Abstract
Ubiquitin conjugation typically requires three classes of enzyme: E1, E2, and E3. A fourth type of enzyme (E4), however, was recently shown to be required for the degradation of certain types of substrate in yeast. We previously identified UFD2a (also known as E4B) as an E4 in mammals. UFD2a is exclusively expressed in cardiac muscle during mouse embryonic development, but it is abundant in neurons of adult mice and is implicated in the pathogenesis of neurodegenerative disease. The precise physiological function of this enzyme has remained largely unknown, however. Here, we show that mice lacking UFD2a die in utero, manifesting marked apoptosis in the developing heart. Polyubiquitylation activity for an E4 substrate was greatly reduced in Ufd2a−/− mouse embryonic fibroblasts. Furthermore, Ufd2a+/− mice displayed axonal dystrophy in the nucleus gracilis, as well as degeneration of Purkinje cells accompanied by endoplasmic reticulum stress. These animals also developed a neurological disorder. UFD2a thus appears to be essential for the development of cardiac muscle, as well as for the protection of spinocerebellar neurons from degeneration induced by endoplasmic reticulum stress.
The ubiquitin (Ub)-proteasome system for protein degradation plays a pivotal role in many cellular processes (10, 38). The ubiquitylation of proteins serves to mark them for degradation by the proteasome, an ATP-dependent multiprotease complex. Protein ubiquitylation is achieved by a multistep mechanism involving several enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-protein ligase (E3). A new class of ubiquitylation enzyme, a ubiquitin chain assembly factor (E4), was recently discovered and shown to be required for the degradation of certain types of substrate, including a fusion protein with an NH2-terminal ubiquitin moiety, by a ubiquitin fusion degradation pathway, designated UFD (17, 21). Ufd2 of Saccharomyces cerevisiae, the prototype E4 enzyme, binds to oligoubiquitylated UFD substrate and catalyzes extension of the ubiquitin chain. Lysine-29 of the ubiquitin moiety of the UFD substrate is required for polyubiquitylation by E4. A UFD substrate molecule with a short chain of Lys-29-linked ubiquitin molecules is thus presented by the Cdc48-Ufd1-Npl4 complex to Ufd2, which then mediates further elongation of the oligoubiquitin chain in a manner dependent on Lys-48 of ubiquitin. The polyubiquitylated UFD substrate is recognized by the UBL-UBA proteins Rad23 and Dsk2 and is then degraded by the 26S proteasome (40). In yeast, Ufd2 is implicated in proteolysis by the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway.
Ufd2 and its homologs in other eukaryotes share a conserved domain known as the U box. The U box of Ufd2 mediates the interaction of this protein with ubiquitin-conjugated targets and therefore appears to be an essential functional domain for E4 activity. We recently showed that mammalian U-box proteins, including UFD2a (also known as E4B), also possess E3 activity and that E4 activity might reflect a specialized type of E3 activity that targets oligoubiquitylated proteins for further ubiquitylation (9). UFD2a interacts with VCP, an AAA-type ATPase and a mammalian ortholog of yeast Cdc48, the latter of which interacts with Ufd2 in yeast. The UFD2a-VCP (Ufd2-Cdc48) system thus appears to be well conserved throughout evolution.
UFD2a is expressed predominantly in the neuronal tissues of adult mice (19). We previously identified UFD2a as a mammalian ortholog of yeast Ufd2 and showed that it participates as an E4 in the ubiquitin-dependent degradation of ataxin-3 (27). The abnormal expansion of a polyglutamine tract in this latter protein is responsible for spinocerebellar ataxia type 3 (SCA3) in humans. We found that overexpression of UFD2a in mammalian cells promoted degradation of a pathological form of ataxin-3. In contrast, a dominant negative mutant of UFD2a inhibited degradation of this form of ataxin-3, resulting in the formation of intracellular aggregates. Expression of UFD2a suppressed the neurodegeneration induced by an ataxin-3 mutant in a Drosophila melanogaster model of SCA3. UFD2a is also implicated in the process of Wallerian degeneration of neurons (3, 26). The precise physiological function of this enzyme has remained largely unknown, however.
We have now generated mice deficient in UFD2a and found that this protein is indispensable for cardiac development during embryogenesis. Furthermore, Ufd2a+/− mice developed a neurological disorder, manifesting axonal dystrophy in the nucleus gracilis, as well as degeneration of Purkinje cells accompanied by ER stress. These results suggest that UFD2a plays an essential role in cardiac development and in the protection of neurons from degeneration evoked by ER stress.
MATERIALS AND METHODS
Construction of a Ufd2a targeting vector and generation of Ufd2a−/− mice.
Cloned genomic DNA corresponding to the Ufd2a locus was previously isolated from a 129/Sv mouse genomic library (19). The targeting vector, pUFD2a-KO, was constructed by replacing a 9-kb fragment containing exons 23 to 27 of Ufd2a by a phosphoglycerate kinase (PGK)-neo-poly(A) cassette. The targeting vector thus contained 1.3- and 6.0-kb regions of homology 5′ and 3′ of the neomycin resistance marker, respectively. The PGK-thymidine kinase (tk)-poly(A) cassette was ligated at the 3′ end of the insert. The maintenance, transfection, and selection of embryonic stem cells were performed as previously described (34). The recombination event was confirmed by Southern blot analysis with a 0.9-kb XbaI-EcoRI probe that flanked the 5′ homology region (Fig. 1A). The mutant embryonic stem cells were microinjected into C57BL/6 blastocysts, and the resulting male chimeras were mated with female C57BL/6 mice. The germ line transmission of the mutant allele was confirmed by Southern blot analysis. Heterozygous offspring were intercrossed to produce homozygous mutant animals. For genotyping of embryos, DNA was extracted from the yolk sac at embryonic day 9.5 (E9.5) to E13.5 and analyzed by PCR with the primers 5′-CTG AGG GGA TGA CGA GAA GAG GAT-3′ and 5′-TTG CTA GAA GGA CCA GGC TCA CAG-3′ for the wild-type allele and 5′-CAG CAG CAG CCC AGA GGG AAC TTG-3′ and 5′-GCC TTC TAT CGC CTT CTT GAC GAG-3′ for the mutant allele.
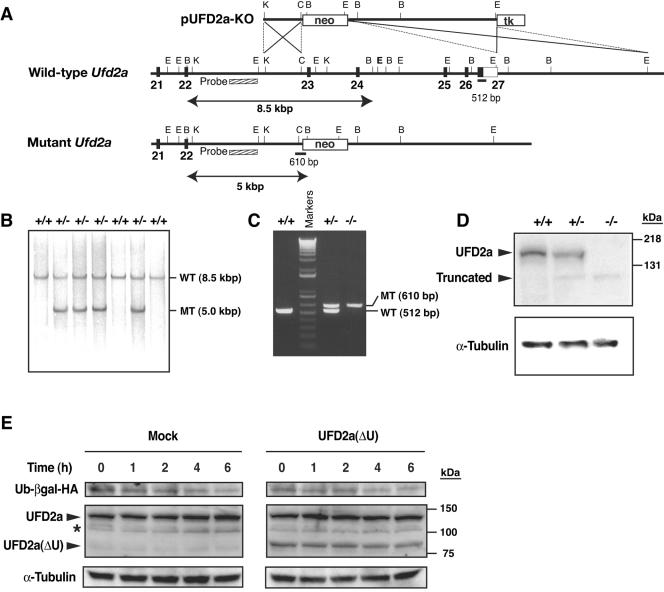
FIG. 1.
