) to mediate hepatocyte-specific deletion of the Foxm1 fl/fl targeted allele, which removed exon 4 through exon 7 encoding the Foxm1 DNA binding and transcriptional activation domains (67). Liver regeneration studies demonstrated that Alb-Cre Foxm1−/− hepatocytes displayed an 80% reduction in DNA replication and a complete inhibition of mitosis (67). The reduction in hepatocyte DNA replication was consistent with reduced Cdk2-cyclin E activity resulting from posttranscriptional increases in nuclear levels of the CDKI proteins p21Cip1 and p27Kip1 (27, 67). However, the mechanism by which these CDKI proteins are degraded by Foxm1 remains to be determined. The inhibition of hepatocyte mitosis was associated with insufficient Cdk1-cyclin B activity resulting from undetectable expression of Cdc25B phosphatase (67), yet the complete block in M-phase progression suggested that Foxm1 may control transcription of other mitotic regulators. Using a well-established liver tumor induction and promotion method, we showed that Alb-Cre Foxm1−/− hepatocytes fail to proliferate and are highly resistant to chemical-induced hepatocellular carcinoma (HCC) formation (13, 27). The mechanism of resistance to HCC development is associated with persistent hepatocyte nuclear accumulation of the CDKI protein p27Kip1 and diminished expression of the Cdk1-activating Cdc25B phosphatase (13, 27). Moreover, we showed that FoxM1 is a novel inhibitory target of the p19ARF tumor suppressor, which inhibits FoxM1 transcriptional activity by targeting it to the nucleolus (13, 27).
We previously developed an inducible osteosarcoma U2OS clone C3 cell line (U2OS C3 cells) in which doxycycline (Dox) treatment induced expression of the green fluorescent protein (GFP)-human FoxM1b fusion protein (27). To identify FoxM1 transcriptional targets mediating progression into S-phase and mitosis, we developed FoxM1 small interfering RNA (siRNA) that, when transfected into the human osteosarcoma U2OS C3 cells, suppressed expression of both endogenous FoxM1 and induced GFP-FoxM1b proteins. Here, we show that both U2OS cells depleted of FoxM1 levels and early-passage Foxm1−/− MEFs were unable to significantly grow in culture due to a block in mitotic progression and accumulated nuclear levels of the CDKI proteins p21Cip1and p27Kip1. Interestingly, these early-passage Foxm1−/− MEFs display premature senescence as evidenced by high levels of senescence-associated β-galactosidase and increased nuclear levels of the senescence markers p19ARF and p16INK4A. Using quantitative chromatin immunoprecipitation (ChIP) and expression assays, we show for the first time that FoxM1 is essential for transcription of the mitotic regulatory genes Cdc25B, Aurora B kinase, survivin, CENPA, and CENPB. Interestingly, both FoxM1- and Aurora B kinase-depleted U2OS cells exhibited identical accumulation of polyploid (8N) cells, supporting the hypothesis that diminished expression of Aurora B kinase contributed to development of a polyploid genotype in FoxM1-deficient cells. Undetectable expression of PLK1 protein was also found in FoxM1-depleted U2OS cells, and recent studies have used FoxM1 cotransfection and ChIP assays to demonstrate that FoxM1 regulates transcription of the PLK1 gene (37). Consistent with reduced S-phase progression, we showed that increased nuclear levels of the CDKI proteins p27Kip1 and p21Cip1 in FoxM1-depleted cells were associated with undetectable levels of the SCF ubiquitin ligase complex proteins Skp2 and Cks1, which are specificity subunits of the SCF ubiquitin ligase complex. Our data imply that during the G1/S transition FoxM1 mediates transcription of Skp2 and Cks1, which are essential for degradation of these CDKI proteins. Our results support the hypothesis that FoxM1 controls gene expression of cell cycle regulatory proteins critical for S-phase and M-phase progression.
MATERIALS AND METHODS
Doxycycline-inducible U2OS C3 cell culture, siRNA transfection, and synchronization.
We previously reported on the generation of an osteosarcoma U2OS clone C3 cell line (U2OS C3 cells) that allowed Dox-inducible expression of the GFP-FoxM1b fusion protein (27). U2OS C3 cells were maintained as a monolayer in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 IU/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, and 50 μg/ml of hygromycin B (Invitrogen). Dharmacon Research synthesized two 21-nucleotide siRNA duplexes specific to human FoxM1 mRNA, named siFoxM1 #1 (CAACAGGAGUCUAAUCAAG) and siFoxM1 #2 (GGACCACUUUCCCUACUUU), and human p27Kip1 siRNA duplex (GUACGAGUGGCAAGAGGUGUU), containing symmetric 2-uracil (U) 3′ overhangs. Human Aurora B siRNA duplex was purchased from Cell Signaling Technology (Beverly, MA). These siRNA duplexes were transfected into U2OS C3 cells using Lipofectamine 2000 reagent (Invitrogen) in serum-free tissue culture medium following the manufacturer's protocol. Four hours after transfection, the cells were fed with complete DMEM containing 10% FCS, 100 IU/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, and 50 μg/ml of hygromycin B. For induced expression of the GFP-FoxM1b fusion protein, we added complete DMEM containing 10% FCS and 1 μg/ml of Dox (Sigma) 4 hours following FoxM1 siRNA transfection. U2OS C3 or U2OS cells were harvested at 48 h after FoxM1 siRNA transfection for total RNA or harvested at 72 h after FoxM1 siRNA transfection to prepare protein extracts for Western blot analysis, flow cytometry, or immunofluorescent staining. For CENPA immunofluorescent staining of mitotic synchronized cells, at 72 h after FoxM1 siRNA 2 transfection, we synchronized U2OS cells at early stages of mitosis with 100 ng/ml nocodazole (Sigma) for 24 h and then released them for 1 h to allow progression into metaphase.
Cell growth rate of FoxM1-depleted U2OS C3 cells by siRNA transfection.
U2OS cells were transfected with siFoxM1 #2 or siRNA p27Kip1 duplexes and then incubated for 2 days to allow siRNA silencing of expression. The FoxM1- or p27Kip1-depleted U2OS cells were then trypsinized, and the cell growth rate was determined in triplicate at 3, 4, 5, or 6 days after siRNA transfection. To determine cell number, U2OS cells were trypsinized and removed from the plate, nonviable cells were identified by 3 min of staining with 0.4% trypan blue (Sigma), and the number of unstained U2OS C3 cells was counted using a hemocytometer. We plotted cell number ± standard deviation (SD) versus days after siRNA transfection. Statistical analysis of experiment data was performed with Microsoft Excel tools.
Generation and growth analysis of Foxm1−/−, Foxm1+/−, and Foxm1+/+ MEFs.
Foxm1+/− mice containing an inactivated Foxm1-targeted allele that deleted essential Foxm1 exons 4 through 7, which encode the Foxm1 DNA binding domain and C-terminal transcriptional activation domain, were described previously (36, 67). We bred Foxm1+/− mice to generate 13.5-day embryos with Foxm1+/+ (wild type), Foxm1+/− (heterozygous), or Foxm1−/− (knockout) genotypes for isolation of MEFs. To isolate MEFs from the embryos, the liver, heart, and head were removed from the embryos and then the remaining embryo was digested with 0.25% trypsin in 2.21 mM EDTA (Cellgro, Herndon, VA) to isolate single-cell suspensions of MEFs using standard procedures described by Hogan et al. (23). The heart tissue was used to generate DNA for PCR genotyping of the embryos as described previously (36). The MEFs were grown in DMEM supplemented with 10% FCS, 100 IU/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, 0.1 mM MEM nonessential amino acids, and 55 mM 2-mercaptoethanol in a humidified 9% CO2 incubator under conditions described by Zindy et al. (76). To measure cell population doublings, MEFs were trypsinized and then 3 × 105 cells were replated and fed with DMEM containing 10% FCS. Following 3 days of growth at 37°C and 9% CO2, we counted the number of cells as described above. Population doubling during each passage was calculated according to the formula log[(cell number after 3 days of growth)/(3 × 105)]/log 2.
Flow cytometry assays to determine cell cycle profiles.
U2OS C3 cells were transfected with 100 nM of either siFoxM1 #2, p27Kip1, or Aurora B kinase siRNA duplexes, and then 72 h after transfection the cells were subjected to flow cytometry to analyze their cell cycle profile. Wild-type (WT) Foxm1+/+, heterozygous Foxm1+/−, or knockout Foxm1−/− MEFs were trypsinized at passage 4 to analyze their cell cycle profile by flow cytometry. For flow cytometry, cells were trypsinized and fixed in 70% ethanol for 2 h at 4°C. Cells were incubated with 40 μg/ml propidium iodide and 100 μg/ml RNase A (Sigma) in phosphate-buffered saline (PBS) for 1 h at 37°C. After washing, cells were resuspended in PBS for further analysis. Data were acquired using a Beckman Coulter EPICS Elite ESP apparatus (Hialeah, FL) and then analyzed using Multicycle AV (Phoenix Flow Systems, San Diego, CA). The flow cytometry and analysis were performed in the Research Resource Center at the University of Illinois at Chicago.
Description of antibodies used for Western blot analysis to determine protein expression levels.
To prepare protein extracts, U2OS cells depleted of FoxM1 or Aurora B kinase or Foxm1+/+, Foxm1+/−, or Foxm1−/− MEFs were harvested in ice-cold PBS, pelleted by centrifugation, and used to make whole-cell protein extracts using the NP-40 lysis buffer as described previously (42). Cytoplasmic and nuclear protein extracts were made using a nuclear/cytosol fractionation kit (K266-100; BioVision) following protocols provided by the manufacturer. Protein concentrations were determined by the Bradford method with the Bio-Rad protein assay reagent. Equal amounts of proteins in whole-cell extracts or nuclear and cytoplasmic extracts from each set of experiments were fractionated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membrane (Bio-Rad). The membrane was subjected to Western blot analysis with antibodies against proteins of interest as described previously (27, 42). The signals from the primary antibody were amplified by horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG; Bio-Rad, Hercules, CA) and detected with enhanced chemiluminescence (ECL Plus; Amersham Pharmacia Biotech, Piscataway, NJ).
The following commercially available antibodies and dilutions were used for Western blotting: mouse anti-Plk-1 (F-8; 1:500); mouse anti-Cdc25A (F-6; 1:300); rabbit anti-Cdk2 (M2; 1:1,500), and mouse anti-cyclin A (H-432; 1:2,000) (Santa Cruz Biotechnology, Inc.); rabbit anti-INCENP (1:5,000), mouse anti-β-actin (AC-15; 1:5,000) (Sigma); mouse anti-Cdc25B (1:250), mouse anti-Aurora B kinase/AIM-1 (1:250), mouse anti-Kip1/p27 (1:3,000), mouse anti-Cip1/p21 (1:3,000), and mouse anti-human cyclin B1 antibody (GNS-11; 1:500) (BD Biosciences); mouse anti-GFP (JL-8; 1:1,000; Clontech, Franklin Lakes, NJ); rabbit anti-CENPA (1:200; Upstate), rabbit anti-Aurora A/AIK (1:1,000), and mouse antisurvivin (6E4; 1:500) (Cell Signaling Technology); and mouse anti-p45/Skp2 (2B12; 1:500) and rabbit anti-Cks1 (C-term; 1:500) (Zymed, South San Francisco, CA). The rabbit anti-Cullin 4A (Cul4A) antibody (1:1,000) was a gift from P. Raychaudhuri (University of Illinois at Chicago).
Generating rabbit antisera specific to the human C-terminal FoxM1b protein region.
We cloned the human FoxM1b 365-748 amino acid protein into a His-tagged expression vector. The His-tagged FoxM1b 365-748 amino acid protein was expressed in Escherichia coli and affinity purified by nickel chromatography following the manufacturer's protocol (Invitrogen). To generate a rabbit FoxM1 antibody, we provided Genemed Synthesis, Inc. (South San Francisco, CA) with affinity-purified His-tagged FoxM1b 365-748 amino acid protein as an antigen to immunize two rabbits, and the subsequent antibody production consisted of initial immunization followed by six boosts with the His-tagged FoxM1b 365-748 antigen. For Western blot analysis we used the Genemed-generated rabbit anti-FoxM1 antibody at a 1:5,000 dilution using procedures described above.
Procedure for immunofluorescent staining of U2OS cells and MEFs.
U2OS C3 cells or MEFs were fixed with 10% buffered formalin (Fisher) for 20 min at room temperature, rinsed with PBS, and permeabilized with PBS supplemented with 1% bovine serum albumin (BSA; Sigma) and 0.2% Triton X-100 (Fisher). After washing in PBS with 1% BSA, proteins of interest were visualized by staining cells with specific antibodies in PBS containing 0.5% BSA at 25°C for 16 h. The following commercially available antibodies and dilutions were used for immunofluorescent staining: mouse anti-Aurora B kinase/AIM-1 (1:25), mouse anti-p27Kip1 (1:200), and mouse anti-p21Cip1 (1:200) (BD Transduction Laboratories); rabbit anti-CENPA (1:75) and rabbit anti-phospho-histone H3 (Ser10; 1:100) (Upstate); mouse anti-α-tubulin (1:1,000; Sigma); anti-P16Ink4A (1:100; Santa Cruz); and anti-p19ARF (1:100; Abcam). After being washed with PBS, cells were incubated with tetramethyl rhodamine isocyanate-conjugated polyclonal anti-mouse immunoglobulins (1:100) or fluorescein isothiocyanate-conjugated polyclonal anti-mouse immunoglobulins (1:100; DakoCytomation, Denmark) or Texas Red-conjugated anti-mouse IgG antibody (1:150; Vector Laboratories) in PBS containing 0.5% BSA at 25°C for 30 min. The slides were washed with PBS, and coverglasses were mounted with Vectashield mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI; H-1200; Vector Laboratories). Immunofluorescence with primary antibodies followed by secondary antibodies conjugated to either tetramethyl rhodamine isocyanate or fluorescein isothiocyanate or Texas Red was detected using an Axioplan 2 microscope (Carl Zeiss).
Procedure for senescence-associated β-galactosidase staining of MEFs.
