Abstract
Nrf2 is a transcription factor critical for the maintenance of cellular redox homeostasis. We have previously found that Nrf2 is a labile protein, and its activation in cells under stress involves mechanisms leading to its stabilization. As a modular protein, Nrf2 possesses distinct transactivation and DNA binding domains essential for its transcriptional activity. In this study, we found that the C-terminal “Neh3” domain of Nrf2 is also important for its activity. Deletion of the last 16 amino acids of the protein completely abolishes its ability to activate both reporter and endogenous gene expression. Using site-directed mutagenesis, we have identified a stretch of amino acids within this region that are essential for its activity and that are found to be conserved across species and among other members of the CNC-bZIP family. Importantly, deletion of the final 16 amino acids of Nrf2 does not influence its dimerizing capability, DNA binding activity, or subcellular localization, although it does increase the half-life of the protein. In addition, this region was found to be important for interaction with CHD6 (a chromo-ATPase/helicase DNA binding protein) in a yeast two-hybrid screen. RNA interference-mediated knockdown of CHD6 reduced both the basal and tert-butylhydroquinone-inducible expression of NQO1, a prototypical Nrf2 target gene. These data suggest that the Neh3 domain may act as a transactivation domain and that it is possibly involved in interaction with components of the transcriptional apparatus to affect its transcriptional activity.
A major mechanism by which cells defend against oxidative insult is mediated by Nrf2, a cap ‘n’ collar (CNC) basic leucine zipper (bZIP) transcription factor. Nrf2 is a labile protein that is stabilized in cells under oxidative stress and that upregulates cytoprotective genes, such as those for NAD(P)H-quinone oxidoreductase (NQO1), glutathione S-transferases (GSTs), glutamate cysteine ligase (GCL), and heme oxygenase 1 (HO-1) (1, 10, 17, 18, 22, 24, 32, 34). Mice lacking Nrf2 are deficient in this coordinated genetic program and so are susceptible to various oxidative-stress-related pathologies, including chemical carcinogenesis, acetaminophen toxicity, hyperoxia, and hemolytic anemia (3, 6, 16, 26).
Nrf2 binds to antioxidant response elements (AREs) in the promoters of its target genes to activate transcription. The ARE has the core sequence 5′-TGACNNNGC-3′ and responds to a wide variety of chemicals with potential to cause oxidative damage (27). The ARE and Nrf2 also drive the constitutive expression of many antioxidant and phase II drug-metabolizing enzymes presumably providing a level of protection required for normal cellular activities (7, 17, 27).
Based upon the homology of cross-species orthologues, Nrf2 has been divided into six domains, Neh1 to Neh6 (11) (Fig. 1A). Neh1 is located in the C-terminal half of the molecule and constitutes the basic DNA binding domain and the leucine zipper for dimerization. The Neh2 domain is located in the proximal N terminus and represents the region through which the repressor protein Keap1 recognizes and targets Nrf2 to a cullin 3 (cul3)-based E3 ubiquitin ligase for ubiquitylation and subsequent degradation by the 26S proteasome (5, 8, 11, 15, 37). Electrophiles disrupt the interaction between Keap1 and the Neh2 domain either by inducing phosphorylation of Nrf2 or by directly modifying cysteine residues in Keap1 (9, 33). Alternatively, it has also been reported that Keap1 may remain bound to the Neh2 domain during oxidative stress but cul3 is displaced from the complex (37). Either way, the result is that Nrf2 is stabilized, leading to an enhanced transcriptional response. Adjacent to Neh2 are the Neh4 and Neh5 regions, which were identified by deletion analysis as transcriptional activation domains (TADs). Although the overall mechanism by which Nrf2 transactivates its target genes is not well understood, it appears to be partly reliant on the abilities of the Neh4 and Neh5 domains to cooperatively recruit CREB binding protein (CBP) to ARE-regulated genes (13). Within the central part of Nrf2 lies the Neh6 domain, which has been shown to function as a degron, mediating the destabilization of the molecule only under conditions of oxidative stress (19).
FIG. 1.
Nrf2CTΔ16 is unable to activate transcription of ARE reporter genes. (A) Domain structure of Nrf2. The numbers shown represent the positions in the rat protein sequence. (B) Alignment of Neh3 domains from the indicated species. The position of the first amino acid in each sequence is shown in parentheses. The 16-amino-acid deletion of Nrf2 is shown below the aligned sequences. (C) Q293 cells were cotransfected with either GSTA2-ARE-CAT (left; 6-well plates) or −1016/nqo15′-luc (right; 96-well plates) and the indicated dose of HA-Nrf2 or HA-Nrf2CTΔ16 plasmid. CAT and luciferase assays were performed using standard procedures. Activity is shown as activation (n-fold) over the level obtained when reporter constructs were transfected alone (Ctrl), and results are the mean plus standard deviation of three independent experiments. (D) CAT lysates from panel C were resolved by SDS-polyacrylamide gel electrophoresis, blotted, and then probed with antibodies against either the HA epitope or GAPDH. (E) Q293 cells were cotransfected with either GSTA2-ARE-CAT (left; 6-well plates) or −1016/nqo15′-luc (right; 96-well plates), and the indicated dose of HA-Nrf2 plasmid in either the presence or absence of increasing doses of vector encoding HA-Nrf2CTΔ16, as indicated. Assays were performed and results are presented as described for panel C.