Targeted disruption of mouse Ufd2a. (A) Structures of the targeting vector (pUFD2a-KO), the mouse Ufd2a locus, and the mutant allele resulting from homologous recombination. The coding exons and coding portion of exon 27 are depicted by filled boxes, with the open box indicating the noncoding portion of exon 27. A genomic fragment used as a probe for Southern blot analysis is shown as a striped box, and the expected sizes of the BamHI fragments (arrows) that hybridize with the probe are indicated. The positions (solid bars) and sizes of PCR products used for screening are also indicated. neo, neomycin transferase gene linked to the PGK gene promoter; tk, thymidine kinase gene derived from herpes simplex virus linked to the PGK gene promoter. The orientations of both neo and tk are the same as that of Ufd2a. Restriction sites: E1, EcoRI; B, BamHI; C, ClaI; K, KpnI. Not all restriction sites are shown. (B) Southern blot analysis of genomic DNA extracted from the tail of adult mice. The DNA was digested with BamHI and subjected to hybridization with the probe shown in panel A. The positions and sizes of bands corresponding to the wild-type (WT) and mutant (MT) alleles are indicated, as are the genotypes of the analyzed mice. (C) PCR analysis of genomic DNA extracted from the yolk sacs of embryos of the indicated genotypes at E11.5. Amplification products corresponding to the black bars in panel A are indicated. (D) Immunoblot analysis of E11.5 embryo lysates with antibodies to UFD2a (top) and to α-tubulin (loading control) (bottom). The positions of full-length and truncated forms of UFD2a are indicated. (E) Cycloheximide chase analysis of the UFD substrate. HEK293T cells were transfected with an expression plasmid encoding Ub-βgal tagged with HA at its COOH terminus (Ub-βgal-HA) either alone (Mock) or together with a vector for UFD2a(ΔU). The cells were then treated with cycloheximide for the indicated times, lysed, and subjected to immunoblot analysis with anti-HA, anti-UFD2a, and anti-α-tubulin. The asterisk indicates a nonspecific band.
Preparation of embryo lysate and immunoblot analysis.
Frozen embryos were cut into small pieces and then homogenized for 5 min in an ice-cold solution containing 50 mM Tris-HCl (pH 7.5), 0.25 M sucrose, 1 mM EDTA, aprotinin (10 μg ml−1), leupeptin (10 μg ml−1), and 1 mM phenylmethylsulfonyl fluoride. The homogenate was diluted with an equal volume of 2× radioimmunoprecipitation assay buffer (0.3 M NaCl, 20 mM Tris-HCl [pH 7.5], 2% NP-40, 0.2% sodium deoxycholate, 0.2% sodium dodecyl sulfate, aprotinin [10 μg ml−1], leupeptin [10 μg ml−1], and 1 mM phenylmethylsulfonyl fluoride), incubated on ice for 15 min, and centrifuged at 15,000 × g for 15 min. The resulting supernatant (50 μg of protein) was then subjected to immunoblot analysis as previously described (19) with rabbit polyclonal antibodies to UFD2a (1 μg ml−1) (9) and mouse monoclonal anti-α-tubulin (1 μg ml−1; TU-01, Zymed).
Histological, immunohistochemical, and immunofluorescence analyses.
For histology, embryos were fixed in 4% paraformaldehyde for 24 h, embedded in paraffin, and sectioned at a thickness of 5 μm; sections were stained with hematoxylin-eosin. For immunohistochemistry, frozen sections (thickness, 5 μm) were prepared from embryos and stained with polyclonal anti-UFD2a (1 μg ml−1) as described previously (19). For immunofluorescence analysis, sections were stained with anti-UFD2a (1 μg ml−1) and rat monoclonal anti-platelet endothelial cell adhesion molecule 1 (1 μg ml−1; MEC13.3, BD Biosciences PharMingen) or with mouse monoclonal anti-sarcomeric α-actinin (15 μg ml−1; EA-53, Sigma-Aldrich). Immune complexes were detected with Alexa488-conjugated goat antibodies to rabbit or mouse immunoglobulin (Ig) (Molecular Probes) and Cy3-conjugated goat anti-rat IgG (Amersham Pharmacia Biotech). For preparation of brain sections, adult mice were subjected to deep anesthesia by ether inhalation and then perfused transcardially first with phosphate-buffered saline and subsequently with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). The brain was then removed and reexposed to the fixative. Paraffin-embedded sections were prepared and subjected either to hematoxylin-eosin or Kluver-Barrera staining or to immunohistochemistry with rabbit polyclonal anti-ERp72 (1:200 dilution; Calbiochem) or goat polyclonal anti-Grp78 (1 μg ml−1; Santa Cruz Biotechnology); immune complexes were detected with biotinylated goat anti-rabbit IgG or rabbit anti-goat IgG (0.2 μg ml−1; Vector Laboratories), respectively, and horseradish peroxidase-conjugated streptavidin (Vector Laboratories).
Electron microscopy.
Mice were anesthetized as described above and then perfused transcardially first with phosphate-buffered saline and subsequently with 3% glutaraldehyde and 1% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). The brain was removed, immersed in the same fixative for an additional 16 h at 4°C, exposed to 1% osmium tetroxide, dehydrated with a graded series of ethanol solutions, and embedded in Epon 812 resin (Polysciences). Ultrathin sections were prepared, stained with uranyl acetate and lead citrate, and examined with a Hitachi-7100 electron microscope. For immunoelectron microscopy, brain tissue was fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4), dehydrated with a graded series of dimethylformamide solutions, and embedded in LR White resin (London Resin Company). Ultrathin sections were cut, mounted on nickel grids, and incubated consecutively with 10% normal goat serum, rabbit anti-ERp72 (1:40 dilution), and goat anti-rabbit IgG conjugated to 15-nm gold particles (1:40 dilution; British BioCell International). They were then incubated with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) before being stained with uranyl acetate and lead citrate and examined with the electron microscope. For examination of vascular sections of embryos, the yolk sac and brain were removed and immersed for 2 h at room temperature in 0.1 M cacodylate buffer (pH 7.4) containing 2.5% glutaraldehyde, 0.1 M sucrose, and 3 mM CaCl2. The tissue was then exposed to 1% osmium tetroxide for 1 h at 4°C, dehydrated with a graded series of ethanol solutions and with propylene oxide, and embedded in Epon 812 resin. Ultrathin sections were prepared, stained with 2% uranyl acetate for 10 min and lead acetate for 15 min, and then examined with a JEM 2000EX (JEOL, Tokyo, Japan) electron microscope.
TUNEL assay.
Transverse sections of cardiac tissue were subjected to the terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) assay for the detection of apoptotic nuclei. Sections were incubated at 37°C first for 15 min with proteinase K (20 μg ml−1) and then for 1 h with a solution containing 0.1 M potassium cacodylate (pH 7.2), 2 mM cobalt chloride, terminal deoxynucleotidyl transferase (500 U ml−1; Invitrogen), and 30 μM biotinylated dUTP (Boehringer Mannheim). Incorporation of biotin-dUTP was then detected with a streptavidin-biotin-peroxidase detection kit (Vector Laboratories) and diaminobenzidine (Wako).
Expression plasmids.
The plasmid encoding the ubiquitin-β-galactosidase fusion protein (Ub-βgal; UFD substrate) was kindly provided by E. S. Johnson (17). A cDNA for Ub-βgal tagged at its COOH terminus with the hemagglutinin (HA) epitope and the His6 epitope were generated by PCR and subcloned into pCI-neo (Promega) and pBacPAC9 (Clontech). Complementary DNAs for the K29R and K48R mutants of the UFD substrate were constructed with the use of a Quick Change kit (Stratagene) and mutated oligonucleotide primers corresponding to each site. Construction of pcDNA3-FLAG-UFD2a(ΔU) was described previously (9).
Transfection, immunoprecipitation, and immunoblot analysis.
HEK293T cells were transfected by the calcium phosphate method and subjected to immunoprecipitation as described previously (35). Immunoblot analysis was performed with anti-HA (1 μg ml−1; HA.11/16B12, Babco) and anti-ubiquitin (1 μg ml−1; FK2, Nippon Bio-Test Laboratories).
Cycloheximide chase analysis.