In situ SA-β-Gal activity was detected as described elsewhere (55, 70) with minor modifications. Passage 3 Foxm1+/+ (WT), Foxm1+/−, or Foxm1−/− MEFs were washed with PBS and fixed with 2% formaldehyde, 0.2% glutaraldehyde in PBS for 15 min at room temperature, washed twice with PBS, and stained for 16 h at 37°C with 1 mg/ml 5-bromo-4-chloro- 3-indolyl-β-galactoside (X-Gal) (pH 6.0) in 40 mM citric acid-sodium phosphate buffer containing 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM MgCl2, and 150 mM NaCl. Micrographs of β-galactosidase-stained MEFs were taken at 200× magnification using an Axioplan 2 microscope (Carl Zeiss).
Primers used for real-time reverse transcriptase PCR (RT-PCR) to determine mRNA expression levels.
U2OS cells or MEFs were harvested at 48 h following siRNA transfection for preparation of total RNA using RNA-STAT-60 (Tel-Test B Inc., Friendswood, TX). Following DNase I (RNase free; New England BioLabs) digestion of total RNA to remove contaminating genomic DNA, we used the Bio-Rad cDNA synthesis kit containing both oligo(dT) and random hexamer primers to synthesize cDNA from 10 μg of total RNA. The following reaction mixture was used for all PCR samples: 1× IQ SybrGreen supermix (Bio-Rad, Carlsbad, CA), 100 to 200 nM of each primer, and 2.5 μl of cDNA in a 25-μl total volume. Reactions were amplified and analyzed in triplicate using a MyiQ single-color real-time PCR detection system (Bio-Rad, Carlsbad, CA).
The following sense (S) and antisense (AS) primer sequences and annealing temperatures (Ta) were used to amplify and measure the amount of human mRNA by real-time RT-PCR: FoxM1-S, 5′-GGA GGA AAT GCC ACA CTT AGC G-3′, and FoxM1-AS, 5′-TAG GAC TTC TTG GGT CTT GGG GTG-3′ (Ta, 55.7°C); CENPA-S, 5′-CTT CCT CCC ATC AAC ACA GTC G-3′, and CENPA-AS, 5′-TGC TTC TGC TGC CTC TTG TAG G-3′ (Ta, 54°C); survivin-S, 5′-TCA AGG ACC ACC GCA TCT CTA-3′, and survivin-AS, 5′-TGA AGC AGA AGA AAC ACT GGG C-3′ (Ta, 61°C); CENPB-S, 5′-ATT CAG ACA GTG AGG AAG AGG ACG-3′, and CENPB-AS, 5′-CAT CAA TGG GGA AGG AGG TCA G-3′ (Ta, 58°C); Skp2-S, 5′-GGT GTT TGT AAG AGG TGG TAT CGC-3′, and Skp2-AS, 5′-CAC GAA AAGGGC TGA AATGTT C-3′ (Ta, 62°C); Cks1-S, 5′-GAA TGG AGG AAT CTT GGC GTT C-3′, and Cks1-AS, 5′-TCT TTG GTT TCT TGG GTA GTG GG-3′ (Ta, 55.7°C); KPC1-S, 5′-CCT GAA CCG CAT CTT TTC CTC-3′, and KPC1-AS, 5′-CTC ATT GTC CAC CTG TAG CAA CTG-3′ (Ta, 62.0°C); KPC2-S, 5′-GCC GTA TTT TTA GCA TCT GAC AGG-3′, and KPC2-AS, 5′-TTC AAG CAG CAA CTC AAG AGC C-3′ (Ta, 60.1°C); JAB1-S, 5′-ATC GGG AGG CAA CTT GGA AG-3′, and JAB1-AS, 5′-GCA TTT ACT CGG GTT TCA GTG-3′ (Ta, 61.3°C). These real-time RT-PCR RNA levels were normalized to human cyclophilin mRNA levels, and these primers are as follows: cyclophilin-S, 5′-GCA GAC AAG GTC CCA AAG ACA G-3′, and cyclophilin-AS, 5′-CAC CCT GAC ACA TAA ACC CTG G-3′ (Ta, 55.7°C).
The following sense and antisense primer sequences and annealing temperatures were used to amplify and measure the amount of mouse mRNA by real-time RT-PCR: FoxM1-S, 5′-CAC TTG GAT TGA GGA CCA CTT −3′, and FoxM1-AS, 5′-GTC GTT TCT GCT GTG ATT CC-3′ (Ta, 57.5°C); CENPA-S, 5′-AGC CGT GGT GTG TTT T-3′, and CENPA-AS, 5′-TCG GAT TCT CCT GGT CAA CT-3′ (Ta, 57.5°C); Cdc25B-S, 5′-CCC TTC CCT GTT TTC CTT TC-3′, and Cdc25B-AS, 5′-ACA CAC ACT CCT GCC ATA GG-3′ (Ta, 61.7°C); Skp2-S, 5′-GTA TGT TAG GGA ACC ATT TGC GAG-3′, and Skp2-AS, 5′-TTA GAA GGG CAC TTG GAA GAG TT-3′ (Ta, 55.7°C); Cks1-S, 5′-GAC CTC AAA GCC CTC GTG T-3′, and Cks1-AS, 5′-TGA AAC ATA AAT CCA TAA GTC ATC A-3′ (Ta, 55°C); p19ARF-S, 5′-AGA GCG GGG ACA TCA AGA C-3′, and p19ARF-AS, 5′-CTG AGG CCG GAT TTA GCT C-3 (Ta, 58.7°C). These real-time RT-PCR RNA levels were normalized to mouse cyclophilin mRNA levels, and these primers were as follows: cyclophilin-S, 5′-GGC AAA TGC TGG ACC AAA CAC-3′, and cyclophilin-AS, 5′-TTC CTG GAC CCA AAA CGC TC-3′ (Ta, 57.5°C).
Cdk2 and Cdk1 immunoprecipitation kinase reactions.
To measure the amount of Cdk2-cyclin E/A or Cdk1-cyclin B activity, FoxM1-depleted or untreated U2OS cells were lysed in NP-40 lysis buffer (42), and 500 μg of total cellular protein extract was immunoprecipitated (IP) using a monoclonal antibody for Cdk2 or Cdk1 (Santa Cruz) for 16 h with gentle rocking at 4°C. The antibody-antigen complex was IP with 25% protein A-Sepharose for 2 h at 4°C. Immunoprecipitates were then washed twice in lysis buffer followed by an additional two washes in ADBI kinase assay buffer (42). We used the Cdk2 or Cdk1 IP protein complexes for kinase reactions with 10 μg of the Cdk2 substrate RB amino acids 773 to 928 fusion protein or Cdk1 substrate histone H1 protein (Upstate) and radioactively labeled with [γ-32P]ATP. Kinase reactions were performed for 30 min at 30°C followed by 15% SDS-PAGE. The gel was then fixed in 10% methanol and 10% acetic acid for 4 h, during which we changed the wash three times. The fixed SDS-PAGE gel was dried for 45 min, the radioactively labeled RB or histone H1 protein was visualized by scanning with the Storm 860 PhosphorImager, and phosphorylated bands were quantitated with the ImageQuant program (Amersham-Pharmacia Biotech).
Transfection of Aurora B kinase promoter-luciferase constructs into U2OS cells for dual luciferase assays.
We used PCR of human U2OS genomic DNA to amplify −749 bp of the human Aurora B promoter region. This PCR-amplified promoter region was cloned in the correct orientation in the pGL3-Basic Luciferase reporter plasmid (Promega). The following PCR primers were used to amplify the Aurora B kinase promoter region: forward, 5′-CGGAATACAAAGTCTAGA-3′; reverse, 5′-GTCCAAGGCACTGCTACT-3′. The Aurora B kinase promoter region was confirmed by DNA sequencing (University of Chicago Sequencing Facility).
We used Fugene 6 reagent (Roche) to transfect U2OS cells with 200 ng of either cytomegalovirus (CMV) FoxM1B expression construct or CMV empty vector with 1.5 μg of −749 bp Aurora B promoter luciferase reporter with 10 ng of CMV-Renilla luciferase, which served as an internal control. Twenty-four hours after transfection, cells were harvested and protein extracts were prepared for dual luciferase assays (Promega) as described previously, and luciferase levels were normalized to Renilla luciferase activity (42). Promoter expression was expressed as the fold induction of transcriptional activity by the FoxM1b expression vector ± the SD, where promoter activity resulting from transfection with CMV empty vector was set at 1. Experiments were performed in triplicate, and statistical analysis was performed with Microsoft Excel tools.
ChIP assay.
FoxM1-depleted or untreated U2OS cells were processed for ChIP assay 3 days after siRNA transfection using published methods with additional modifications (71). Briefly, FoxM1-depleted or untreated U2OS cells were cross-linked in situ by addition of 37% formaldehyde (Fisher Scientific) to a final concentration of 1% (wt/vol) and incubated at 25°C for 10 min with gentle swirling. The cross-linking reaction was stopped by the addition of 2.5 M glycine to a final concentration of 0.125 M followed by an additional 5 min of gentle swirling. Cells were washed once with 4°C sterile PBS and then collected by adding 1 ml of 4°C sterile PBS containing protease inhibitors (Roche, Mannheim, Germany). Cells were scraped from the dish with a razor blade and transferred into an Eppendorf tube, which was centrifuged at 2,000 × g for 10 min. The cell pellet was then resuspended in a 2× pellet volume of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1) and placed on ice for 10 min.
The resulting extract was sonicated using a Misonix 600W sonicator (Misonix Inc., Farmingdale, NY) fitted with a 3-mm stepped microtip for 10 pulses of 15 seconds at a power setting of 30%. Between each pulse, the extract was incubated on ice for 1 min. At this stage, the processing of all experimental samples and total input was carried out according to the Upstate Cell ChIP assay protocol (catalog no. 17-295; Lake Placid, NY). For the immunoprecipitation, specific amounts of antibody as indicated were added to the precleared and clarified sample, which was incubated at 4°C with rotation for 12 to 16 h and washed according to the Upstate ChIP assay protocol. The following antibodies were used in the indicated amounts: 10, 25, or 50 μl of rabbit antiserum specific for FoxM1 protein (amino acids 365 to 748), 2 μg of rabbit serum (Vector Laboratories), and 2 μg of either rabbit CBP antibody (sc-369 [A-22]; Santa Cruz Biotechnology) or RNA polymerase II antibody (sc-899 [N-20]; Santa Cruz Biotechnology, Santa Cruz, CA). Cross-links were reversed on all samples, including 20% input, by addition of 100 μl TE (1 mM EDTA, 10 mM Tris-HCl, pH 7.4) containing 10 μg of RNase A and then incubated for 15 min at 25°C. Proteinase K (10 μg) and NaCl (4 μl of 5 M solution) were then added, and samples were digested for 16 h at 65°C. DNA was extracted from the digested samples using PCR purification columns following the manufacturer's instructions (QIAGEN, Maryland). We then used 2.5 μl of this ChIP DNA sample in the subsequent 25-μl real-time PCR mixture. The total input sample was diluted 1:10, and 2.5 μl was used for real-time PCR (10% total input).
PCR primers and reaction conditions for ChIP assay.
The primers used to amplify the following human gene promoter fragments are annotated with the binding position upstream of the transcription start site, annealing temperature, and whether in the sense or antisense orientation: Cdc25B −92S, 5′-AAG AGC CCA TCA GTT CCG CTT G-3′, and Cdc25B +120AS, 5′-CCC ATT TTA CAG ACC TGG ACG C-3′ (Ta, 62°C); Aurora B kinase (AurkB) −856S, 5′-GCA ACG AAA GGT CTA TTG GTG G-3′, and AurkB −611AS, 5′-TCT AAC TTC TCT GCC CGA TGG AG-3′ (Ta, 58°C); survivin −1531S, 5′-GGA GGA AGA AGC AGA GAG TGA ATG-3′, and survivin −1373AS, 5′-CTG GGA TTA CAG ATG TGA GCC AC-3′ (Ta, 65°C); CENPA −6695S, 5′-CCT TGG TGT TAT GCT CTG GGA AG-3′, and CENPA −6607AS, 5′-GGG CTG TTA CTG TTT TCT CAG GTT G-3′ (Ta, 60°C); CENPB −1036S, 5′-CCC AGA AAG TGA CAC ACC AAG ATG-3′, and CENPB −817AS, 5′-GGT AAG TAT CAT TGC CAA CAC GC-3′ (Ta, 60°C); Skp2 −7554S, 5′-AAT TAG CCA GGT GTG GTA GCG G-3′, and Skp2 −7428AS, 5′-CAG GCT TCA GTG TAA TGG CAC G-3′ (Ta, 62°C); Cks1 −213S, 5′-GTG AGA ACT GCC CTC CAA TAA GG-3′, and Cks1 −64AS, 5′-GTG AGA ACT GCC CTC CAA TAA GG-3′ (Ta, 64°C); transthyretin (TTR) −308S, 5′-TTA GTG CAC GCA GTC ACA CA-3′, and TTR −54AS, 5′-GCT TAT CCC TGC CAA TCT GAC TG-3′ (Ta, 62°C). The following reaction mixture was used for all PCR samples: 1× IQ SybrGreen Supermix (Bio-Rad, Carlsbad, CA), 100 nM of each primer, and 2.5 μl of each purified ChIP extract in a 25-μl total volume. Reactions were amplified and analyzed in triplicate using a MyiQ single-color real-time PCR detection system (Bio-Rad, Carlsbad, CA). Normalization was carried out using the ΔΔCt method. Briefly, IP samples and total input threshold cycles (Ct) for each treatment were subtracted from the Ct of the corresponding serum control IP (rabbit serum). The resulting corrected value for the total input was then subtracted from the corrected experimental IP value (ΔΔCt), and these values were raised to the power of 2 (2ΔΔCt). These values were then expressed as a relative promoter binding ± SD.
Statistical analysis.
We used the Microsoft Excel program to calculate the SD and statistically significant differences between samples using the Student t test. The asterisks in each graph indicate statistically significant changes, with P values calculated by the Student t test. P values of <0.05 were considered statistically significant.
RESULTS
Transfection of FoxM1 siRNA into U2OS cells effectively diminishes expression of induced GFP-FoxM1b and endogenous FoxM1 and Cdc25B proteins.