By contrast to these regions, the C terminus of Nrf2 (the Neh3 domain), which presumably harbors a critical activity, since it is highly conserved (Fig. 1B), has not been well characterized. In this report, we demonstrate that the Neh3 domain is important for the transcriptional activity of the protein. Deletion of the final 16 amino acids of Nrf2 gives rise to a molecule that is transcriptionally silent but localizes normally to the nucleus and binds DNA. We used site-directed mutagenesis to identify a short stretch of amino acids (the VFLVPK motif) that is essential for the activity of Neh3. This region was found to be important for protein-protein interaction in a yeast two-hybrid screen, and our data suggest that this motif may be involved in recruiting components of the transcriptional complex.
MATERIALS AND METHODS
Chemicals and antibodies.
Cycloheximide and tert-butylhydroquinone (t-BHQ) were obtained from Sigma. MG-132 was obtained from Calbiochem, and doxycycline (Dox) was supplied by BD Biosciences. Antibodies against the hemagglutinin (HA) epitope were obtained from Roche Applied Science, and anti-V5 antibodies used in Western blotting were obtained from Invitrogen. Immobilized anti-V5 antibodies used for immunoprecipitation were obtained from Sigma. Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies were obtained from Research Diagnostics Inc. Blasticidin and hygromycin B were both obtained from Invitrogen.
Plasmid constructs.
Plasmids encoding full-length Nrf2 or Nrf2CTΔ16 tagged with the HA epitope at their N termini were generated by inserting the PCR-amplified rat cDNA into pHM6 (Roche Applied Science). To generate expression constructs containing HA epitope-tagged alanine substitution mutants of Nrf2, site-directed mutagenesis was performed using the QuikChange kit (Stratagene) with pHM6 containing the full-length rat Nrf2 cDNA as a template. For generation of 293-FlpIn TRex cell lines, Nrf2 or Nrf2CTΔ16 tagged with the HA epitope was PCR amplified and cloned into pcDNA5/FRT/TO (Invitrogen). MafK and Keap1 expression plasmids, along with ARE-CAT and ARE-luciferase reporter constructs, were described previously (9, 21, 24, 27). Gal4-DNA binding domain Nrf2 fusion constructs were obtained by inserting the relevant PCR-amplified Nrf2 cDNA into pGBKT7 (BD Biosciences). The V5-tagged CHD6 expression construct was generated by amplifying a short fragment of the corresponding cDNA (548 to 1,153 bp) and cloning it into pCDNA3.1/V5-His (Invitrogen). All plasmid constructs were verified by DNA sequencing.
Cell culture.
Q293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, both of which were obtained from Invitrogen. 293/HA-Nrf2, 293/HA-Nrf2CTΔ16, and 293/CAT were generated by cotransfecting pcDNA5/FRT/TO containing cDNAs encoding HA-Nrf2, HA-Nrf2CTΔ16, or CAT with pOG44 (Invitrogen), which encodes Flp recombinase, into the parental FlpIn TREx 293 cell line (Invitrogen). The parental cells contain a single Flp recombinase target (FRT) site located at a transcriptionally active genomic locus to which a gene of interest can be targeted. Stable transfectants were selected and maintained in Dulbecco's modified Eagle's medium containing 10% tetracycline-reduced fetal bovine serum (BD Biosciences), 15 μg/ml blasticidin, and 150 μg/ml hygromycin B. Transfections were performed using Lipofectamine 2000 reagent (Invitrogen) as specified by the manufacturer.
Immunocytochemistry.
Q293 cells were seeded on coverslips in six-well dishes and transfected as described above. Forty-eight hours posttransfection, the cells were fixed with formaldehyde (3.7%; 10 min), permeabilized with Triton X-100 (1%; 5 min), and then blocked with 0.1% (wt/vol) bovine serum albumin in phosphate-buffered saline for 20 min. The cells were subsequently incubated with anti-HA for 1 h and at the same time treated with RNase A. After being washed, the cells were incubated with Alexafluor 488-conjugated secondary antibodies (Invitrogen) in the presence of propidium iodide for 1 h. After being washed, coverslips were air dried and mounted using Prolong Gold antifade reagent (Invitrogen). Fluorescent images were obtained by confocal laser scanning microscopy (DM IRBE; Leica).
Immunoprecipitation and immunoblot analysis.