HEK293T cells were transfected with 9 μg of pCI-neo-Ub-βgal-HA and either 1 μg of pcDNA3 or 1 μg of pcDNA3-FLAG-UFD2a(ΔU) with the use of the FuGene 6 reagent (Roche). After 24 h, the cells were treated with cycloheximide (100 μg ml−1) for 0 to 6 h and then subjected to immunoblot analysis with anti-HA (1 μg ml−1), anti-UFD2a (1 μg ml−1), and anti-α-tubulin (1:2,000 dilution).
Microarray analysis.
Total RNA was extracted from embryos by the guanidinium thiocyanate-phenol-chloroform method and purified. Microarray analysis was performed according to the protocol recommended by the array manufacturer (Agilent). In brief, 5 μg of total RNA was converted to double-stranded cDNA, which was then used to generate cRNA labeled with cyanin 3-CTP or cyanin 5-CTP (Perkin-Elmer) with the use of an Agilent Fluorescent Linear Amplification kit. Linearly amplified Cy3- or Cy5-labeled cRNA was purified, fragmented, and subjected to hybridization with Agilent Mouse Development Oligonucleotide Microarrays with the use of an in situ hybridization kit (Agilent). The arrays were then washed first with nonstringent buffer (6× standard saline citrate and 0.005% Triton X-102) and then with stringent buffer (0.1× standard saline citrate and 0.005% Triton X-102) and were then scanned with an Agilent DNA Microarray scanner. The initial absolute and comparative analyses were performed with images obtained from the scanned arrays with Agilent Feature Extraction software. Pairwise comparisons between individual mice were made with Excel software as recommended by Agilent. We analyzed two sets of Cy3-labeled wild-type cRNA and Cy5-labeled Ufd2a−/− cRNA, as well as two sets of color-swapped cRNA, for a total of four independent analyses. Reproducible differences of >1.5-fold in gene expression were considered significant and are included below (see Table 2).
TABLE 2.
Selected genes whose expression level differs between Ufd2a−/− and wild-type mouse embryos at E11.5 as revealed by microarray analysis
| Accession no. | Gene product | Change in expressiona |
|---|---|---|
| Transcription factors | ||
| AF179425.1 | GATA-6 | − |
| NM_013601.1 | Homeobox, msh-like 2 (Msx2) | − |
| H3050G01-5 | Zinc finger protein 68 (Zfp68) | − |
| H3147E12-5 | Zfp207 | − |
| H3085C10-3 | Zfp238 | − |
| H3094D05-3 | Msx-interacting Zfp | − |
| H3128C07-3 | Zfp36, C3H type-like 1 | − |
| U43714.1 | Helix-loop-helix transcription factor (Hxt) | − |
| H3146G01-3 | Dp 1 | − |
| H3152G11-3 | Leucine zipper protein 1 | − |
| Signal transduction | ||
| H3014C07-3 | Brain natriuretic peptide | − |
| AF286725.1 | Platelet-derived growth factor, C polypeptide | − |
| L0504D06-3 | Atrial natriuretic factor | + |
| K0336D02-3 | Interferon-activated gene 204 | + |
| K0439H07-3 | Interferon-activated gene 202A | + |
| Stress response K0424F03-3 | DnaJ (Hsp40) homolog, subfamily C, member 1 | + |
| Matrix or structural proteins | ||
| C0503D01-3 | Protocadherin beta 17 | − |
| H3121E07-5 | Protocadherin 18 | − |
| Others | ||
| H3047D02-3 | Albumin | − |
| H3114E11-3 | Histone deacetylase 1 | − |
| H3009F07-3 | Kinesin family member 5B | − |
| C0624G12-3 | Kinesin family member 21A | − |
| K0430G01-3 | Dynactin 4 | − |
| H3026C09-3 | Ubiquitin-specific protease 23 | − |
Genes are grouped according to the function of the encoded protein. + and − indicate genes whose expression was up- or down-regulated, respectively, in Ufd2a−/− embryos compared with wild-type embryos.
qRT-PCR analysis.
Total RNA was extracted as described above, and 1 μg was used for cDNA synthesis primed with random hexanucleotide primers (ReverTra Ace α; Toyobo). The cDNA was added to a quantitative reverse transcription-PCR (qRT-PCR) mixture that contained 1× SYBR Green PCR master mix (Applied Biosystems) and 100 nM gene-specific primers. Assays were performed in triplicate with an ABI Prism 7700 sequence detector (Applied Biosystems). The PCR protocol comprised incubations at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles, each consisting of 95°C for 15 s and 60°C for 1 min. The expression level of each target gene was normalized by that of the glyceraldehyde-3-phosphate dehydrogenase gene. Specific PCR primer pairs (5′ to 3′) were as follows: glyceraldehyde-3-phosphate dehydrogenase, GCC TGG AGA AAC CTG CCA AGT ATG and GAG TGG GAG TTG CTG TTG AAG TCG; GATA4, CAG AAA ACG GAA GCC CAA GAA CCT and GAG TTA CCG CTG GAG GCA CCA CTG; GATA6, GCA GGC CCC TCA TCA AGC CAC AGA AGC and AAG CAT TGC ACA CAG GCT CAC CCT CAG; ANF, TTC CTC GTC TTG GCC TTT TGG CTT and CCT CAT CTT CTA CCG GCA TCT TCT; Msx2, ACC GAA GGG CTA AGG GGA AAA GAC and CAT AGA GTC CAA CAG GCG GGA TGG; α-myosin heavy chain, GCT CCC TCA ATG ACT TCA CCA CAC and CCT TCC TCC TCC AGT TGC CTC TTG; myosin light chain 2a, CCA GGG GGT GGT GAA CAA GGA AGA GTT and TCA GGC ACA GAG TTT ATT GAG GTG ACC; and myosin light chain 2v, GGG AGA TGC TGA CCA CAC AAG CAG and AGG CTG TGG TTC AGG GCT CAG TCC.
In vitro ubiquitylation assay.
Recombinant Ub-βgal-HA-His6, Ub(K29R)-βgal-HA-His6, and Ub(K48R)-βgal-HA-His6 were expressed and purified with the use of a baculovirus expression system as described previously (9). Primary mouse embryonic fibroblasts (MEFs) were obtained from embryos at E12.5 and cultured as previously described (34). Confluent MEFs in 15-cm culture dishes were harvested and lysed in 100 μl of a solution containing 20 mM Tris-HCl (pH 7.4) and 0.1 mM dithiothreitol. After several freeze-thaw cycles, the lysate was centrifuged at 100,000 × g for 4 h, and the resulting supernatant (S100 lysate) was used as a source of E3 and E4. The in vitro ubiquitylation assay was performed as described with some modifications (24). In brief, reaction mixtures (20 μl) containing 0.5 μg of UFD substrate, 4 μg of S100 lysate protein, 0.1 μg of recombinant rabbit E1 (Boston Biochem), 1 μl of a crude lysate of Escherichia coli expressing human UbcH5C, 0.5 U of phosphocreatine kinase, 1 μg of bovine ubiquitin (Sigma-Aldrich), 25 mM Tris-HCl (pH 7.5), 120 mM NaCl, 2 mM ATP, 1 mM MgCl2, 0.3 mM dithiothreitol, and 1 mM creatine phosphate were incubated for 1.5 h at 30°C. Samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 6% gel and subjected to immunoblot analysis with anti-HA (1 μg ml−1).
Behavioral testing.
For footprint analysis, front and hind paws of the test animals were dipped in red and black nontoxic water-soluble paint, respectively, and the mice were allowed to walk on a replaceable strip of white paper. Animals were also tested with a Rotamex Rotarod (Ugo Basile) on four consecutive days during the light phase of a 12-h light, 12-h dark cycle. We performed four trials each day, with breaks of at least 1 h between tests. In each trial, four mice were placed in separate chambers on the resting rod before rotation was initiated. After 5 s of constant rotation at 4 rpm, the speed was increased gradually over the course of 5 min to 40 rpm. The timer was stopped either automatically if the mouse fell from the rod or manually if the mouse gripped the rod and started rotating with it. We analyzed five mice of each age and averaged the results.