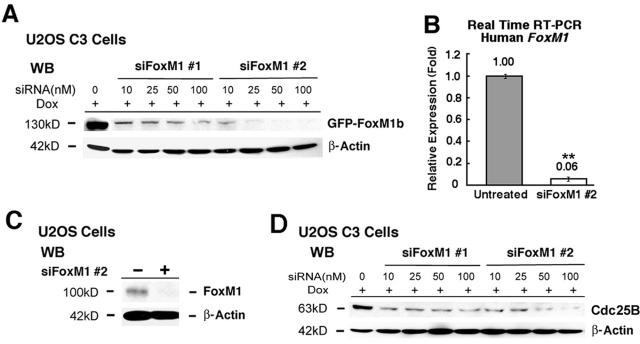
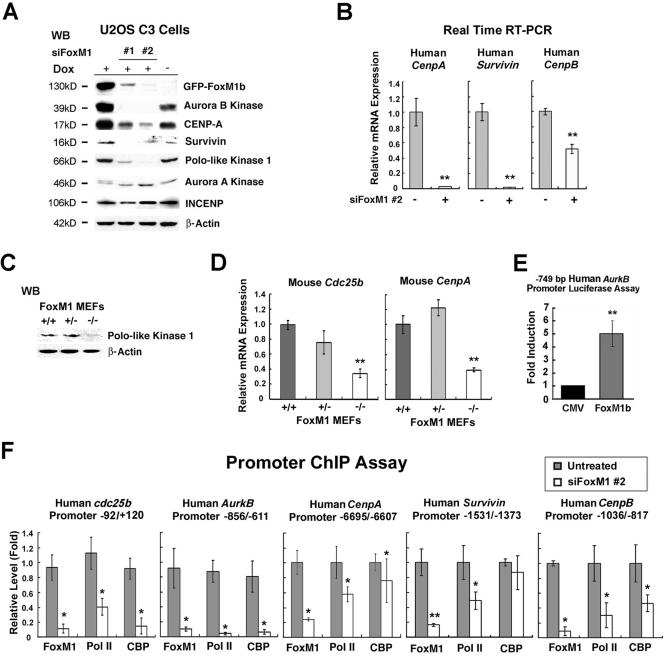
We previously reported on the generation of an osteosarcoma U2OS clone C3 cell line (U2OS C3 cells) which could be induced for expression of the GFP-FoxM1b protein by Dox treatment (27). In order to inhibit expression of the human FoxM1 protein in this cancer cell line, two 21-nucleotide siRNA duplexes were synthesized to target the human FoxM1 mRNA (designated siFoxM1 #1 and #2). We first performed a dose-response curve by transfecting increasing amounts of siFoxM1 #1 or #2 duplex into Dox-induced U2OS C3 cells, preparing protein extracts 72 h after transfection and then assessing for GFP-FoxM1b protein levels by Western blot analysis with anti-GFP monoclonal antibody (Fig. 1A). Transfection of siFoxM1 #1 or siFoxM1 #2 duplex diminished expression of induced GFP-FoxM1b protein in a dose-dependent manner, and siFoxM1 #2 was more effective in abolishing expression of GFP-FoxM1b protein than the siFoxM1 #1 duplex (Fig. 1A). Transfection of U2OS cells with siFoxM1 #2 duplex inhibited expression of endogenous FoxM1 mRNA (Fig. 1B) and FoxM1 protein (Fig. 1C) as determined by quantitative real-time RT-PCR (QRT-PCR) and Western blot analysis, respectively. Moreover, transfection of Dox-induced U2OS C3 cells with increasing amounts of siFoxM1 #1 or #2 duplexes demonstrated that they reduced expression of Cdc25B protein, a known FoxM1 target gene (36, 67), in a dose-dependent manner (Fig. 1D).
FIG. 1.
Transfection of FoxM1 siRNA into U2OS cells effectively diminishes expression of induced GFP-FoxM1b and endogenous FoxM1 and Cdc25B proteins. (A) Dose response; FoxM1 siRNA transfections into U2OS C3 cells effectively diminish expression of induced GFP-FoxM1b protein. We transfected increasing amounts of siFoxM1 #1 or #2 into U2OS C3 cells, which were then induced for GFP-FoxM1b expression by Dox treatment. Protein extracts were prepared 72 h after transfection and then analyzed for GFP-FoxM1b protein levels by Western blot analysis using an anti-GFP monoclonal antibody. (B and C) Diminished expression of endogenous FoxM1 mRNA (B) (**, P = 0.003) or FoxM1 protein (C) in U2OS cells transfected with siFoxM1 #2 duplex as determined by QRT-PCR and Western blot analysis with a rabbit anti-FoxM1 serum, respectively (see Materials and Methods). (D) Dose response for FoxM1 siRNA transfections into Dox-induced U2OS C3 cells, which effectively diminishes expression of endogenous Cdc25B protein.
FoxM1 is required for growth of U2OS cells and MEFs in culture.
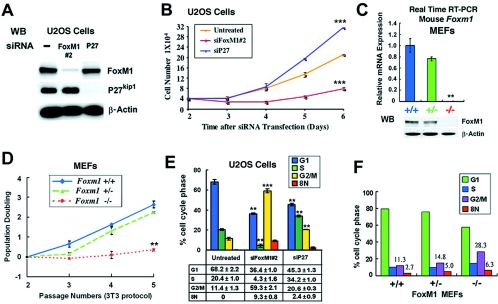
To determine the growth rate of FoxM1-depleted cells, U2OS cells were transfected with siFoxM1 #2 duplex and then incubated for 2 days to allow siRNA depletion of FoxM1 expression. These U2OS cells were then trypsinized, and the cell growth rate was determined in triplicate at 3, 4, 5, or 6 days after siRNA transfection (Fig. 2B). As a control for siRNA silencing of an unrelated gene, we also transfected U2OS cells with siRNA specific to the p27Kip1 gene and examined growth of the p27Kip1-depleted U2OS cells as described above. Western blot analysis showed that U2OS cells transfected with siFoxM1 #2 or siRNA p27Kip1 duplexes only inhibit their own endogenous protein expression (Fig. 2A). U2OS cells depleted of FoxM1 failed to display significant growth in culture compared to untransfected U2OS cells (Fig. 2B). However, the FoxM1-depleted U2OS cells began to show a slight increase in growth rate at 5 and 6 days after transfection with siFoxM1 #2 duplex, when the siRNA silencing of FoxM1 expression was beginning to wane (Fig. 2B). U2OS cells depleted for p27Kip1 displayed an increased growth rate compared to untreated controls (Fig. 2B), indicating that the reduction in growth of FoxM1-depleted cells was not due to nonspecific effects resulting from siRNA gene silencing. These results demonstrated that expression of the FoxM1 transcription factor is essential for growth of U2OS cells.
FIG. 2.
FoxM1-deficient cells fail to exhibit significant growth in culture and display reduced mitotic progression with accumulation of polyploid cells. (A and B) FoxM1-depleted U2OS cells fail to exhibit significant growth in culture. U2OS cells were transfected with siFoxM1 #2 or siRNA p27Kip1 duplexes and then incubated for 2 days to allow siRNA silencing of their expression; cells were then trypsinized, 4 × 104 cells were plated in each culture dish (in triplicate), and the number of cells were then determined at 3, 4, 5, or 6 days after siRNA transfection. (A) Western blot analysis shows that U2OS cells transfected with siFoxM1 #2 or si-p27Kip1 duplexes only inhibit their own endogenous expression. (B) Graphic representation of mean number of cells at indicated days following transfection ± SD. ***, P = 0.001. (C) Foxm1−/− MEFs do not express detectable levels of Foxm1 mRNA or protein. MEFs containing WT Foxm1+/+, Foxm1+/−, or Foxm1−/− genotypes were used to prepare total RNA or protein at passage 4 in culture. Foxm1−/− MEFs do not express detectable levels of the FoxM1 mRNA or FoxM1 protein as determined by quantitative real-time RT-PCR (**, P = 0.004) and Western blot analysis, compared to WT (+/+) or Foxm1+/− MEFs. (D) Foxm1−/− MEFs are unable to grow in culture. We used the 3T3 protocol to measure cell doubling rates of the MEFs at each passage by plating 3 × 105 of either Foxm1+/+, Foxm1+/−, or Foxm1−/− MEFs in 60-mm plates (in triplicate) and then counted the number of cells after 3 days of growth to determine population doubling ± SD as described in Materials and Methods. **, P = 0.006. (E) Flow cytometry analysis of FoxM1-depleted U2OS cells shows decreased S-phase and accumulation of G2/M (4N) and polyploid (8N) cells. U2OS cells were transfected as described for panel A and then subjected to flow cytometry analysis. Graphically shown is the statistically significant change in the percentage of either FoxM1- or p27Kip1-depleted U2OS cells accumulating in G1, S, G2/M (4N), and 8N compared to untransfected U2OS cells ± SD from triplicate plates. The asterisks in panels B to E indicate statistically significant changes, with the following P values calculated by the Student t test: *, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (F) Flow cytometry analysis of passage 4 Foxm1+/+, Foxm1+/−, or Foxm1−/− MEFs.
In order to confirm this growth suppression observed with FoxM1-depleted U2OS cells, we examined the growth properties of Foxm1-deficient MEFs. These Foxm1−/− MEFs contained a functionally inactive Foxm1-targeted allele that deleted the essential Foxm1 exons 4 through 7 encoding both the DNA binding and C-terminal transcriptional activation domains (36, 42). These Foxm1−/− MEFs do not express detectable levels of the FoxM1 mRNA or FoxM1 protein compared to WT Foxm1+/+ or Foxm1+/− MEFs as determined by QRT-PCR and Western blot analysis (Fig. 2C). We used the 3T3 protocol to measure cell doubling rate at each passage by plating 3 × 105 of either Foxm1+/+, Foxm1+/−, or Foxm1−/− MEFs on 60-mm plates (in triplicate) and then counting the number of cells after 3 days of growth. We found that passage 3 Foxm1+/− MEFs initially showed diminished plating efficiency, but they exhibited growth rates similar to WT MEFs thereafter (Fig. 2D). In contrast, Foxm1−/− MEFs failed to exhibit significant growth in culture after passage 2 compared to WT and Foxm1+/− MEFs (Fig. 2D), demonstrating that FoxM1 expression is essential for MEFs to grow in culture.
FoxM1-depleted U2OS cells exhibited diminished mitotic progression with accumulation of polyploid cells.
To demonstrate that FoxM1-depleted U2OS cells exhibited diminished cell cycle progression, U2OS cells were transfected with either siFoxM1 #2 or si-p27Kip1 duplexes or left untransfected, and then 72 h after transfection the cells were subjected to flow cytometry analysis. Suppression of FoxM1 levels in U2OS cells caused an 80% reduction in S-phase cells and significant decreases in mitotic progression as evidenced by a fivefold increase in G2/M-phase cells (4N DNA content) with an accumulation of polyploid (8N) cells compared to untreated U2OS cells (Fig. 2E, siFoxM1 #2). In contrast, U2OS cells depleted for p27Kip1 levels exhibited only a slight accumulation of polyploid cells and increased the number of cells entering both S-phase and G2/M-phase transition (Fig. 2E, siP27). Furthermore, no detectable sub-G1 peak was observed in U2OS cells transfected with siFoxM1 #2 duplex, indicating that this siRNA transfection did not induce apoptosis (data not shown). These findings suggest that depletion of FoxM1 in U2OS cells caused significant decreases in S-phase and mitotic progression with accumulation of polyploid cells. We next identified the cell cycle profile of Foxm1+/+, Foxm1+/−, and Foxm1−/− MEFs harvested at passage 4 by flow cytometry (Fig. 2F). Foxm1−/− MEFs exhibited defects in mitotic progression as determined by a twofold increase in accumulation of G2/M (4N) cells with a slight increase in polyploid (8N) cells compared to control MEFs (Fig. 2F). The difference in the accumulation of G2/M cells between U2OS cells and MEFs most likely reflects differences in cell cycle progression between a transformed cell (U2OS) and primary cell (MEFs). Taken together, these results suggest that Foxm1-depleted U2OS cells and Foxm1−/− MEFs display defects in mitotic progression.
FoxM1-deficient cells exhibited reduced PH3 staining and failed to progress beyond the prophase stage of mitosis.
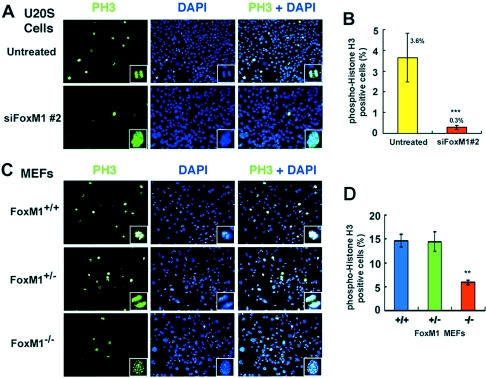
We used immunofluorescent staining for mitotic-specific phosphorylation of histone H3 (PH3) to determine whether FoxM1-depleted U2OS cells or Foxm1−/− MEFs exhibited diminished mitotic progression. U2OS cells depleted in FoxM1 expression or early-passage Foxm1−/−, Foxm1+/−, or WT MEFs were fixed and immunofluorescently stained with the PH3 antibody, and nuclei were counterstained with DAPI. This analysis determined that both FoxM1-depleted U2OS cells and Foxm1−/− MEFs failed to progress beyond the prophase stage of mitosis, while all stages of mitosis were visible in untransfected U2OS cells or control MEFs (Fig. 3A and C; see magnified image in lower righthand corners). Furthermore, U2OS cells with suppressed FoxM1 levels or early-passage Foxm1−/− MEFs displayed a significant reduction in PH3-positive cells compared to controls (Fig. 3B and D), suggesting that FoxM1-deficient cells were unable to efficiently enter mitosis. Taken together, these studies indicated that FoxM1-deficient cells exhibited reduced mitotic entry and were inhibited in their progression through mitosis.
FIG. 3.
FoxM1-depleted U2OS cells and Foxm1−/− MEFs show significant decreases in mitotic phospho-histone H3 staining and fail to progress beyond prophase. Results show reduced mitotic progression in FoxM1-depleted U2OS cells (A) or Foxm1−/− MEFs (C) as evidenced by diminished phospho-histone H3 (PH3) staining. Control or FoxM1-depleted U2OS cells or Foxm1+/+, Foxm1+/−, or Foxm1−/− MEFs were immunofluorescently stained with the PH3 antibody. FoxM1-depleted U2OS cells (A, siFoxM1 #2) or Foxm1−/− MEFs (C) do not progress beyond the prophase stage of mitosis as evidenced by PH3 and DAPI staining. Shown are micrographs taken at 100× magnification, and the enlargement of a cell undergoing mitosis in the righthand corner box was taken from micrographs taken at 400× magnification. (B and D) The PH3-positive cells were quantitated in FoxM1-depleted and untransfected U2OS cells (B) and in Foxm1+/+, Foxm1+/−, and Foxm1−/− MEFs (D) and are shown graphically as the percent PH3-positive cells ± SD. This analysis revealed a statistically significant decrease in percent PH3-positive cells in FoxM1-depleted U2OS cells compared to control cells (B) (***, P = 0.00002) and a statistically significant reduction in the percent PH3-positive cells in Foxm1−/− MEFs compared to control MEFs (D) (**, P = 0.003).