RESULTS
To elucidate the physiological function of UFD2a, we generated mice deficient in this protein. The Ufd2a gene was disrupted in mouse embryonic stem cells by replacement of the last five exons, which encode the U-box domain (1, 19), with a neo cassette (Fig. 1A). The ratio of heterozygous mice to wild-type mice appeared normal at birth, but no homozygous mutants were detected among 234 newborn animals produced from heterozygote crosses (Fig. 1B). Ufd2a−/− embryos were detected in utero (Fig. 1C) and appeared normal in morphology until E10.5 or E11.5; most of the mutant embryos were grossly abnormal after E12.5, however (Table 1). The mutation thus appeared to be lethal in the homozygous state. Immunoblot analysis of embryo lysates at E11.5 revealed the presence of UFD2a in Ufd2a+/+ and Ufd2a+/− embryos but not in Ufd2a−/− embryos (Fig. 1D). However, a faint band corresponding to a protein of <131 kDa was detected with both Ufd2a+/− and Ufd2a−/− embryos but not with Ufd2a+/+ embryos, suggesting that a truncated UFD2a protein was produced from the mutant allele. Given that the abundance of the truncated form of UFD2a appeared much less (<10%) than that of the wild-type protein in Ufd2a+/− embryos, it is likely unstable. To confirm that the truncated form of UFD2a did not affect the function of the wild-type protein, we examined whether the stability of the UFD substrate (Ub-βgal) was influenced by expression of the UFD2a(ΔU) mutant, which lacks the U-box domain and mimics the truncated form of UFD2a, in HEK293T cells. The turnover of Ub-βgal in cycloheximide-treated cells appeared unaffected by expression of UFD2a(ΔU) at a level about one-third of that of the endogenous UFD2a protein (Fig. 1E). These data suggest that the truncated form of UFD2a does not act in a dominant negative manner.
TABLE 1.
Frequency of genotypes in offspring of Ufd2a+/− intercrossesa
| Stage | Genotype
|
Total | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Live born | 75 | 159 | 0 | 234 |
| Embryos | ||||
| E9.5 | 3 | 5 | 6 (0) | 14 |
| E10.5 | 12 | 28 | 15 (0) | 55 |
| E11.5 | 23 | 55 | 19 (7) | 97 |
| E12.5 | 24 | 77 | 31 (25) | 132 |
| E13.5 | 11 | 19 | 6 (6) | 36 |
| E14.5 | 5 | 9 | 3 (3) | 17 |
Numbers in parentheses indicate embryos that were grossly abnormal.
Immunohistochemical analysis of wild-type embryos at E10.5 showed that UFD2a was largely restricted to cardiac tissue (Fig. 2A), being especially abundant in cardiomyocytes (Fig. 2B and C). UFD2a was not expressed in the neural tube of wild-type embryos at E10.5 (Fig. 2A), whereas it was highly expressed in the adult brain (19). We also performed in situ hybridization to examine the distribution of UFD2a mRNA in wild-type embryos at E10.5 and E12.5 (unpublished data). In contrast to the restricted expression of the UFD2a protein in cardiac muscle, UFD2a mRNA was detected in many organs, including the heart, and was also abundant in limb buds, the branchial arch, and brain. Given that UFD2a undergoes autoubiquitylation (9, 37), its abundance in different organs is thus likely regulated at the level of protein stability.
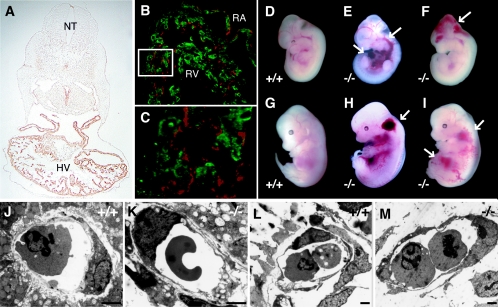
FIG. 2.
Vascular abnormalities of Ufd2a−/− embryos. (A) Immunohistochemical analysis of UFD2a expression in a wild-type embryo at E10.5. NT, neural tube; HV, heart ventricle. (B and C) Immunofluorescence analysis of UFD2a (green) and platelet endothelial cell adhesion molecule 1 (a marker for endothelial cells; red) in cardiac tissue of a wild-type embryo at E10.5. The boxed region in panel B is shown at higher magnification in panel C. RA, right atrium; RV, right ventricle. (D to I) Gross morphology of wild-type embryos at E12.5 (D) and E13.5 (G), as well as of Ufd2a−/− embryos at E12.5 (E and F) and E13.5 (H and I). Arrows indicate the various regions of hemorrhage apparent in Ufd2a−/− embryos. (J to M) Electron microscopic examination of vascular sections of the yolk sac (J and K) and brain (L and M) from Ufd2a+/+ (J and L) and Ufd2a−/− (K and M) embryos at E11.5. Scale bars, 2 μm.
Ufd2a−/− embryos at E12.5 or E13.5 manifested dilation of veins, as well as massive hemorrhage in the brain ventricles and in cervical and abdominal subcutaneous tissue (Fig. 2D to I). Electron microscopic examination of vascular sections of the brain and yolk sac at E11.5 revealed no abnormalities in the structure of endothelial cell-cell junctions or in investment by supporting cells (Fig. 2J to M). These data thus exclude the possibility that vascular integrity is lost in the mutant embryos. The heart of the mutant embryos was also enlarged and associated with pericardial effusion, indicative of congestive heart failure. Histopathologic examination revealed that the heart of Ufd2a−/− embryos also exhibited reduced trabeculation, as well as an undeveloped and compact myocardial layer (Fig. 3A to D). The pericardial effusion was first evident at E11.5 (Fig. 3A and B), and the marked defect in the thickness of the myocardial layer was apparent at E12.5 (Fig. 3C and D). The TUNEL assay revealed that most cardiomyocytes underwent apoptosis in Ufd2a−/− embryos at E13.5 (Fig. 3E to H). Consistent with the expression pattern of UFD2a, tissues other than the heart did not exhibit increased levels of apoptosis in Ufd2a−/− embryos. Incorporation of bromodeoxyuridine into cardiac tissue of Ufd2a−/− embryos appeared similar to that in wild-type embryos (data not shown), suggesting that the impaired growth of cardiac muscle is attributable to apoptotic cell death rather than to decreased cell proliferation.
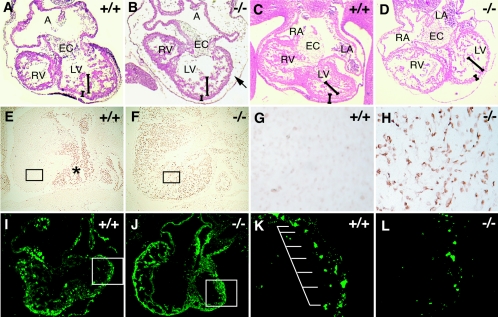
FIG. 3.
Impaired development of cardiac muscle in Ufd2a−/− embryos. (A to D) Hematoxylin-eosin staining of transverse cardiac sections of wild-type (A and C) and Ufd2a−/− (B and D) embryos at E11.5 (A and B) or E12.5 (C and D). The arrow in panel B indicates edema and bleeding in the pericardial space. Long bars indicate the trabecular layer, and short bars indicate the ventricular myocardial layer. A, atrium; EC, endocardial cushion; LV, left ventricle; LA, left atrium. (E to H) TUNEL staining of transverse cardiac sections of wild-type (E and G) and Ufd2a−/− (F and H) embryos at E13.5. Boxed regions (E and F) are shown at higher magnifications in panels G and H, respectively. The asterisk (E) indicates blood cells with endogenous peroxidase activity. (I to L) Immunofluorescence staining of cardiac sections of wild-type (I and K) and Ufd2a−/− (J and L) embryos at E11.5 with anti-sarcomeric α-actinin. Boxed regions (I and J) are shown at higher magnification in panels K and L. Sarcomeric α-actinin immunoreactivity was arrayed with regular spacing in the heart of wild-type embryos but was sparse and irregularly distributed in that of Ufd2a−/− embryos.