Early-passage Foxm1−/− MEFs displayed premature senescence as evidenced by increased expression of senescence-associated markers.
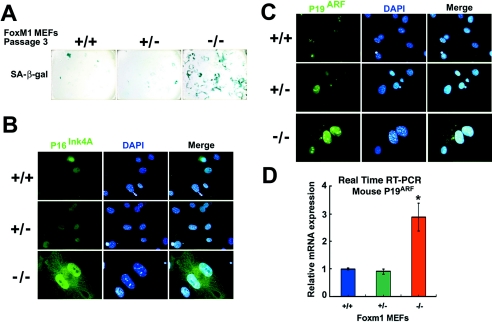
Because early-passage Foxm1−/− MEFs failed to grow in culture and progress through mitosis, we next sought to determine whether they had undergone premature cellular senescence by staining for expression of the senescence-associated β-galactosidase, p16INK4A, and p19ARF proteins (53-55, 70). We fixed passage 3 Foxm1+/+, Foxm1+/−, or Foxm1−/− MEFs and used X-Gal substrate to stain them for senescence-associated β-galactosidase enzyme activity or performed immunofluorescent staining with antibodies specific to the senescence marker p16INK4A and p19ARF proteins. Indicative of premature senescence, we found that early-passage Foxm1−/− MEFs exhibited strong positive staining for the SA-β-Gal enzyme, whereas only a few cells stained positive for the SA-β-Gal enzyme in Foxm1+/− or WT MEF controls (Fig. 4A). In addition, early-passage Foxm1−/− MEFs exhibited enlarged nuclei with high levels of nuclear p16INK4A staining compared to Foxm1+/− or WT MEF controls (Fig. 4B). Likewise, Foxm1−/− MEFs exhibited strong nuclear and nucleolar staining for the p19ARF tumor suppressor protein compared to low nuclear/nucleolar expression levels in a subset of Foxm1+/− or WT MEF controls (Fig. 4C). Furthermore, QRT-PCR analysis of mRNA demonstrated that Foxm1−/− MEFs exhibited a significant threefold increase in expression of p19ARF mRNA compare to that of Foxm1+/− or WT MEF controls (Fig. 4D). These results suggest that early-passage Foxm1−/− MEFs have undergone premature cellular senescence as evidenced by high expression of senescence marker proteins. Recent studies report that MEFs cultured in vitro, under atmospheric oxygen concentrations, show rapid “cellular senescence,” and passage 3 MEFs exhibit an increase in mutations and undergo immortalization (8, 25, 72). An alternative interpretation of our increased SA-β-Gal staining and expression of senescence marker proteins in early-passage Foxm1−/− MEFs is that they fail to undergo immortalization at passage 3 compared to WT and Foxm1+/− MEFs.
FIG. 4.
Early-passage Foxm1−/− MEFs express high levels of senescence-associated β-galactosidase, p16INK4A, and p19ARF proteins. Passage 3 Foxm1−/− MEFs express high levels of senescence-associated β-galactosidase enzyme (A) and increased nuclear levels of CDKI p16INK4A (B) and p19ARF tumor suppressor (C) proteins. Passage 3 Foxm1+/+ (WT), Foxm1+/−, or Foxm1−/− MEFs were stained for senescence-associated β-galactosidase enzyme activity with X-Gal substrate (55, 70) or immunofluorescently stained with antibody specific to either p16INK4A or p19ARF protein. In panels B and C, the MEF nuclei were counterstained with DAPI and merged with the immunofluorescent staining. β-Galactosidase enzyme and immunofluorescent staining micrographs are taken at 200× and 400× magnification, respectively. (D) Foxm1−/− MEFs express significantly higher levels of p19ARF mRNA compared to control WT and Foxm1+/− MEFs. RNAs isolated from Foxm1−/−, Foxm1+/−, and WT MEFs were analyzed for p19ARF mRNA levels by quantitative real-time RT-PCR analysis using primers specific to the mouse p19ARF gene, and Foxm1−/− MEFs displayed a statistically significant increase in p19ARF mRNA expression compared to control MEFs (*, P = 0.02).
FoxM1 is essential for expression of the mitotic regulators Aurora B kinase, survivin Polo-like kinase 1, CENPA, and CENPB.
The mitotic defect found in FoxM1-depleted U2OS cells and Foxm1−/− MEFs (Fig. 2 to 4) indicated that FoxM1 regulated transcription of essential mitotic regulatory genes. We therefore examined whether siRNA silencing of FoxM1 levels in U2OS cells could reduce expression of the mitotic regulators Aurora B kinase, Aurora A kinase, PLK1, survivin, INCENP, and CENPA. Dox-induced U2OS C3 cells were transfected with either siFoxM1 #1 or #2 duplexes or left untransfected, protein extracts were isolated 72 h after transfection, and Western blot analysis was performed to measure protein levels of these mitotic regulators (Fig. 5A). Western blot analysis demonstrated that FoxM1 siRNA transfection suppressed expression of the GFP-FoxM1b fusion and endogenous levels of the FoxM1 protein (Fig. 5A and data not shown).
FIG. 5.
FoxM1 regulates transcription of the mitotic regulators Aurora B kinase, survivin, CENPA, and CENPB. (A) FoxM1 is required for expression of mitotic regulatory proteins. U2OS C3 cells were transfected with 100 nM of FoxM1 siRNA #1 or #2 and induced for GFP-FoxM1b with Dox, and protein extracts were isolated 72 h after transfection. Western blot analysis was used to measure siFoxM1 suppression of induced GFP-FoxM1b protein and Aurora B kinase, CENPA, Plk1, survivin, Aurora A kinase, or INCENP. (B) FoxM1 regulates mRNA levels of CENPA, survivin, and CENPB. RNA was isolated from siFoxM1 #2-transfected or untransfected U2OS cells and used for QRT-PCR analysis with the indicated human gene primers. This analysis demonstrated that FoxM1-depleted U2OS cells exhibited a statistically significant decrease in mRNA expression of CENPA (**, P = 0.008), survivin (**, P = 0.004), and CENPB (**, P = 0.01). (C and D) Compared to control MEFs, Foxm1−/− MEFs expressed reduced levels of PLK1 protein as determined by Western blotting (C) and reduced mRNA levels of Cdc25B (**, P = 0.004) and CENPA (**, P = 0.007) as determined by quantitative real-time RT-PCR (D) analysis. (E) FoxM1b activates transcription of the Aurora B kinase promoter in cotransfection assays. U2OS cells were cotransfected with CMV-FoxM1b expression vector and the −749 bp Aurora B kinase (AurkB) promoter fused to the luciferase reporter and, 24 h following transfection, protein extracts were prepared and analyzed for dual luciferase activity as described previously (42). Triplicate plates were used to calculate the mean fold induction of transcriptional activity by CMV-FoxM1B transfection ± SD (**, P = 0.004). Promoter expression with CMV-empty vector transfection was set at 1. (F) FoxM1 regulates transcription of Cdc25B, Aurora B kinase, CENPA, survivin, and CENPB promoters as determined by quantitative ChIP assays. FoxM1-depleted or untreated U2OS cells were processed for ChIP assay as described in Materials and Methods. The cross-linked and sonicated human chromatin was IP with antibodies specific to FoxM1, CBP, RNA polymerase II, or rabbit serum (control), and the amount of promoter DNAs associated with the IP chromatin was quantitated by real-time PCR with primers specific to the indicated regions of the human Cdc25B, Aurora B kinase, CENPA, survivin, and CENPB promoter. FoxM1 ChIP promoter binding in untreated U2OS cells was set at 1 ± SD. FoxM1-depleted U2OS cells showed diminished binding of FoxM1, RNA polymerase II, or CBP coactivator protein to the endogenous human promoter regions of Cdc25b, AurkB, CENPA, survivin, and CENPB genes. The asterisks in panels B, D, E, and F indicate statistically significant changes, with P values calculated by the Student t test as follows: *, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. The IgG control ChIP assays produced 0.1 of the FoxM1 binding levels observed with untreated U2OS cells and similar to the FoxM1-depleted U2OS cells.
Dox induction of the GFP-FoxM1b protein caused increased protein expression of Aurora B kinase and CENPA compared to unstimulated U2OS C3 cells (Fig. 5A, compare data in the absence versus presence of Dox). Depletion of FoxM1 protein levels in Dox-induced U2OS C3 cells resulted in undetectable levels of the Aurora B kinase, PLK1, and survivin proteins and significant reduction in expression of CENPA protein compared to untransfected Dox-induced U2OS C3 cells (Fig. 5A). In contrast, suppression of FoxM1 levels was unable to inhibit expression of either Aurora A kinase or INCENP (Fig. 5A). U2OS cells depleted for FoxM1 showed significant reduction in mRNA expression of survivin and CENPA as determined by QRT-PCR analysis (Fig. 5B). FoxM1-depleted U2OS cells also exhibited a 50% reduction in CENPB mRNA levels compared to untransfected U2OS controls (Fig. 5B). Consistent with the FoxM1-deficient U2OS cells, Foxm1−/− MEFs exhibited undetectable expression of PLK1 protein as determined by Western blot analysis (Fig. 5C) and significant reduction in mRNA levels of Cdc25B and CENPA compared to WT or Foxm1+/− MEFs as determined by QRT-PCR (Fig. 5D). These studies indicate that the Foxm1 transcription factor is essential for expression of the mitotic regulatory proteins Aurora B kinase, survivin, PLK1, CENPA, and CENPB.
To determine whether FoxM1 regulates transcription of the Aurora B kinase promoter, the luciferase reporter gene was linked to the −749 bp Aurora B kinase promoter region, which contained two potential FoxM1 binding sites at −730 to −742 bp and −652 to −640 bp (73, 75). We performed cotransfection assays with the CMV-FoxM1b expression vector and the −749 bp Aurora B kinase promoter luciferase plasmid, prepared protein extracts from U2OS cells at 24 h following transfection, and used them to measure dual luciferase enzyme activity. Cotransfection of FoxM1b expression vector caused a fivefold increase in Aurora B kinase promoter activity (Fig. 5E), demonstrating that FoxM1b protein can transcriptionally activate this Aurora B kinase promoter region.
Cdc25B, Aurora B kinase, survivin, CENPA, and CENPB genes are direct transcriptional targets of FoxM1 as determined by quantitative ChIP assays.
Recent studies have used FoxM1 cotransfection and ChIP assays to demonstrate that FoxM1 regulates transcription of the PLK1 gene (37). We next used quantitative ChIP assays (71) to determine whether suppressing FoxM1 expression in U2OS cells by transfection of siFoxM1 #2 duplex prevented FoxM1 binding to the endogenous human Cdc25B, Aurora B kinase, survivin, CENPA, or CENPB promoter regions. The cross-linked and sonicated human chromatin was IP with antibodies specific to either FoxM1 or RNA polymerase II or rabbit serum (control), and the amount of promoter DNA associated with the IP chromatin was quantitated by QRT-PCR with primers specific to the human Cdc25B, Aurora B kinase, survivin, CENPA, or CENPB promoter regions. These ChIP PCR primers were made to DNA sequences situated near the potential FoxM1 binding sites in the human Aurora B kinase (−730 to −742 bp and −652 to −640 bp), CENPA (−6531 to −6512 bp), survivin (−1491 to −1476 bp), and CENPB (−552 to −537 bp) promoter regions. Because the FoxM1 protein stimulates transcription by recruiting the CBP transcriptional coactivator (42), we also performed ChIP analysis with the CBP antibody. This quantitative ChIP assay showed that FoxM1 binds to the endogenous human Cdc25B, Aurora B kinase, CENPA, and CENPB promoters, while depleting FoxM1 significantly reduced association of FoxM1 protein, CBP coactivator, and RNA polymerase II to these endogenous promoter regions compared to untransfected U2OS cells (Fig. 5F). These quantitative ChIP assays also revealed that FoxM1 binds to the endogenous human survivin promoter region and that U2OS cells with depleted FoxM1 levels exhibited a significant reduction in binding of FoxM1 protein and RNA polymerase II to the endogenous survivin promoter regions (Fig. 5F). However, suppression of FoxM1 levels did not diminish recruitment of CBP to the endogenous survivin promoter region (Fig. 5F), presumably due to another transcription factor that also recruits the CBP coactivator. We also performed control ChIP assays with cross-linked extracts prepared from U2OS cells and FoxM1 antibody or control mouse IgG serum, and the IP genomic DNA was analyzed for the presence of the liver-specific human TTR promoter region by QRT-PCR. Consistent with the specificity of our ChIP assays, this control ChIP experiment demonstrated that neither the FoxM1 antibody nor IgG serum immunoprecipitated significant levels of this proximal TTR promoter region from either untransfected or FoxM1-depleted U2OS cell extracts (data not shown). Furthermore, we transfected U2OS cells with siRNA specific to the p27Kip1 (siP27) gene and performed ChIP assays with FoxM1 antibody and primers specific to FoxM1 target promoters to demonstrate that this control siRNA transfection does not reduce binding of FoxM1 to the Cdc25B, Aurora B Kinase, survivin, or CENPA promoter regions (data not shown). Taken together, results from these quantitative ChIP and expression assays demonstrated that Cdc25B, Aurora B kinase, survivin, CENPA, and CENPB promoter regions are direct transcriptional targets of FoxM1.
Immunofluorescent staining of FoxM1-depleted U2OS cells for Aurora B kinase and CENPA expression.
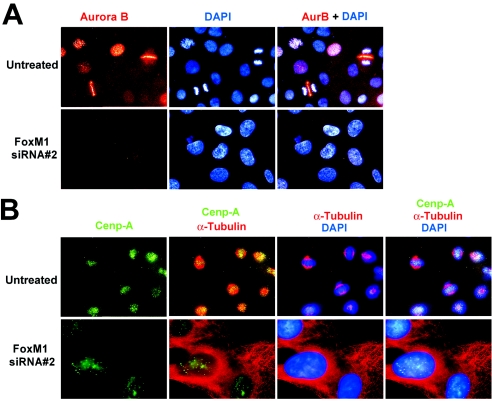
To determine whether the expression patterns of the Aurora B kinase or CENPA proteins were detectable in FoxM1-depleted U2OS cells, we performed immunofluorescent staining with antibodies specific to these proteins and counterstained nuclei with DAPI. Aurora B kinase staining is punctated in U2OS cells during prophase, consistent with its localization to the inner centromere region, while Aurora B kinase relocalizes to the spindle midzone region situated between the separating chromosomes during anaphase (Fig. 6A, untreated) (3, 31). Consistent with a block in mitotic progression, depletion of FoxM1 expression in U2OS cells completely eliminated detectable Aurora B kinase protein staining (Fig. 6A, siRNA #2). We next examined immunofluorescent staining of CENPA protein in FoxM1-depleted U2OS cells synchronized at the beginning of mitosis. To synchronize U2OS cells at early stages of mitosis, they were treated for 24 h with nocodazole, an inhibitor of spindle microtubule polymerization, and then released for 1 hour to allow progression into mitosis (28). The CENPA protein is a histone variant that replaces histone H3 protein in the nucleosomes of the centromere region and, therefore, its fluorescence tracks with the chromosomes in prophase (punctated) and metaphase (midzone), as shown in Fig. 6B (2, 5, 47). U2OS cells depleted in FoxM1 levels displayed reduced intensity of punctated CENPA staining and were unable to proceed into the metaphase stage of mitosis (Fig. 6B).
FIG. 6.
Diminished Aurora B kinase levels contribute to the polyploid phenotype found in FoxM1-deficient cells. (A) FoxM1-depleted U2OS cells exhibited undetectable immunofluorescent staining of Aurora B kinase. Nuclei were counterstained with DAPI and then merged with immunofluorescent staining. (B) FoxM1-depleted U2OS cells displayed reduced intensity of punctated CENPA staining. We examined immunofluorescent staining of CENPA protein in cells synchronized at the beginning of mitosis. To synchronize U2OS cells at early stages of mitosis, they were treated for 24 h with nocodazole, an inhibitor of spindle microtubule polymerization, and then released for 1 h to allow progression into mitosis (28). Synchronized cells were immunofluorescently stained with antibodies specific to either CENPA or α-tubulin, and nuclei were counterstained with DAPI and merged using the indicated combinations. Shown are 400× magnifications.
Diminished Aurora B kinase levels contributed to the polyploid phenotype found in FoxM1-deficient cells.
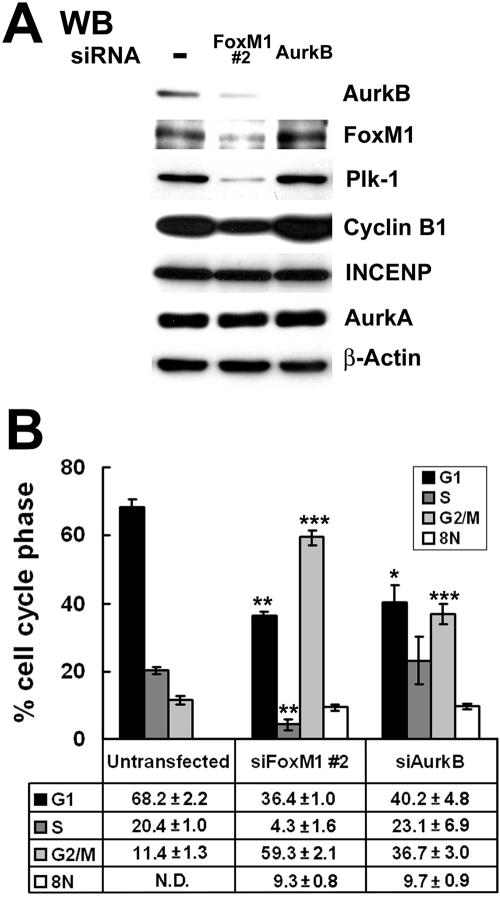
Aurora B kinase-depleted HeLa cells displayed significant reduction in PH3 staining and accumulation of polyploid cells due to endoreduplication (14), which are mitotic defects found in FoxM1-deficient U2OS cells (Fig. 2 and 3). We therefore examined whether reduced levels of Aurora B kinase in FoxM1-deficient U2OS cells were responsible for defects in G2/M progression and accumulation of polyploid cells. U2OS cells were transfected with siRNA duplexes specific to either the Aurora B kinase or FoxM1 or left untransfected, and then 72 h after transfection the cells were harvested to examine the cell cycle profile by flow cytometry. Western blot analysis demonstrated that expression of Aurora B kinase protein was significantly diminished by transfection with either the Aurora B kinase siRNA or siFoxM1 #2 duplex (Fig. 7A). Aurora B kinase-depleted U2OS cells exhibited wild-type expression of FoxM1, PLK1, cyclin B1, INCENP, and Aurora A kinase proteins (Fig. 7A). In contrast, FoxM1-depleted U2OS cells displayed significant reduction in levels of PLK1 and cyclin B1 proteins, but FoxM1 deficiency did not influence expression of the INCENP and Aurora A kinase proteins (Fig. 7A). Flow cytometry analysis demonstrated that Aurora B kinase-depleted U2OS cells exhibited a statistically significant threefold increase in G2/M (4N) cells, compared to a fivefold increase in G2/M phase cells with FoxM1-deficient U2OS cells (Fig. 7B) (G2/M, 36.7% ± 3.0% versus 59.3% ± 2.1%). This result indicates that Aurora B-deficient U2OS cells exhibit a less severe defect in mitotic progression than FoxM1-depleted cells, a finding consistent with the role of FoxM1 in regulating expression of other mitotic regulators, Cdc25B, PLK1, survivin, CENPA, and CENPB. In contrast to FoxM1-depleted U2OS cells, normal levels of S-phase cells were found in Aurora B kinase-deficient U2OS cells (Fig. 7B), a finding consistent with a restricted role of Aurora B Kinase in mitotic progression. Interestingly, both FoxM1- and Aurora B kinase-depleted U2OS cells exhibited identical accumulation of polyploid (8N) cells (Fig. 7B), suggesting that they have undergone endoreduplication. Consistent with these findings, published studies have demonstrated that inhibition of Aurora B kinase activity leads to a polyploid genotype resulting from a failure in the mitotic spindle checkpoint causing premature mitotic exit and reinitiation of DNA replication (28, 48, 50, 51). These studies support the hypothesis that diminished expression of Aurora B kinase contributed to the development of the polyploid genotype in FoxM1-depleted U2OS cells.
FIG. 7.
Aurora B kinase-depleted U2OS cells accumulate polyploid (8N) cells similar to that found in FoxM1-depleted U2OS cells. U2OS cells were either transfected with Aurora B kinase siRNA duplex, siFoxM1 #2 duplex, or left untransfected and 72 h after transfection cells were harvested for flow cytometry analysis (in triplicate) to determine cell cycle profile. (A) Western blot analysis demonstrated that expression of Aurora B kinase protein was significantly diminished by siRNA specific to either Aurora B kinase or FoxM1. We also performed Western blot analysis with antibodies specific to FoxM1, Plk1, cyclin B1, Aurora A kinase, and INCENP. (B) Aurora B kinase contributes to accumulation of polyploid (8N) cells in FoxM1-deficient cells. Depicted graphically is the percentage of cells accumulating in G1, S, G2/M (4N), and 8N in either FoxM1- or Aurora B kinase (AurkB)-depleted U2OS cells compared to untransfected U2OS cells ± SD from triplicate plates. FoxM1- or Aurora B kinase-depleted U2OS cells showed a statistically significant change in cells accumulating in G1, S, G2/M (4N), and 8N. The asterisks in panel B indicate statistically significant changes with P values calculated by the Student t test as follows: *, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
FoxM1 is required for normal levels of DNA replication in U2OS cells.
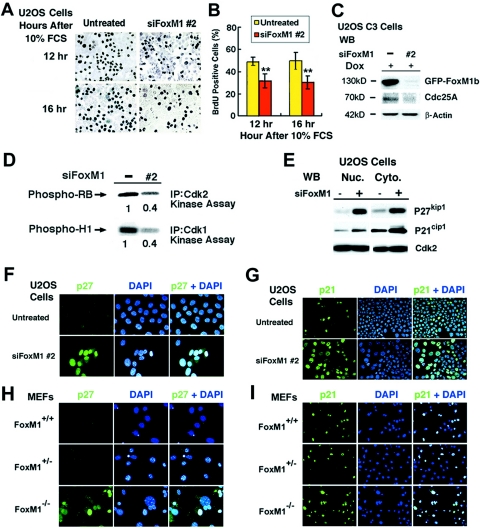
In order to determine whether inhibiting expression of FoxM1 influenced progression into DNA replication, U2OS cells were transfected with siFoxM1 #2 duplex or left untransfected, serum starved for 48 h, and then stimulated to reenter the cell cycle with the addition of 10% FCS. We subjected the cells to a 1-hour pulse-label with bromodeoxyuridine (BrdU) prior to harvesting them at 12 and 16 h after serum stimulation, which represents the period of DNA replication in U2OS cells (42). DNA replication rates in U2OS cells were determined by measuring the amount of BrdU incorporation using immunohistochemical staining (Fig. 8A). Quantitation of the BrdU incorporation rates demonstrated that depletion of FoxM1 expression in U2OS cells caused significant decreases in DNA replication compared to untransfected U2OS cells (Fig. 8B), suggesting that FoxM1 contributes to S-phase progression.
FIG. 8.
FoxM1-depleted U2OS cells and Foxm1−/− MEFs exhibit increased nuclear levels of CDKI proteins. (A) Diminished BrdU incorporation rates in serum-stimulated FoxM1-depleted U2OS cells. FoxM1-depleted or untreated U2OS cells were serum starved for 48 h and then stimulated to reenter the cell cycle with the addition of 10% fetal calf serum and cells at 12 and 16 h after serum stimulation and a 1-hour pulse-label with BrdU before harvesting the cells. (B) Graph quantitating BrdU incorporation rates in serum-stimulated FoxM1-depleted and control U2OS cells. We counted the number of BrdU-positive cells from three distinct 200× fields at 12 and 16 h after serum stimulation (in triplicate), and this was used to calculate the percentage of cells with BrdU incorporation ± the SD as shown graphically. Statistically significant decreases in the percentage of BrdU incorporation were found in serum-stimulated FoxM1-depleted U2OS as determined by the Student t test (**, P < 0.01). (C) FoxM1-depleted U2OS cells displayed reduced levels of Cdc25A phosphatase protein as determined by Western blot analysis. (D) FoxM1-depleted U2OS cells exhibited diminished Cdk1 and Cdk2 kinase activities. FoxM1-depleted U2OS cells were IP with Cdk2 or Cdk1 antibodies and used for radioactive kinase assays with either recombinant Cdk2 substrate RB protein or Cdk1 substrate histone H1 protein. The radioactively labeled phosphorylated substrates were analyzed by SDS-PAGE followed by autoradiography and then quantitated by the Kodak BioMax 1D program. (E) FoxM1-depleted U2OS cells exhibited increased nuclear and cytoplasmic levels of the CDKI proteins p27Kip1 and p21Cip1 as determined by Western blot analysis. FoxM1-depleted or untreated U2OS cells were used to prepare nuclear and cytoplasmic protein extracts for Western blot analysis with antibodies specific to either the p27Kip1 or p21Cip1 proteins. Both FoxM1-depleted U2OS cells (F) and early-passage Foxm1−/− MEFs (H) exhibited increased nuclear staining of the CDKI protein p27Kip1 compared to low levels in untransfected U2OS cells and WT or Foxm1+/− control MEFs. The number of nuclei expressing high levels of the CDKI protein p21Cip1 was significantly increased in FoxM1-depleted U2OS cells (G) and early-passage Foxm1−/− MEFs (I) compared to untransfected U2OS cells and WT or Foxm1+/− MEF controls. Passage 3 Foxm1+/+ (WT), Foxm1+/−, or Foxm1−/− MEFs and FoxM1-depleted or untreated U2OS cells were immunofluorescently stained with monoclonal antibody specific to either p27Kip1 (400×) or p21Cip1 proteins (200×), nuclei were counterstained with DAPI, and this was merged with CDKI immunofluorescent staining.
FoxM1 is required for expression of Cdc25A and full activity of the Cdk2-cyclin E/A and Cdk1-cyclin B complexes.
Liver regeneration studies with Alb-Cre Foxm1 fl/fl mice demonstrated that regenerating Foxm1−/− hepatocytes exhibited diminished expression of the Cdk2 activator Cdc25A phosphatase and reduced activation of the Cdk2-cyclin E/A or Cdk1-cyclin B complexes as determined by IP kinase assays (67). Consistent with these regenerating Foxm1−/− liver studies, Western blot analysis demonstrated that FoxM1-depleted U2OS cells exhibited reduced expression of Cdc25A phosphatase (Fig. 8C). Furthermore, U2OS cells depleted in FoxM1 levels exhibited a 60% reduction in activity of both Cdk2-cyclin E/A and Cdk1-cyclin B complexes compared to untransfected U2OS cells, as evidenced by IP kinase assays using either the Cdk2 substrate, RB protein, or the Cdk1 substrate, histone H1 protein (Fig. 8D). These studies demonstrated that siRNA silencing of FoxM1 levels in U2OS cells caused diminished activation of the S-phase-promoting Cdk2-cyclin E/A complex and M-phase-promoting Cdk1-cyclin B complex.
FoxM1-depleted U2OS cells and early-passage Foxm1−/− MEFs exhibit increased nuclear levels of CDKI proteins p21Cip1 and p27Kip1.