In Caenorhabditis elegans, UFD-2 contributes to myosin assembly through regulation of the myosin-directed chaperone UNC-45 (14). We examined myofibril assembly in cardiac muscle of wild-type and Ufd2a−/− embryos at E11.5 by immunofluorescence staining with antibodies to sarcomeric α-actinin, a well-characterized marker for myofibril assembly (5, 6, 13). In the heart of wild-type embryos, staining for sarcomeric α-actinin was detected in a periodic punctate pattern (7), whereas most signals appeared to be disorganized and irregular in the heart of Ufd2a−/− embryos (Fig. 3I to L). These data suggest that regulation of the abundance of mouse UNC-45 is impaired in Ufd2a−/− embryos, resulting in defective assembly of myosin in cardiac muscle cells. The defect in myofibril assembly may lead to the massive apoptosis apparent in cardiac muscle of Ufd2a−/− embryos (Fig. 3E to H). Abnormalities in heart development were not observed with Ufd2a+/− mice (data not shown). The de novo formation of blood vessels that gives rise to the primary capillary plexus and the process of angiogenic remodeling appeared to occur normally in Ufd2a−/− embryos (data not shown). The defect in cardiac development thus likely accounts for the midgestation death of UFD2a-deficient mice.
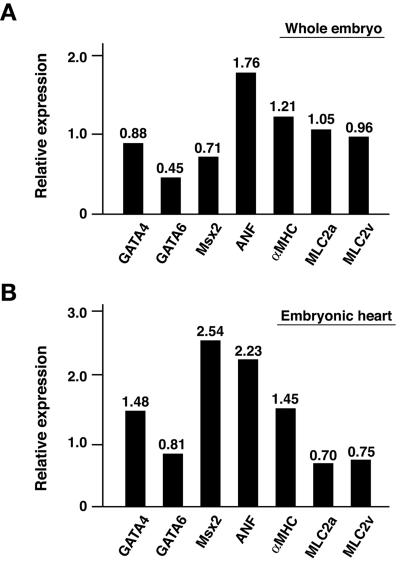
Mouse embryos at E11.5, a stage at which wild-type and Ufd2a−/− embryos were indistinguishable by gross morphology, were subjected to microarray analysis of gene expression. Of a total of 20,371 genes analyzed, the expression of 169 genes (0.83%) was significantly increased (93 genes) or decreased (76 genes) by a factor of >1.5 in Ufd2a−/− embryos compared with wild-type embryos (Table 2). The affected genes included that for GATA6, which is a member of the GATA family of zinc finger transcription factors and which has been implicated in regulation of myocardial differentiation during cardiogenesis (31, 39). Quantitative RT-PCR analysis confirmed that the amount of GATA6 mRNA in Ufd2a−/− embryos was only 45% of that in wild-type embryos (Fig. 4A). Microarray and qRT-PCR analyses also showed that the expression of Msx2, a homeobox gene related to the Drosophila muscle segment homeobox gene (a marker of the proximal ventricular conduction system) (4, 22), was decreased by 29% in Ufd2a−/− embryos. Although the mechanism responsible for these differences is unclear, the altered expression of these and other genes that contribute to cardiac development may underlie the defect in this process in Ufd2a−/− embryos. Similar qRT-PCR analysis of the embryonic heart yielded results that were overall consistent with the data from whole embryos, although differences in the expression of some genes, including Msx2, were apparent between the two sources of RNA (Fig. 4B).
FIG. 4.
Quantitative RT-PCR analysis of gene expression in Ufd2a−/− embryos. Expression of genes for the indicated proteins was determined for E11.5 whole embryos (A) or in cardiac tissue derived therefrom (B). Data are expressed relative to the corresponding values for wild-type embryos and are means obtained from six embryos.
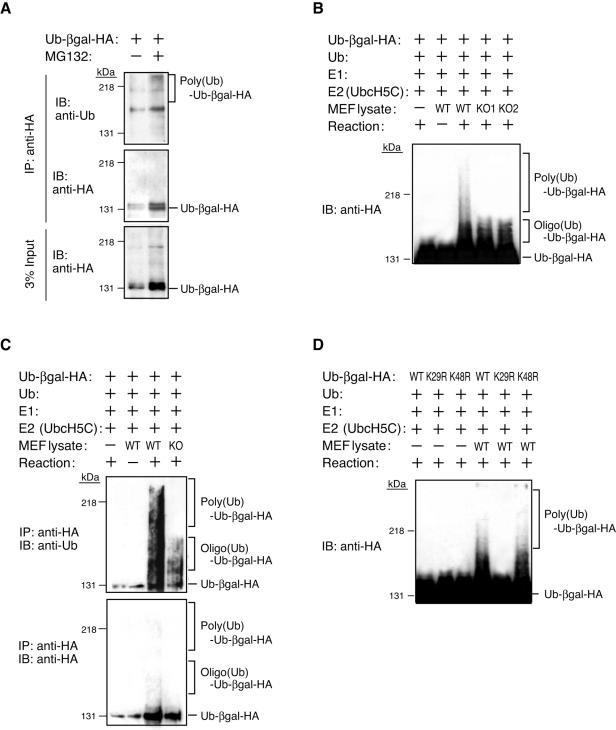
The UFD pathway was identified for S. cerevisiae (17), but whether this pathway is conserved in mammals has remained unknown. We transfected HEK293T cells with an expression vector for HA-tagged UFD substrate and then cultured the cells in the absence or presence of the proteasome inhibitor MG132. The UFD substrate was immunoprecipitated from cell lysates with antibodies to HA and then subjected to immunoblot analysis with anti-ubiquitin. The immunoprecipitate from the MG132-treated cells yielded a smeared signal with anti-ubiquitin (Fig. 5A), indicating that it contained polyubiquitylated UFD substrate. To determine whether the UFD pathway is conserved in mammals, we subjected the HA-tagged UFD substrate to an in vitro ubiquitylation assay with recombinant ubiquitin, E1, and E2, as well as an extract of MEFs as a source of E3 and E4 (Fig. 5B). The UFD substrate underwent marked polyubiquitylation in the presence of an extract of wild-type MEFs. In contrast, an extract of Ufd2a−/− MEFs did not support polyubiquitylation of the UFD substrate, retaining only the ability to mediate its oligoubiquitylation. To confirm that the smeared signal obtained with anti-HA and the reaction mixture containing the extract of wild-type MEFs was attributable to ubiquitylation of the UFD substrate, we subjected the various reaction mixtures to immunoprecipitation with anti-HA before immunoblot analysis with anti-ubiquitin. The immunoprecipitate obtained from the reaction mixture containing the wild-type MEF extract yielded a smeared signal with anti-ubiquitin, whereas the Ufd2a−/− MEF extract again was found to mediate oligoubiquitylation but not polyubiquitylation of the UFD substrate (Fig. 5C).
FIG. 5.
UFD2a is required for polyubiquitylation of the UFD substrate in mammals. (A) HEK293T cells were transiently transfected with an expression vector for HA-tagged UFD substrate (Ub-βgal-HA) and subsequently incubated in the absence or presence of 10 μM MG132 for 6 h, after which cell lysates were subjected to immunoprecipitation (IP) with anti-HA. The resulting precipitates were subjected to immunoblot analysis (IB) with either antiubiquitin or anti-HA. A portion (3%) of the input lysates was also subjected directly to immunoblot analysis with anti-HA. (B) The HA-tagged UFD substrate was subjected to an in vitro ubiquitylation assay with ubiquitin (Ub), E1, E2, and an extract of wild-type (WT) or Ufd2a−/− (KO1 and KO2) MEFs. The reaction mixtures were subjected to immunoblot analysis with anti-HA. The positions of polyubiquitylated and oligoubiquitylated forms of the substrate are indicated. (C) The in vitro ubiquitylation assay was performed as in panel B, but the reaction mixtures were subjected to immunoprecipitation with anti-HA before immunoblot analysis with either antiubiquitin or anti-HA. (D) The in vitro ubiquitylation assay was performed as in panel B, but K29R and K48R mutant forms of the HA-tagged UFD substrate were used in addition to the wild-type form.