Liver regeneration studies with Alb-Cre Foxm1 fl/fl mice demonstrated that regenerating Foxm1−/− hepatocytes displayed posttranscriptional increases in nuclear levels of the CDKI proteins p21Cip1 and p27Kip1 (27, 67, 68). Using Western blot analysis with fractionated nuclear and cytoplasmic protein extracts, we showed that siRNA silencing of FoxM1 in U2OS cells caused increased expression of the CDKI proteins p27Kip1 or p21Cip1 compared to untransfected U2OS cell extracts (Fig. 8E). Consistent with these findings, both FoxM1-depleted U2OS cells (Fig. 8F) and early-passage Foxm1−/− MEFs (Fig. 8H) exhibited increased nuclear staining of p27Kip1 protein compared to low levels found in untreated U2OS cells or WT or Foxm1+/− control MEFs. Likewise, the number of nuclei expressing high levels of p21Cip1 protein was significantly increased in FoxM1-depleted U2OS cells (Fig. 8G) and early-passage Foxm1−/− MEFs (Fig. 8I) compared to untransfected U2OS cells and WT or Foxm1+/− MEF controls. These studies indicate that FoxM1 deficiency caused increased nuclear levels of CDKI proteins in both U2OS cells and early-passage MEFs.
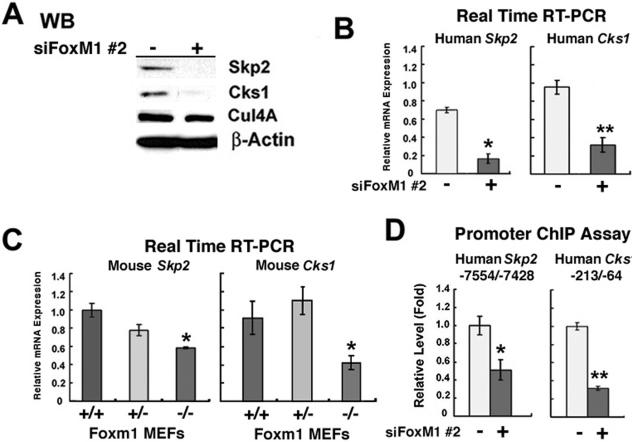
FoxM1 is essential for transcription of SCF ubiquitin ligase complex Skp2 and Cks1 genes.
CDKI proteins p27Kip1 and p21Cip1 phosphorylated by the Cdk2-cyclin E complex are recognized by the specificity subunits Skp2 and Cks1 of the SCF ubiquitin ligase complex, which targets them for ubiquitin-mediated proteasome degradation (11, 19, 49, 63). In order to examine whether FoxM1 regulates Skp2 and Cks1 expression, protein extracts or total RNA was prepared from FoxM1-depleted (+) or untreated (−) U2OS cells and then used to measure Skp2 or Cks1 expression levels. Western blot analysis demonstrated that FoxM1 is essential for detectable expression of both the Skp2 and Cks1 proteins, whereas levels of the Cullin 4A (Cul4A) protein were unchanged in FoxM1-depleted U2OS cells (Fig. 9A). QRT-PCR analysis of mRNA demonstrated that siRNA silencing of FoxM1 expression caused significant reduction of Skp2 and Cks1 mRNA levels, suggesting that FoxM1 regulates transcription of these genes (Fig. 9B). Likewise, Foxm1−/− MEFs displayed reduced mRNA levels of Skp2 and Cks1 compared to WT and Foxm1+/− MEF controls as determined by QRT-PCR (Fig. 9C). Furthermore, RNA isolated from FoxM1-depleted and untreated U2OS cells was used for used QRT-PCR analysis to determine that FoxM1 does not control mRNA expression of other regulators of CDKI protein stability (data not shown), such as Jab1, Kip1 ubiquitination-promoting complex 1 (KPC1), and KPC2 (30, 64, 65). These results indicate that FoxM1 is essential for regulating expression of the specificity subunits Skp2 and Cks1 of the SCF ubiquitin ligase complex, which are critical for targeting these CDKI proteins for degradation during the G1/S transition.
FIG. 9.
FoxM1 regulates transcription of Skp2 and Cks1 genes as determined by quantitative ChIP and expression assays. (A) FoxM1-depleted U2OS cells exhibit undetectable expression of Skp2 and Cks1 proteins, whereas Cullin 4A protein expression was unchanged in FoxM1-depleted U2OS cells as determined by Western blot analysis. (B) FoxM1-depleted U2OS cells exhibit diminished Skp2 and Cks1 mRNA levels as determined by quantitative real-time RT-PCR with primers specific to either the Skp2 or Cks1 gene. FoxM1-depleted U2OS cells exhibit a statistically significant decrease in mRNA levels of Skp2 and Cks1 compared to untransfected controls. (C) Foxm1−/− MEFs display reduced Skp2 and Cks1 mRNA levels compared to WT (+/+) or Foxm1+/− MEFs as determined by real-time RT-PCR. Foxm1−/− MEFs exhibit a statistically significant decrease in mRNA levels of Skp2 and Cks1 compared to control MEFs. (D) Depletion of FoxM1 in U2OS cells by siFoxM1 #2 transfection inhibits binding of FoxM1 protein to the endogenous Skp2 or Cks1 promoter regions. FoxM1-depleted (siFoxM1 #2) or untreated U2OS cells were processed for ChIP assay as described in Materials and Methods. The cross-linked and sonicated human chromatin was IP with antibodies specific to FoxM1 or rabbit serum (control), and the amount of Human Skp2 or Cks1 promoter DNA associated with the IP chromatin was quantitated by real-time PCR with primers specific to the indicated proximal promoter regions. Untreated U2OS cell levels of FoxM1 promoter binding as determined by ChIP assay were set at 1 ± the SD. FoxM1-depleted U2OS cells showed diminished binding of FoxM1 protein to the endogenous human promoter regions of Skp2 and Cks1. The asterisks in panels B to D indicate statistically significant changes with P values calculated by the Student t test, as follows: *P < 0.05; **P ≤ 0.01.
In order to determine whether FoxM1 regulates transcription of the Skp2 and Cks1 genes, FoxM1-depleted or untreated U2OS cells were processed for quantitative ChIP assays. The cross-linked and sonicated human chromatin was IP with antibodies specific to FoxM1 or rabbit serum (control), and the amount of human Skp2 or Cks1 promoter DNA associated with the IP chromatin was quantitated by real-time PCR. These ChIP assays demonstrated that FoxM1 protein associated near the endogenous −7,500 bp Skp2 promoter region and the −200 bp proximal Cks1 promoter region and that siRNA silencing FoxM1 expression in U2OS cells significantly reduced binding of FoxM1 protein to these endogenous human promoter regions (Fig. 9D). Taken together, these studies demonstrate that FoxM1 regulates transcription of the Skp2 and Cks1 genes, which encode specificity subunits of the SCF ubiquitin ligase complex, and that their reduced expression in FoxM1-deficient cells contributes to increased nuclear levels of the CDKI proteins p27Kip1 and p21Cip1.
DISCUSSION
We previously used liver regeneration studies with Alb-Cre Foxm1 fl/fl mice to demonstrate that Foxm1-deficient hepatocytes exhibited a block in mitotic progression and reduced DNA replication due to posttranscriptional increases in nuclear levels of the CDKI proteins (27, 67). However, the FoxM1 transcriptional target genes mediating mitotic progression and degradation of CDKI proteins remain uncharacterized. In our current study, we showed that both human osteosarcoma U2OS cells depleted in FoxM1 levels by siRNA transfection and early-passage Foxm1−/− MEFs were unable to significantly grow in culture due to a failure to progress beyond the prophase stage of mitosis and accumulated nuclear levels of CDKI proteins p21Cip1 and p27Kip1. We provide evidence that FoxM1 is essential for transcription of Skp2 and Cks1, which are specificity subunits of the SCF ubiquitin ligase complex that targets these CDKI proteins for degradation during the G1/S transition. FoxM1-depleted U2OS cells were blocked in mitotic progression as evidenced by a 5-fold increase in G2/M (4N) cells, a 10-fold decrease in mitotic phosphorylation of histone H3 (PH3) protein, and an accumulation of polyploid (8N) cells. The block in mitotic progression was due to undetectable expression of the mitotic regulators Cdc25B, Aurora B kinase, survivin, and PLK1 and the fact that FoxM1 deficiency caused reduced levels of cyclin B1, CENPA, and CENPB (Fig. 5 and 7A). Interestingly, both FoxM1- and Aurora B kinase-depleted U2OS cells exhibited identical accumulation of polyploid (8N) cells (Fig. 7B), supporting the hypothesis that diminished expression of Aurora B kinase contributed to development of a polyploid genotype in FoxM1-deficient cells. Quantitative ChIP and expression assays in FoxM1-deficient or control U2OS cells demonstrated that FoxM1 is essential for transcription of the mitotic regulators Cdc25B, Aurora B kinase, survivin, CENPA, and CENPB (Fig. 5), suggesting that FoxM1 regulates the transcriptional network of genes essential for mitotic progression (Fig. 10). Moreover, early-passage Foxm1−/− MEFs displayed premature senescence as evidenced by high expression of the senescence-associated β-galactosidase enzyme and increased nuclear levels of p19ARF and p16INK4A proteins, the latter of which are cell cycle inhibitor proteins associated with cellular senescence (1, 20, 53, 54).
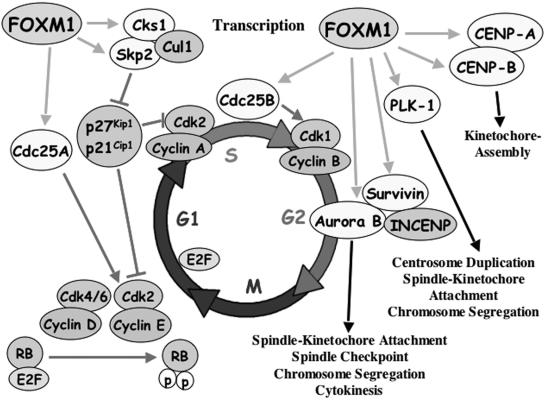
FIG. 10.
Model summarizing Foxm1 target genes involved in regulating G1/S and G2/M progression. FoxM1 protein is necessary for expression of Cdc25A phosphatase, which is required to dephosphorylate and stimulate Cdk2 kinase activity (52). Foxm1 regulates transcription of the Skp2 and Cks1 genes, which are specificity subunits essential for recognition of phosphorylated CDKI proteins p27Kip1 and p21Cip1 by the Skp1-Cullin1-F-box (SCF) ubiquitin ligase complex to target these CDKI proteins for ubiquitin-mediated proteasome degradation (11, 19, 49, 59, 63). Diminished nuclear levels of CDKI proteins p27Kip1 and p21Cip1 are required for stimulating Cdk2-cyclin E complex activity that cooperates with Cdk4/6-cyclin D to phosphorylate the RB protein and activates E2F to stimulate transcription of S-phase genes (45). For progression into mitosis, Foxm1 transcriptionally activates Cdc25B phosphatase (67), which is required to dephosphorylate and activate Cdk1 kinase (52). FoxM1 regulates transcription of PLK1 (37) or Aurora B kinase and survivin, which forms a complex with INCENP and regulates numerous stages of mitosis (48). FoxM1 regulates transcription of CENPA and CENPB, both of which are essential for kinetochore assembly.
FoxM1 regulates transcription of Skp2 and Cks1 proteins required for targeting CDKI proteins for degradation during G1/S phase transition.
Liver regeneration studies with genetically altered transgenic and knockout Foxm1 mice demonstrated that FoxM1 protein regulates posttranscriptional nuclear levels of the CDKI proteins p21Cip1 and p27Kip1 and that S-phase progression required a FoxM1-mediated decrease in nuclear levels of these CDKI proteins (27, 68, 69). In our current study, both FoxM1-depleted U2OS cells and early-passage Foxm1−/− MEFs also exhibited this posttranscriptional increase in nuclear levels of the CDKI proteins p27KIP1 and p21Cip1. We used FoxM1-depleted U2OS cells to determine the molecular mechanism behind this increase in nuclear levels of these CDKI proteins. We found that FoxM1 is required for detectable expression of the Skp2 and Cks1 proteins, which are specificity subunits of the SCF ubiquitin ligase complex that are essential for the recognition of the phosphorylated p27KIP1 and p21Cip1 proteins. The SCF ubiquitin ligase complex associates and targets these phosphorylated CDKI proteins for ubiquitin modification and subsequent proteasome degradation during the G1/S transition of the cell cycle (7, 11, 19, 49, 59, 63). This degradation of CDKI proteins prevents inhibition of the Cdk2-cyclin E complex and allows phosphorylation of RB protein and dissociation of E2F transcription factor to stimulate expression of genes required for S-phase progression (45). Quantitative ChIP and expression assays showed diminished binding of FoxM1 to the endogenous Skp2 and Cks1 promoter region in FoxM1-depleted U2OS cells, and this correlated with significant decreases in their mRNA and protein expression (Fig. 9). This implies that at the G1/S transition of the cell cycle, expression of FoxM1 protein is required for transcription of Skp2 and Cks1, which are essential for proteasome degradation of the CDKI proteins p27KIP1 and p21Cip1, thus facilitating progression of cells into S-phase (Fig. 10).
Diminished activation of Cdk2-cyclin E/A and Cdk1-cyclin B complexes in FoxM1-depleted U2OS cells.
The reduction in S-phase progression in FoxM1-depleted U2OS cells is also associated with decreased expression of Cdc25A phosphatase (Fig. 10), which activates Cdk2 through dephosphorylation of inhibitory Thr14 and Tyr15 residues (6). Consistent with these findings, diminished Cdc25A levels correlate with a 60% reduction in the S-phase-promoting Cdk2-cyclin E/A kinase activity in FoxM1-depleted U2OS cells. Reduced Cdk2-cyclin E/A kinase activity in FoxM1-deficient cells also correlates with increased nuclear levels of CDKI proteins p27Kip1 and p21Cip1, which associate with and inhibit Cdk activity. A 60% reduction in Cdk1-cyclin B kinase activity in FoxM1-depleted U2OS cells contributes to significant decrease in mitotic entry through reduced expression of Cdc25B phosphatase (Fig. 10), which is essential to stimulate Cdk1 kinase activity in late S-phase through dephosphorylation (52). FoxM1 was also shown to regulate transcription of the cyclin B1 promoter (37, 38, 69), and therefore diminished cyclin B1 expression may also contribute to reduced Cdk1-cyclin B activity in FoxM1-deficient cells. Moreover, decreased PLK1 levels in FoxM1-deficient cells prevent phosphorylation and activation of Cdc25C phosphatase in early G2 phase (4), so that Cdc25C phosphatase is unavailable to compensate for reduced Cdc25A and Cdc25B protein expression. These results suggest that FoxM1 is critical for regulating expression of cell cycle genes that mediate activation of the Cdk-cyclin complexes for G1/S and G2/M transitions.
The mitotic defect in FoxM1-deficient cells is associated with significant reduction in expression of PLK1, Aurora B kinase, survivin, CENPA, and CENPB.
Undetectable expression of PLK1 and the chromosome passenger proteins Aurora B kinase and survivin in FoxM1-depleted U2OS cells contribute to their failure to progress past the prophase stage of mitosis (Fig. 10). ChIP assays with FoxM1-depleted and untreated U2OS cells enabled us to determine that FoxM1 specifically binds to the endogenous human Aurora B kinase and survivin promoter regions, demonstrating that these promoter regions are direct transcriptional targets of FoxM1. Recent studies have used FoxM1 cotransfection and ChIP assays to demonstrate that FoxM1 regulates transcription of the PLK1 gene (37). Aurora B kinase forms a complex with survivin and INCENP, and formation of this complex is required for Aurora B kinase activity (48). During the prophase-to-metaphase-anaphase transition the Aurora B-survivin-INCENP complex localizes to the inner centromere region, and during anaphase this complex relocalizes to the midzone spindle region followed by redistribution to the midbody region during telophase (3, 31). Survivin plays an essential role in appropriate localization of the Aurora B kinase-INCENP complex, and undetectable levels of survivin in FoxM1-depleted cells therefore inhibit proper localization and mitotic function of Aurora B kinase (3). Disruption of the Aurora B kinase-survivin-INCENP complex inhibits phosphorylation of the histone H3 protein involved in chromosome condensation and prevents cytokinesis due to loss of phosphorylated proteins on the cleavage furrow that are essential for cytokinesis (3, 10, 31, 48). Loss of Aurora B kinase and PLK1 activity causes chromosome alignment defects, because these kinases are critical for bipolar spindle microtubule attachment to each of the sister chromatid kinetochores and inhibit chromosome segregation because they phosphorylate and dissociate the cohesin protein complex, which holds the sister chromosomes together (4, 22, 66). Loss of PLK1 expression also inhibits duplication of centrosomes, bipolar protein complexes that attach spindle microtubules originating from the chromosomal kinetochores, which are essential for chromosomal separation (4, 66). Undetectable expression of Aurora B kinase and PLK1 proteins is therefore predicted to inhibit progression past the prophase stage of mitosis and to cause reduced phosphorylation of histone H3, which are mitotic defects observed in FoxM1-depleted cells.
Interestingly, U2OS cells deficient in Aurora B kinase expression exhibited accumulation of polyploid cells similar to that found in FoxM1-deficient cells. These results suggest that diminished expression of Aurora B kinase contributes to endoreduplication, causing development of the polyploid genotype in FoxM1-depleted U2OS cells. Aurora B Kinase also regulates localization of the spindle assembly checkpoint proteins BubR1, Mad2, and CENP-E to the centromeric kinetochores (18), which function to inhibit the onset of anaphase until all sister chromosomal kinetochores have bipolar attachment to spindle microtubules (4, 48). Published studies show that Aurora B kinase deficiency causes endoreduplication and a polyploid genotype due to a failure in the spindle assembly checkpoint, resulting in premature mitotic exit during prophase and reinitiation of S-phase (18, 21, 28, 48, 50, 51). This is further supported by the fact that embryonic Foxm1−/− hepatoblasts and vascular smooth muscle cells are also severely polyploid and display significant reduction in expression of Aurora B kinase protein (32, 36). Taken together, these studies suggest that diminished levels of Aurora B kinase in FoxM1-deficient cells contribute significantly to development of a polyploid genotype.
Significant decreases in expression of CENPA and CENPB in FoxM1-depleted cells lead to defects in kinetochore assembly and may therefore contribute to the defect in mitotic progression (Fig. 10). ChIP assays showed diminished binding of FoxM1 to the endogenous CENPA and CENPB promoter regions in FoxM1-depleted U2OS cells, and this correlated with significant decreases in their expression levels. CENPA replaces histone H3 in centromeric nucleosomes, and its incorporation in the nucleosomes of centromeres is necessary for recruitment of the CENPB and CENPC proteins to centromeres and is a prerequisite for assembly of the kinetochore protein complex (2, 5, 47). Cenpa-deficient cells display severe mitotic defects and chromosome abnormalities because they are unable to assemble kinetochores on centromeres due to a failure to localize CENPB and CENPC proteins to centromeres (24). Given the important role of CENPA and CENPB in establishing centromeric kinetochores, their diminished expression in FoxM1-depleted U2OS cells may elicit abnormal assembly of centromeric kinetochores, thus further inhibiting mitotic progression. Interestingly, the CENPB gene is located adjacent to the FoxM1 target gene Cdc25B, and these genes are transcribed in opposing orientations and are separated by 7-kb and 10-kb DNA sequences in the mouse and human genome, respectively (http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?TAXID=10090&QSTR=CDC25B&QUERY=uid(331447)&CHR=2&MAPS=genes[130692393.50%3A130720858.50]&ZOOM=0.1000 and http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?TAXID=9606&QSTR=CDC25B&QUERY=uid(917)&CHR=20&MAPS=genes[3716604.00%3A3742544.00]&ZOOM=0.1000). Based on the close proximity of the CENPB and Cdc25B promoters on the mammalian genome, it is tempting to speculate that FoxM1 may coordinately regulate transcription of the CENPB and Cdc25B genes during the G2 phase of the cell cycle.
Early-passage Foxm1−/− MEFs failed to grow in culture and progress through mitosis and undergo premature cellular senescence.
We generated MEFs from Foxm1−/− embryos that died in utero between 13.5 and 17.5 days of gestation due to severe defects in liver development and exhibited a 75% reduction in the number of Foxm1−/− hepatoblasts compared to WT embryonic livers (36, 42). These Foxm1−/− embryos contained a functionally inactive Foxm1-targeted allele that deleted essential Foxm1 exons 4 through 7 encoding the Foxm1 DNA binding and C-terminal transcriptional activation domains (36, 42). In our current study, we showed that these early-passage Foxm1−/− MEFs failed to divide in culture and did not proceed beyond the prophase stage of mitosis, which was consistent with diminished expression of several mitotic regulators identified as FoxM1 transcriptional target genes. Furthermore, they also undergo premature cellular senescence as evidenced by high expression levels of senescence-associated β-galactosidase enzyme and increased nuclear levels of the p19ARF tumor suppressor and CDKI proteins p16INK4A, p27Kip1, and p21Cip1. The expression of these cell cycle inhibitors in early-passage Foxm1−/− MEFs and the fact that they fail to grow in culture is consistent with premature cellular senescence and is similar to the phenotype observed in late-passage primary MEFs that have undergone replicative senescence (1, 20, 53, 54).
During preparation of the manuscript, Medema and colleagues reported significant growth of Foxm1−/− MEFs in culture isolated from embryos containing a completely different Foxm1-targeted allele that inserted the PGK-Neo selection cassette 50 nucleotides upstream of the sequences encoding the FoxM1 DNA binding domain (MAMIQFAI) (43) without deleting any coding sequences (35, 37). They showed that their Foxm1−/− MEFs grew at passage 4 in culture, displayed reduced G2/M progression, and yet were able to complete mitosis, producing daughter cells with a variety of different aneuploid genotypes resulting from defective chromosomal segregation (37). This delay in G2/M progression in Foxm1−/− MEFs was associated with diminished transcription of PLK1, cyclin B, Nek2, Gas-1, and CENPF genes and with the fact that chromosome segregation defects were due to inhibition of the spindle assembly checkpoint caused by reduced CENPF expression (37). In contrast, our Foxm1−/− MEFs were unable to grow in culture due to a severe mitotic defect and had undergone premature cellular senescence. One explanation most consistent with this difference in growth properties is that the Foxm1−/− MEFs generated by Medema and colleagues are expressing hypomorphic levels of Foxm1 protein, which allowed sufficient levels of mitotic regulators to facilitate aberrant progression through mitosis with defective chromosome segregation. Our ideas regarding this hypomorphic Foxm1-targeted allele are supported by the fact that no embryonic lethality was reported in Foxm1−/− (Trident−/−) embryos generated by Medema and colleagues (35), a finding which differs significantly from the complete embryonic lethality observed with our Foxm1−/−-targeted allele (36). The PGK-Neo Foxm1-targeted allele generated by Medema and colleagues is capable of expressing a functional N-terminal-truncated Foxm1 protein containing an intact DNA binding domain and C-terminal transcriptional activation domain. Our preliminary studies indicate that deletion of this FoxM1 N-terminal region caused a greater than 20-fold increase in FoxM1 transcriptional activity compared to full-length FoxM1 protein (H. J. Park, M. L. Major, and R. H. Costa, unpublished data). This implies that low-level expression of this N-terminal truncated FoxM1 protein from this PGK-Neo-targeted Foxm1 allele could be sufficient to provide hypomorphic levels of Foxm1 transcriptional activity.
The importance of this difference in growth properties of Foxm1−/− MEFs observed in the two studies is highlighted when one considers the role of FoxM1 in tumor cell proliferation. It is interesting that Foxm1−/− MEFs generated by Medema and colleagues exhibit aneuploid genotypes (37), which are a hallmark of neoplastic transformation (58, 62). However, no liver tumor formation was observed in the absence of the Foxm1 gene in response to hepatic carcinogens (27). In a recent publication, we clearly showed that our Alb-Cre Foxm1−/− hepatocytes failed to proliferate and are resistant to developing HCC by using a well-established tumor initiation/progression protocol (13, 27). The mechanism of resistance to HCC development is associated with persistent hepatocyte nuclear accumulation of CDKI protein p27Kip1, diminished expression of the Cdk1-activating Cdc25B phosphatase, and the fact that Foxm1−/− hepatocytes failed to complete mitosis, acquiring a polyploid genotype. Taken together, these studies suggest that both the Alb-Cre Foxm1−/− mouse hepatocytes and cultured Foxm1−/− MEFs fail to complete mitosis and divide to produce daughter cells. We believe that the Foxm1−/− MEFs used by Medema and colleagues (37) contained a potentially hypomorphic Foxm1-targeted allele, allowing aberrant progression through mitosis and causing a variety of aneuploid genotypes, and that they do not provide an accurate depiction of the role of Foxm1 in both normal and tumor cell proliferation.
Decreased expression of FoxM1 during aging is responsible for reduced cell proliferation.
Flow cytometry analysis of proliferating fibroblasts isolated from elderly humans revealed diminished G2/M progression, which resulted in accumulation of cells with a polyploid (8N) genotype (41). This published study used microarray analysis comparing gene expression profiles of proliferating fibroblasts from young versus old humans and allowed us to determined that the decrease in cellular proliferation in aging fibroblasts is associated with diminished expression of Foxm1 transcription factor and its cell cycle target genes (41, 74). In published studies, we tested the hypothesis that reduced FoxM1 expression contributes to the proliferation defects observed in aging by using the −3 kb TTR promoter to maintain hepatocyte expression of the FoxM1b transgene in 12-month-old (old-aged) transgenic mice during liver regeneration (69). We showed that maintaining hepatocyte expression of FoxM1b alone in old-aged TTR-FoxM1b transgenic mice is sufficient to restore regenerating hepatocyte DNA synthesis and mitosis to levels found in young regenerating mouse liver (69). Furthermore, FoxM1b-mediated stimulation of hepatocyte proliferation was associated with increased expression of genes critical for both DNA replication and mitosis (41, 69). We also showed that increased FoxM1 expression reduced nuclear levels of the CDKI proteins p21Cip1 and p27Kip1 protein in old regenerating livers (68, 69). This finding is consistent with the essential role of FoxM1 in regulating expression of the SCF (Skp2/Cks1) ubiquitin ligase complex proteins required to target these CDKI proteins for degradation. The facts that early-passage Foxm1−/− MEFs undergo premature cellular senescence and that increased FoxM1b expression restores proliferation of regenerating hepatocytes in old-aged mice suggest that loss of Foxm1 expression may be the primary defect that leads to reduced cell cycle progression during aging.
In summary, both FoxM1-depleted U2OS cells and Foxm1−/− MEFs are unable to grow in culture due to a block in mitotic progression (Fig. 2 and 3) and accumulate nuclear levels of the CDKI proteins p27Kip1 and p21Cip1 (Fig. 8), and Foxm1−/− MEFs express senescence-associated marker proteins (Fig. 4). We show that FoxM1 is required for expression of the mitotic regulatory genes Cdc25B, Aurora B kinase, survivin, PLK1, CENPA, and CENPB (Fig. 5). FoxM1-deficient cells express undetectable levels of the Skp2 and Cks1 proteins, which are specificity subunits of the SCF ubiquitin ligase complex that are essential for targeting the phosphorylated CDKI proteins p27Kip1 and p21Cip1 for degradation (Fig. 9). Our current study further supports the hypothesis that the FoxM1 transcription factor regulates expression of cell cycle proteins that are essential for G1/S and G2/M progression (Fig. 10).
Acknowledgments
This work was supported by U.S. Public Health Service grants DK 54687-07 from NIDDK and RO1 AG 21842-03 from NIA (to R.H.C.).
We thank K. L. Hagen (Director of the FACS Laboratory, Research Resource Center, UIC) for the flow cytometry and analysis and M.-S. Chen (NHRI, Taiwan) for helpful discussions. We also thank H. Yoder and A. Monson for excellent technical assistance and B. Merrill, P. Raychaudhuri, X. Liao, and V. Kalinichenko for helpful discussions and critical review of the paper. We also thank members of S. A. Duncan's laboratory for advice and protocols for ChIP assays.