Given that Lys-29 of the ubiquitin moiety of the UFD substrate is required for its polyubiquitylation by yeast Ufd4 (E3) and Ufd2 (E4) (21), we examined whether this was also the case with mammalian E3 and E4. We generated mutants of the UFD substrate (K29R and K48R) in which Lys-29 or Lys-48 of the ubiquitin moiety was replaced by arginine and then subjected them to an in vitro ubiquitylation assay with a wild-type MEF extract. The extent of polyubiquitylation of the UFD substrate appeared unaffected by the K48R mutation, whereas the K29R mutant did not undergo ubiquitylation (Fig. 5D). These data suggest that the mechanism of ubiquitylation of the UFD substrate is conserved in mammals and that UFD2a is a functional homolog of yeast Ufd2.
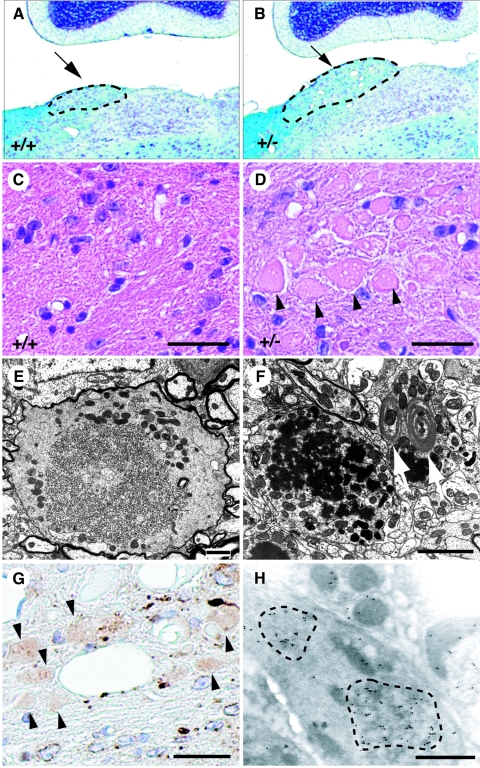
Given that UFD2a is abundant in neurons of adult mice (19), we next examined the brain of Ufd2a+/− mice at ∼12 months of age. Histological analysis revealed that the nucleus gracilis of Ufd2a+/− mice was markedly enlarged compared with that of control mice (Fig. 6A and B). Furthermore, numerous axonal spheroids of various sizes were readily identified on the basis of their eosinophilic profiles and shape in the nucleus gracilis of heterozygous animals (Fig. 6C and D). Although axonal spheroids were sometimes observed with the nucleus gracilis of older wild-type mice, the size of these structures was larger and they were detected as early as 7 months of age with Ufd2a+/− mice. Electron microscopy showed that the enlargement of dystrophic axons in Ufd2a+/− mice was most pronounced in the terminal segment. Many of the affected axons contained abundant neurofilaments, electron-dense bodies, multivesicular profiles, and tubular structures (Fig. 6E and F). Structures resembling Hirano bodies (Fig. 6F), which have been identified in the hippocampal pyramidal cell layers of individuals with various neurodegenerative disorders (11, 12), were also apparent in the dystrophic axons of Ufd2a+/− mice. In addition, we observed a dotlike pattern of staining with antibodies to ERp72, an ER-resident chaperone protein, in the nucleus gracilis of the heterozygotes (Fig. 6G and H). A high level of ERp72 immunoreactivity was often detected in association with axonal spheroids, especially in regions showing accumulation of tubular structures. These data suggest that the abnormal neurons of Ufd2a+/− mice are affected by ER stress. Lumbar dorsal roots and their ganglia, peripheral nerves, and muscles of the hind limbs appeared normal in Ufd2a+/− mice (data not shown).
FIG. 6.
Axonal dystrophy in the nucleus gracilis of Ufd2a+/− mice at 13 months of age. (A to D) Sections of the nucleus gracilis of the medulla oblongata of wild-type (A and C) or Ufd2a+/− (B and D) mice were subjected to Kluver-Barrera (A and B) or hematoxylin-eosin (C and D) staining. The outlined areas indicated by arrows (A and B) represent the nucleus gracilis. Arrowheads (D) indicate eosinophilic spheroids. (E and F) Electron microscopy of dystrophic axons in the nucleus gracilis of Ufd2a+/− mice. Arrows (F) indicate Hirano body-like structures. (G) Immunohistochemical staining with anti-ERp72 of spheroids (arrowheads) in the nucleus gracilis of a Ufd2a+/− mouse. (H) Immunoelectron microscopic detection of ERp72 in aggregations (outlined areas) present within spheroids of the nucleus gracilis of a Ufd2a+/− mouse. Scale bars: 25 μm (C, D, and G), 5 μm (E and F), and 1 μm (H).
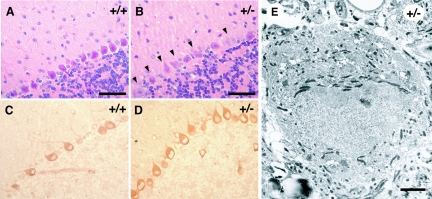
We also observed many degenerated Purkinje cells with a reduced number of dendritic processes projecting into the molecular layer in 13-month-old Ufd2a+/− mice (Fig. 7A and B). Purkinje cells of Ufd2a+/− mice exhibited increased expression of Grp78 (Fig. 7D), a member of the Hsp70 protein family associated with the ER stress response, compared with that apparent in age-matched wild-type mice (Fig. 7C). Electron microscopy also revealed numerous electron-dense bodies in the ER of the dystrophic Purkinje cells of Ufd2a+/− mice (Fig. 7E). These observations suggested that degeneration of Purkinje cells in the heterozygous animals might be induced by ER stress, which in turn might be attributable to an accumulation of misfolded proteins in the ER that results from a reduced rate of degradation regulated by UFD2a-dependent ubiquitylation.
FIG. 7.
Degeneration of Purkinje cells in Ufd2a+/− mice at 13 months of age. (A and B) Hematoxylin-eosin staining of sections of the cerebellum of wild-type (A) or Ufd2a+/− (B) mice. Arrowheads (B) indicate degenerated Purkinje cells. (C and D) Immunohistochemical analysis of Grp78 expression in the cerebellum of wild-type (C) or Ufd2a+/− (D) mice. (E) Electron microscopy of degenerated Purkinje cells in a Ufd2a+/− mouse. Scale bars, 25 μm (A and B) and 1 μm (E).
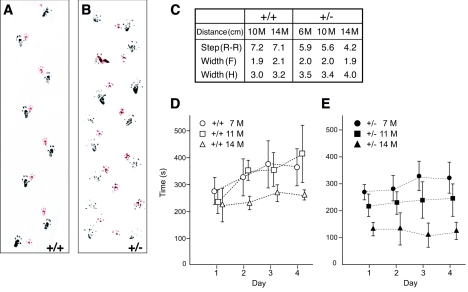
Ufd2a+/− mice developed various neurological symptoms with age. Hind-limb gait abnormalities were apparent from 7 months and were followed at later ages by dragging of the hind legs. These motor symptoms became severe at 12 to 14 months. The abnormal gait of Ufd2a+/− mice was characterized by gait analysis with the use of paint to mark the paws. At 14 months of age, wild-type animals placed their hind paws in almost the same spots as those occupied by the preceding fore paws during walking (Fig. 8A), whereas the steps of Ufd2a+/− mice were irregular (Fig. 8B); the distance between the hind paws was increased from 3.2 cm in wild-type mice to 4.0 cm in the heterozygotes, and the forward step of the right hind paw was reduced from 7.1 to 4.2 cm (Fig. 8C). Ufd2a+/− mice also showed impaired performance in the Rotarod test from 11 months of age compared with age-matched wild-type mice (Fig. 8D and E).
FIG. 8.