REFERENCES
- 1.Alcorta, D. A., Y. Xiong, D. Phelps, G. Hannon, D. Beach, and J. C. Barrett. 1996. Involvement of the cyclin-dependent kinase inhibitor p16INK4a in replicative senescence of normal human fibroblasts. Proc. Natl. Acad. Sci. USA 93:13742-13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amor, D. J., P. Kalitsis, H. Sumer, and K. H. Choo. 2004. Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol. 14:359-368. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, P. D., E. Knatko, W. J. Moore, and J. R. Swedlow. 2003. Mitotic mechanics: the auroras come into view. Curr. Opin. Cell Biol. 15:672-683. [DOI] [PubMed] [Google Scholar]
- 4.Barr, F. A., H. H. Sillje, and E. A. Nigg. 2004. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5:429-440. [DOI] [PubMed] [Google Scholar]
- 5.Black, B. E., D. R. Foltz, S. Chakravarthy, K. Luger, V. L. Woods, Jr., and D. W. Cleveland. 2004. Structural determinants for generating centromeric chromatin. Nature 430:578-582. [DOI] [PubMed] [Google Scholar]
- 6.Borgne, A., and L. Meijer. 1996. Sequential dephosphorylation of p34cdc2 on Thr-14 and Tyr-15 at the prophase/metaphase transition. J. Biol. Chem. 271:27847-27854. [DOI] [PubMed] [Google Scholar]
- 7.Bornstein, G., J. Bloom, D. Sitry-Shevah, K. Nakayama, M. Pagano, and A. Hershko. 2003. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 278:25752-25757. [DOI] [PubMed] [Google Scholar]
- 8.Busuttil, R. A., M. Rubio, M. E. Dolle, J. Campisi, and J. Vijg. 2003. Oxygen accelerates the accumulation of mutations during the senescence and immortalization of murine cells in culture. Aging Cell 2:287-294. [DOI] [PubMed] [Google Scholar]
- 9.Carlsson, P., and M. Mahlapuu. 2002. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 250:1-23. [DOI] [PubMed] [Google Scholar]
- 10.Carmena, M., and W. C. Earnshaw. 2003. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4:842-854. [DOI] [PubMed] [Google Scholar]
- 11.Carrano, A. C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193-199. [DOI] [PubMed] [Google Scholar]
- 12.Clark, K. L., E. D. Halay, E. Lai, and S. K. Burley. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364:412-420. [DOI] [PubMed] [Google Scholar]
- 13.Costa, R. H., V. V. Kalinichenko, M. L. Major, and P. Raychaudhuri. 2005. New and unexpected: forkhead meets ARF. Curr. Opin. Genet. Dev. 15:42-48. [DOI] [PubMed] [Google Scholar]
- 14.Crosio, C., G. M. Fimia, R. Loury, M. Kimura, Y. Okano, H. Zhou, S. Sen, C. D. Allis, and P. Sassone-Corsi. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datta, A., A. Nag, W. Pan, N. Hay, A. Gartel, O. Colamonici, Y. Mori, and P. Raychaudhuri. 2004. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J. Biol. Chem. 279:36698-36707. [DOI] [PubMed] [Google Scholar]
- 16.Datta, A., A. Nag, and P. Raychaudhuri. 2002. Differential regulation of E2F1, DP1, and the E2F1/DP1 complex by ARF. Mol. Cell. Biol. 22:8398-8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai, A., S. Rybina, T. Muller-Reichert, A. Shevchenko, A. Hyman, and K. Oegema. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ditchfield, C., V. L. Johnson, A. Tighe, R. Ellston, C. Haworth, T. Johnson, A. Mortlock, N. Keen, and S. S. Taylor. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganoth, D., G. Bornstein, T. K. Ko, B. Larsen, M. Tyers, M. Pagano, and A. Hershko. 2001. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat. Cell Biol. 3:321-324. [DOI] [PubMed] [Google Scholar]
- 20.Hara, E., R. Smith, D. Parry, H. Tahara, S. Stone, and G. Peters. 1996. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol. Cell. Biol. 16:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauf, S., R. W. Cole, S. LaTerra, C. Zimmer, G. Schnapp, R. Walter, A. Heckel, J. van Meel, C. L. Rieder, and J. M. Peters. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hauf, S., E. Roitinger, B. Koch, C. M. Dittrich, K. Mechtler, and J. M. Peters. 2005. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol 3:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogan, B., R. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Howman, E. V., K. J. Fowler, A. J. Newson, S. Redward, A. C. MacDonald, P. Kalitsis, and K. H. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itahana, K., J. Campisi, and G. P. Dimri. 2004. Mechanisms of cellular senescence in human and mouse cells. Biogerontology 5:1-10. [DOI] [PubMed] [Google Scholar]
- 26.Kaestner, K. H., W. Knochel, and D. E. Martinez. 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14:142-146. [PubMed] [Google Scholar]
- 27.Kalinichenko, V. V., M. Major, X. Wang, V. Petrovic, J. Kuechle, H. M. Yoder, B. Shin, A. Datta, P. Raychaudhuri, and R. H. Costa. 2004. Forkhead Box m1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 18:830-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallio, M. J., M. L. McCleland, P. T. Stukenberg, and G. J. Gorbsky. 2002. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12:900-905. [DOI] [PubMed] [Google Scholar]
- 29.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 30.Kamura, T., T. Hara, M. Matsumoto, N. Ishida, F. Okumura, S. Hatakeyama, M. Yoshida, K. Nakayama, and K. I. Nakayama. 2004. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27Kip1 at G1 phase. Nat. Cell Biol. 6:1229-1235. [DOI] [PubMed] [Google Scholar]
- 31.Keen, N., and S. Taylor. 2004. Aurora-kinase inhibitors as anticancer agents. Nat. Rev. Cancer 4:927-936. [DOI] [PubMed] [Google Scholar]
- 32.Kim, I. M., S. Ramakrishna, G. A. Gusarova, H. M. Yoder, R. H. Costa, and V. V. Kalinichenko. 2005. The Forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J. Biol. Chem. 280:22278-22286. [DOI] [PubMed] [Google Scholar]
- 33.Korver, W., J. Roose, and H. Clevers. 1997. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 25:1715-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korver, W., J. Roose, K. Heinen, D. O. Weghuis, D. de Bruijn, A. G. van Kessel, and H. Clevers. 1997. The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization. Genomics 46:435-442. [DOI] [PubMed] [Google Scholar]
- 35.Korver, W., M. W. Schilham, P. Moerer, M. J. van den Hoff, K. Dam, W. H. Lamers, R. H. Medema, and H. Clevers. 1998. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor trident. Curr. Biol. 8:1327-1330. [DOI] [PubMed] [Google Scholar]
- 36.Krupczak-Hollis, K., X. Wang, V. V. Kalinichenko, G. A. Gusarova, I.-C. Wang, M. B. Dennewitz, H. M. Yoder, H. Kiyokawa, K. H. Kaestner, and R. H. Costa. 2004. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev. Biol. 276:74-88. [DOI] [PubMed] [Google Scholar]
- 37.Laoukili, J., M. R. Kooistra, A. Bras, J. Kauw, R. M. Kerkhoven, A. Morrison, H. Clevers, and R. H. Medema. 2005. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7:126-136. [DOI] [PubMed] [Google Scholar]
- 38.Leung, T. W., S. S. Lin, A. C. Tsang, C. S. Tong, J. C. Ching, W. Y. Leung, R. Gimlich, G. G. Wong, and K. M. Yao. 2001. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 507:59-66. [DOI] [PubMed] [Google Scholar]
- 39.Lowe, S. W., and C. J. Sherr. 2003. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 13:77-83. [DOI] [PubMed] [Google Scholar]
- 40.Luscher-Firzlaff, J. M., J. M. Westendorf, J. Zwicker, H. Burkhardt, M. Henriksson, R. Muller, F. Pirollet, and B. Luscher. 1999. Interaction of the fork head domain transcription factor MPP2 with the human papilloma virus 16 E7 protein: enhancement of transformation and transactivation. Oncogene 18:5620-5630. [DOI] [PubMed] [Google Scholar]
- 41.Ly, D. H., D. J. Lockhart, R. A. Lerner, and P. G. Schultz. 2000. Mitotic misregulation and human aging. Science 287:2486-2492. [DOI] [PubMed] [Google Scholar]
- 42.Major, M. L., R. Lepe, and R. H. Costa. 2004. Forkhead Box M1B (FoxM1B) transcriptional activity requires binding of Cdk/cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol. Cell. Biol. 24:2649-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marsden, I., C. Jin, and X. Liao. 1998. Structural changes in the region directly adjacent to the DNA-binding helix highlight a possible mechanism to explain the observed changes in the sequence-specific binding of winged helix proteins. J. Mol. Biol. 278:293-299. [DOI] [PubMed] [Google Scholar]
- 44.Martelli, F., T. Hamilton, D. P. Silver, N. E. Sharpless, N. Bardeesy, M. Rokas, R. A. DePinho, D. M. Livingston, and S. R. Grossman. 2001. p19ARF targets certain E2F species for degradation. Proc. Natl. Acad. Sci. USA 98:4455-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Massague, J. 2004. G1 cell-cycle control and cancer. Nature 432:298-306. [DOI] [PubMed] [Google Scholar]
- 46.McCormick, F. 1999. Signalling networks that cause cancer. Trends Cell Biol. 9:M53-M56. [PubMed] [Google Scholar]
- 47.Mellone, B. G., and R. C. Allshire. 2003. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 13:191-198. [DOI] [PubMed] [Google Scholar]
- 48.Meraldi, P., R. Honda, and E. A. Nigg. 2004. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr. Opin. Genet. Dev. 14:29-36. [DOI] [PubMed] [Google Scholar]
- 49.Montagnoli, A., F. Fiore, E. Eytan, A. C. Carrano, G. F. Draetta, A. Hershko, and M. Pagano. 1999. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 13:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murata-Hori, M., M. Tatsuka, and Y. L. Wang. 2002. Probing the dynamics and functions of aurora B kinase in living cells during mitosis and cytokinesis. Mol. Biol. Cell 13:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murata-Hori, M., and Y. L. Wang. 2002. The kinase activity of aurora B is required for kinetochore-microtubule interactions during mitosis. Curr. Biol. 12:894-899. [DOI] [PubMed] [Google Scholar]
- 52.Nilsson, I., and I. Hoffmann. 2000. Cell cycle regulation by the Cdc25 phosphatase family. Prog. Cell Cycle Res. 4:107-114. [DOI] [PubMed] [Google Scholar]
- 53.Palmero, I., B. McConnell, D. Parry, S. Brookes, E. Hara, S. Bates, P. Jat, and G. Peters. 1997. Accumulation of p16INK4a in mouse fibroblasts as a function of replicative senescence and not of retinoblastoma gene status. Oncogene 15:495-503. [DOI] [PubMed] [Google Scholar]
- 54.Palmero, I., C. Pantoja, and M. Serrano. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395:125-126. [DOI] [PubMed] [Google Scholar]
- 55.Palmero, I., and M. Serrano. 2001. Induction of senescence by oncogenic Ras. Methods Enzymol. 333:247-256. [DOI] [PubMed] [Google Scholar]
- 56.Qi, Y., M. A. Gregory, Z. Li, J. P. Brousal, K. West, and S. R. Hann. 2004. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 431:712-717. [DOI] [PubMed] [Google Scholar]
- 57.Quelle, D. E., F. Zindy, R. A. Ashmun, and C. J. Sherr. 1995. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83:993-1000. [DOI] [PubMed] [Google Scholar]
- 58.Rajagopalan, H., and C. Lengauer. 2004. Aneuploidy and cancer. Nature 432:338-341. [DOI] [PubMed] [Google Scholar]
- 59.Sheaff, R. J., M. Groudine, M. Gordon, J. M. Roberts, and B. E. Clurman. 1997. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 11:1464-1478.9192873 [Google Scholar]
- 60.Sherr, C. J. 1998. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12:2984-2991. [DOI] [PubMed] [Google Scholar]
- 61.Sherr, C. J., and F. McCormick. 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103-112. [DOI] [PubMed] [Google Scholar]
- 62.Storchova, Z., and D. Pellman. 2004. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 5:45-54. [DOI] [PubMed] [Google Scholar]
- 63.Sutterluty, H., E. Chatelain, A. Marti, C. Wirbelauer, M. Senften, U. Muller, and W. Krek. 1999. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1:207-214. [DOI] [PubMed] [Google Scholar]
- 64.Tomoda, K., Y. Kubota, Y. Arata, S. Mori, M. Maeda, T. Tanaka, M. Yoshida, N. Yoneda-Kato, and J. Y. Kato. 2002. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J. Biol. Chem. 277:2302-2310. [DOI] [PubMed] [Google Scholar]
- 65.Tomoda, K., Y. Kubota, and J. Kato. 1999. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398:160-165. [DOI] [PubMed] [Google Scholar]
- 66.van Vugt, M. A., B. C. van de Weerdt, G. Vader, H. Janssen, J. Calafat, R. Klompmaker, R. M. Wolthuis, and R. H. Medema. 2004. Polo-like kinase-1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J. Biol. Chem. 279:36841-36854. [DOI] [PubMed] [Google Scholar]
- 67.Wang, X., H. Kiyokawa, M. B. Dennewitz, and R. H. Costa. 2002. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. USA 99:16881-16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, X., K. Krupczak-Hollis, Y. Tan, M. B. Dennewitz, G. R. Adami, and R. H. Costa. 2002. Increased hepatic Forkhead Box M1B (FoxM1B) levels in old-aged mice stimulated liver regeneration through diminished p27Kip1 protein levels and increased Cdc25B expression. J. Biol. Chem. 277:44310-44316. [DOI] [PubMed] [Google Scholar]
- 69.Wang, X., E. Quail, N.-J. Hung, Y. Tan, H. Ye, and R. H. Costa. 2001. Increased levels of Forkhead Box M1B transcription factor in transgenic mouse hepatocytes prevents age-related proliferation defects in regenerating liver. Proc. Natl. Acad. Sci. USA 98:11468-11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wei, W., and J. M. Sedivy. 1999. Differentiation between senescence (M1) and crisis (M2) in human fibroblast cultures. Exp. Cell Res. 253:519-522. [DOI] [PubMed] [Google Scholar]
- 71.Wells, J., and P. J. Farnham. 2002. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods 26:48-56. [DOI] [PubMed] [Google Scholar]
- 72.Wright, W. E., and J. W. Shay. 2002. Historical claims and current interpretations of replicative aging. Nat. Biotechnol. 20:682-688. [DOI] [PubMed] [Google Scholar]
- 73.Yao, K. M., M. Sha, Z. Lu, and G. G. Wong. 1997. Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domain. J. Biol. Chem. 272:19827-19836. [DOI] [PubMed] [Google Scholar]
- 74.Ye, H., A. Holterman, K. W. Yoo, R. R. Franks, and R. H. Costa. 1999. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S-phase. Mol. Cell. Biol. 19:8570-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye, H., T. F. Kelly, U. Samadani, L. Lim, S. Rubio, D. G. Overdier, K. A. Roebuck, and R. H. Costa. 1997. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol. Cell. Biol. 17:1626-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zindy, F., D. E. Quelle, M. F. Roussel, and C. J. Sherr. 1997. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 15:203-211. [DOI] [PubMed] [Google Scholar]