Ataxic gait and locomotive defect of Ufd2a+/− mice. (A to C) Gait analysis of 14-month-old wild-type (A and C) and Ufd2a+/− (B and C) mice. The front and hind paws were painted red and black, respectively, and the animals were allowed to walk on a strip of white paper. Data (C) are means determined from five animals of each genotype at 6, 10, or 14 months of age and represent the distances between consecutive positions occupied by the right hind paw [Step(R-R)], between the positions of the two front paws [Width(F)], and between the positions of the two hind paws [Width(H)]. (D and E) Analysis with an accelerating rod apparatus of wild-type (D) and Ufd2a+/− (E) mice. Animals were tested in four trials per day for four consecutive days by measuring the time spent on the rod. Data are means ± 95% confidence intervals, determined for five animals of each genotype at 7, 11, or 14 months of age.
DISCUSSION
The ubiquitin conjugation system mediated by E1, E2, and E3 enzymes has been well characterized. The role of the ubiquitylating enzyme E4 has remained obscure, however, since its recent discovery in yeast (21). It has thus been unclear whether the E4-dependent system of ubiquitylation is conserved in higher eukaryotes, including mammals, and, if so, whether it is important under physiological conditions. To address these questions, we generated mice deficient in UFD2a.
Two mammalian proteins, UFD2a (E4B) and UFD2b (E4A), were identified as homologs of yeast Ufd2 (9, 19). Although both UFD2a and UFD2b show similar levels of amino acid similarity to yeast Ufd2, VCP (mammalian homolog of yeast Cdc48) interacts only with UFD2a (8). UFD2b associates with another chaperone, DnaJc7, but not with VCP. Furthermore, we have now shown that the lack of UFD2a in mouse cells results in the failure to extend a ubiquitin chain formed on the UFD substrate. Together, these structural and functional observations support the notion that UFD2a is the functional ortholog of yeast Ufd2.
Although few studies have addressed the function of mammalian UFD2a, evidence suggests that it plays an important role in the nervous system. First, the natural mouse mutant C57BL/WldS, in which the onset of Wallerian degeneration after neuronal injury is greatly delayed, was found to harbor a chromosomal translocation that results in the production of a fusion protein containing an NH2-terminal fragment of UFD2a fused to nicotinamide mononucleotide adenylyltransferase (3, 26). Although the chimeric WldS protein is responsible for the delay in Wallerian degeneration, it contains only 70 of the 1,173 amino acids of UFD2a, and these residues are absent from the yeast homolog. The region of UFD2a present in WldS is thus unlikely to confer polyubiquitylation activity but may have a related role. Second, UFD2a functions as an E4 in the polyubiquitylation of a pathological form of ataxin-3 and is a rate-limiting factor in ataxin-3 degradation (27). Furthermore, forced expression of UFD2a both promoted the degradation of the pathological form of ataxin-3 in cultured cells and prevented neurodegeneration in a fly model of SCA3. Third, we have recently identified fasciculation and elongation protein zeta 1 (FEZ1) as a protein that interacts with UFD2a (37). FEZ1 is a mammalian homolog of C. elegans UNC-76, which is required for axonal bundling and elongation in the nematode (2). UFD2a mediated the polyubiquitylation of FEZ1 but did not affect its intracellular stability, suggesting that such modification of FEZ1 is not a signal for its proteolysis. Rather, the ubiquitylation of FEZ1 by UFD2a contributes to neuritogenesis in PC12 cells.
Our present results show that mammalian UFD2a is indispensable for normal development of the heart as well as for protection of neurons from axonal degeneration. A neuronal phenotype similar to that of Ufd2a+/− mice was described for gad mice, which harbor an in-frame deletion in the Uchl1 gene (41); these animals thus exhibit gracile axonal dystrophy (GAD), characterized by the “dying-back” type of axonal degeneration and formation of spheroid bodies in nerve terminals (20, 30, 32, 36). UCH-L1 is a member of the ubiquitin COOH-terminal hydrolysis (UCH) family of proteins, and its expression is restricted to the brain and testis (18, 42). UCH-L1 is present in pathological neuronal structures associated with neurodegenerative diseases (25), and missense mutations in the human gene have been detected in a German family with Parkinson's disease (23). These observations suggest that the ubiquitin system may play an important role in maintenance of the nervous system by protecting neurons from degeneration. The WldS protein was recently found to inhibit the GAD phenotype (29), suggesting the existence of a molecular link between UFD2a and UCH-L1.
In yeast, Ufd2 is thought to contribute to cell survival under stressful conditions through its interaction with the AAA-type ATPase Cdc48 (21, 40). In mammals, UFD2a also binds to VCP, an ortholog of Cdc48 (19, 27). Other mammalian U box-type enzymes in addition to UFD2a, including CHIP, UFD2b (E4A), UIP5, and CYC4, also interact with molecular chaperones or cochaperones (8, 9). U-box proteins thus likely play a general role in the ubiquitylation of misfolded proteins, with some of them being thought to participate in the ERAD pathway (15, 16, 28, 33). Yeast Ufd2 was recently shown to be essential for the ERAD pathway (40). We previously showed that ataxin-3 forms a ternary complex with VCP and UFD2a and undergoes ubiquitylation and degradation by the proteasome (27). Our present data now suggest that, by preventing the accumulation of misfolded or otherwise abnormal proteins, especially those associated with aging, UFD2a maintains the intracellular environment of neurons and protects them from ER and other types of cellular stress.
Acknowledgments
We thank E. S. Johnson for the Ub-βgal cDNA; T. Kamura, R. Tsunematsu, and M. P. Coleman for helpful discussions; Y. Yamada, M. Sasaki, K. Tsunematu, S. Matsushita, R. Yada, N. Nishimura, and other laboratory members for technical assistance; and A. Ota and M. Kimura for help in preparing the manuscript.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2000. The U box is a modified RING finger—a common domain in ubiquitination. Curr. Biol. 10:R132-R134. [DOI] [PubMed] [Google Scholar]
- 2.Bloom, L., and H. R. Horvitz. 1997. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc. Natl. Acad. Sci. USA 94:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conforti, L., A. Tarlton, T. G. Mack, W. Mi, E. A. Buckmaster, D. Wagner, V. H. Perry, and M. P. Coleman. 2000. A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proc. Natl. Acad. Sci. USA 97:11377-11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson, D. 1995. The function and evolution of Msx genes: pointers and paradoxes. Trends Genet. 11:405-411. [DOI] [PubMed] [Google Scholar]
- 5.Du, A., J. M. Sanger, K. K. Linask, and J. W. Sanger. 2003. Myofibrillogenesis in the first cardiomyocytes formed from isolated quail precardiac mesoderm. Dev. Biol. 257:382-394. [DOI] [PubMed] [Google Scholar]
- 6.Ehler, E., B. M. Rothen, S. P. Hammerle, M. Komiyama, and J. C. Perriard. 1999. Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J. Cell Sci. 112:1529-1539. [DOI] [PubMed] [Google Scholar]
- 7.Fritz-Six, K. L., P. R. Cox, R. S. Fischer, B. Xu, C. C. Gregorio, H. Y. Zoghbi, and V. M. Fowler. 2003. Aberrant myofibril assembly in tropomodulin1 null mice leads to aborted heart development and embryonic lethality. J. Cell Biol. 163:1033-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatakeyama, S., M. Matsumoto, M. Yada, and K. I. Nakayama. 2004. Interaction of U-box-type ubiquitin-protein ligases (E3s) with molecular chaperones. Genes Cells 9:533-548. [DOI] [PubMed] [Google Scholar]
- 9.Hatakeyama, S., M. Yada, M. Matsumoto, N. Ishida, and K. I. Nakayama. 2001. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276:33111-33120. [DOI] [PubMed] [Google Scholar]
- 10.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 11.Hirano, A. 1994. Hirano bodies and related neuronal inclusions. Neuropathol. Appl. Neurobiol. 20:3-11. [DOI] [PubMed] [Google Scholar]
- 12.Hirano, A., H. M. Dembitzer, L. T. Kurland, and H. M. Zimmerman. 1968. The fine structure of some intraganglionic alterations. Neurofibrillary tangles, granulovacuolar bodies and “rod-like” structures as seen in Guam amyotrophic lateral sclerosis and parkinsonism-dementia complex. J. Neuropathol. Exp. Neurol. 27:167-182. [PubMed] [Google Scholar]
- 13.Holtzer, H., T. Hijikata, Z. X. Lin, Z. Q. Zhang, S. Holtzer, F. Protasi, C. Franzini-Armstrong, and H. L. Sweeney. 1997. Independent assembly of 1.6 microns long bipolar MHC filaments and I-Z-I bodies. Cell Struct. Funct. 22:83-93. [DOI] [PubMed] [Google Scholar]
- 14.Hoppe, T., G. Cassata, J. M. Barral, W. Springer, A. H. Hutagalung, H. F. Epstein, and R. Baumeister. 2004. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell 118:337-349. [DOI] [PubMed] [Google Scholar]
- 15.Imai, Y., M. Soda, S. Hatakeyama, T. Akagi, T. Hashikawa, K. I. Nakayama, and R. Takahashi. 2002. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol. Cell 10:55-67. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, J., C. A. Ballinger, Y. Wu, Q. Dai, D. M. Cyr, J. Hohfeld, and C. Patterson. 2001. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276:42938-42944. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, E. S., P. C. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442-17456. [DOI] [PubMed] [Google Scholar]
- 18.Kajimoto, Y., T. Hashimoto, Y. Shirai, N. Nishino, T. Kuno, and C. Tanaka. 1992. cDNA cloning and tissue distribution of a rat ubiquitin carboxyl-terminal hydrolase PGP9.5. J. Biochem. 112:28-32. [DOI] [PubMed] [Google Scholar]
- 19.Kaneko, C., S. Hatakeyama, M. Matsumoto, M. Yada, K. Nakayama, and K. I. Nakayama. 2003. Characterization of the mouse gene for the U-box-type ubiquitin ligase UFD2a. Biochem. Biophys. Res. Commun. 300:297-304. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi, T., M. Mukoyama, K. Yamazaki, and H. Moriya. 1990. Axonal degeneration of ascending sensory neurons in gracile axonal dystrophy mutant mouse. Acta Neuropathol. 80:145-151. [DOI] [PubMed] [Google Scholar]
- 21.Koegl, M., T. Hoppe, S. Schlenker, H. D. Ulrich, T. U. Mayer, and S. Jentsch. 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635-644. [DOI] [PubMed] [Google Scholar]
- 22.Kwang, S. J., S. M. Brugger, A. Lazik, A. E. Merrill, L. Y. Wu, Y. H. Liu, M. Ishii, F. O. Sangiorgi, M. Rauchman, H. M. Sucov, R. L. Maas, and R. E. Maxson, Jr. 2002. Msx2 is an immediate downstream effector of Pax3 in the development of the murine cardiac neural crest. Development 129:527-538. [DOI] [PubMed] [Google Scholar]
- 23.Leroy, E., R. Boyer, G. Auburger, B. Leube, G. Ulm, E. Mezey, G. Harta, M. J. Brownstein, S. Jonnalagada, T. Chernova, A. Dehejia, C. Lavedan, T. Gasser, P. J. Steinbach, K. D. Wilkinson, and M. H. Polymeropoulos. 1998. The ubiquitin pathway in Parkinson's disease. Nature 395:451-452. [DOI] [PubMed] [Google Scholar]
- 24.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe, J., H. McDermott, M. Landon, R. J. Mayer, and K. D. Wilkinson. 1990. Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. J. Pathol. 161:153-160. [DOI] [PubMed] [Google Scholar]
- 26.Mack, T. G., M. Reiner, B. Beirowski, W. Mi, M. Emanuelli, D. Wagner, D. Thomson, T. Gillingwater, F. Court, L. Conforti, F. S. Fernando, A. Tarlton, C. Andressen, K. Addicks, G. Magni, R. R. Ribchester, V. H. Perry, and M. P. Coleman. 2001. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci. 4:1199-1206. [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto, M., M. Yada, S. Hatakeyama, H. Ishimoto, T. Tanimura, S. Tsuji, A. Kakizuka, M. Kitagawa, and K. I. Nakayama. 2004. Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 23:659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meacham, G. C., C. Patterson, W. Zhang, J. M. Younger, and D. M. Cyr. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3:100-105. [DOI] [PubMed] [Google Scholar]
- 29.Mi, W., B. Beirowski, T. H. Gillingwater, R. Adalbert, D. Wagner, D. Grumme, H. Osaka, L. Conforti, S. Arnhold, K. Addicks, K. Wada, R. R. Ribchester, and M. P. Coleman. 2005. The slow Wallerian degeneration gene, WldS, inhibits axonal spheroid pathology in gracile axonal dystrophy mice. Brain 128:405-416. [DOI] [PubMed] [Google Scholar]
- 30.Miura, H., K. Oda, C. Endo, K. Yamazaki, H. Shibasaki, and T. Kikuchi. 1993. Progressive degeneration of motor nerve terminals in GAD mutant mouse with hereditary sensory axonopathy. Neuropathol. Appl. Neurobiol. 19:41-51. [DOI] [PubMed] [Google Scholar]
- 31.Morrisey, E. E., Z. Tang, K. Sigrist, M. M. Lu, F. Jiang, H. S. Ip, and M. S. Parmacek. 1998. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 12:3579-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukoyama, M., K. Yamazaki, T. Kikuchi, and T. Tomita. 1989. Neuropathology of gracile axonal dystrophy (GAD) mouse. An animal model of central distal axonopathy in primary sensory neurons. Acta Neuropathol. 79:294-299. [DOI] [PubMed] [Google Scholar]
- 33.Murata, S., Y. Minami, M. Minami, T. Chiba, and K. Tanaka. 2001. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2:1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, D. Y. Loh, and K. I. Nakayama. 1996. Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85:707-720. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama, K., H. Nagahama, Y. A. Minamishima, M. Matsumoto, I. Nakamichi, K. Kitagawa, M. Shirane, R. Tsunematsu, T. Tsukiyama, N. Ishida, M. Kitagawa, K. I. Nakayama, and S. Hatakeyama. 2000. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 19:2069-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oda, K., K. Yamazaki, H. Miura, H. Shibasaki, and T. Kikuchi. 1992. Dying back type axonal degeneration of sensory nerve terminals in muscle spindles of the gracile axonal dystrophy (GAD) mutant mouse. Neuropathol. Appl. Neurobiol. 18:265-281. [DOI] [PubMed] [Google Scholar]
- 37.Okumura, F., S. Hatakeyama, M. Matsumoto, T. Kamura, and K. I. Nakayama. 2004. Functional regulation of FEZ1 by the U-box-type ubiquitin ligase E4B contributes to neuritogenesis. J. Biol. Chem. 279:53533-53543. [DOI] [PubMed] [Google Scholar]
- 38.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 39.Pikkarainen, S., H. Tokola, R. Kerkela, and H. Ruskoaho. 2004. GATA transcription factors in the developing and adult heart. Cardiovasc. Res. 63:196-207. [DOI] [PubMed] [Google Scholar]
- 40.Richly, H., M. Rape, S. Braun, S. Rumpf, C. Hoege, and S. Jentsch. 2005. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120:73-84. [DOI] [PubMed] [Google Scholar]
- 41.Saigoh, K., Y. L. Wang, J. G. Suh, T. Yamanishi, Y. Sakai, H. Kiyosawa, T. Harada, N. Ichihara, S. Wakana, T. Kikuchi, and K. Wada. 1999. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat. Genet. 23:47-51. [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson, K. D., S. Deshpande, and C. N. Larsen. 1992. Comparisons of neuronal (PGP 9.5) and non-neuronal ubiquitin C-terminal hydrolases. Biochem. Soc. Trans. 20:631-637. [DOI] [PubMed] [Google Scholar]