Cells were cultured and transfected as described above. For immunoprecipitation, cells were lysed in immunoprecipitation (IP) buffer (1% NP-40, 150 mM NaCl, 50 mM Tris, pH 7.5) for 30 min on ice. Following clearing of lysates, 10% of the total volume was retained as input, and then the specified antibody was added and reaction mixtures were incubated at 4°C overnight with rotation. IPs were washed three times with IP buffer before complexes were eluted with sodium dodecyl sulfate (SDS) sample buffer. IPs using in vitro-translated proteins were conducted as described previously (9). For immunoblot analysis, cells were lysed directly in SDS sample buffer and resolved through 10% NuPAGE gels (Invitrogen). Western blotting was performed using standard procedures.
TaqMan analyses.
Total RNA was isolated using Trizol reagent (Invitrogen) and then reverse transcribed using Superscript-RT (Invitrogen) and random hexamers (Promega). The relative levels of NQO1 and GCLM mRNAs were determined using the ABI Prism 7700 Sequence Detection System (Applied Biosystems). The following probe-primer sets were used: for NQO1, the forward primer was 5′-GCTGCCATGTATGACAAAGG-3′, the reverse primer was 5′-CACTCTGAATTGGCCAGAGA-3′, and the probe was 5′-[6-carboxyfluorescein (FAM)]-CCACTGGTGGCAGTGGCTCC-[6-carboxytetramethylrhoda mine (TAMRA)]-3′; for GCLM, the forward primer was 5′-GCGAGGAGCTTCATGATTG-3′, the reverse primer was 5′-TGGAAACTCCCTGACCAAAT-3′, and the probe was 5′-[FAM]-TGAATGGAGTTCCCAAATCAACCCA-[TAMRA]-3′; for CHD6, the forward primer was 5′-TCAAACAGCACTGCAACAAA-3′, the reverse primer was 5′-TAGTCGATGTCAGGCAGAGG-3′, and the probe was 5′-[FAM]-CATCCAGCTCCCTGGCAGGA-[TAMRA]-3′. The primer-probe set used for detection of Nrf2 has been described previously (22). A primer-probe set for 23hbp was obtained from Applied Biosystems and used as an internal control.
Gel shifts.
Gel shifts were performed essentially as described previously (20, 21) using a 43-base-pair, double-stranded oligonucleotide containing the ARE sequence of the rat NQO1 gene as a probe. The oligonucleotides were labeled at the 5′ end using [γ-32P]ATP and T4 polynucleotide kinase. In vitro-translated proteins were generated using the TNT-T7 coupled wheat germ extract system (Promega). In competition experiments, unlabeled DNA at a 10-, 20-, or 50-fold molar excess was preincubated with protein mixtures for 20 min at 25°C prior to the addition of the labeled probe. Reactions were fractionated using a 6% nondenaturing polyacrylamide gel in Tris-borate-EDTA.
Yeast two-hybrid screening.
A human liver Gal4 activation domain cDNA library (BD Biosciences) was screened with DBD-Nrf2 as specified by the manufacturer. Positive interactions were screened for on synthetic dropout (SD)-Leu-Trp-His-Ade-5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-α-Gal) plates, and cDNAs were isolated using the BD Biosciences Yeastmaker yeast plasmid isolation kit. In mating experiments, yeast bearing Gal4-activation domain and Gal4-DNA binding domain plasmids were grown overnight together in yeast extract-peptone-dextrose-adenine. Diploid yeasts were selected on SD-Leu-Trp plates and subsequently streaked onto SD-Leu-Trp-His-Ade-X-α-Gal plates to detect interacting proteins.
RNA interference (RNAi).
HeLa cells were seeded in six-well plates at a density of 2.1 × 105 cells/well. After allowing the cells to attach for 2 h, double-stranded RNAs corresponding to CHD6, lamin A/C, or a nonspecific control (all from Dharmacon) were transfected into cells using Lipofectamine 2000 reagent (Invitrogen) as specified by the manufacturer. After 48 h of incubation, the cells were treated with either 0.1% (vol/vol) dimethyl sulfoxide (DMSO) or 50 μM t-BHQ and incubated for a further 24 h prior to being harvested.
RESULTS
The C terminus of Nrf2 is essential for transcriptional activation.
The C-terminal Neh3 domain of Nrf2 is highly conserved between rat, chicken, zebrafish, human, and mouse orthologues (Fig. 1B), but no function has been ascribed to it. To explore a role for this domain in the activity of Nrf2, we deleted the last 16 amino acids (Nrf2CTΔ16) (Fig. 1B) and tested the ability of the mutant protein to activate transcription. We reasoned that since Neh1 and Neh3 are contiguous, this small deletion at the proximal C terminus should cause minimum disruption to the dimerization and DNA binding activities of Nrf2. Human Q293 cells were cotransfected with increasing amounts of plasmids encoding either HA-tagged Nrf2CTΔ16 or full-length Nrf2 and either the GSTA2 ARE-CAT or nqo1 ARE-luciferase reporter gene. As expected, wild-type Nrf2 increased reporter gene activity in a dose-dependent manner (Fig. 1C). Remarkably, even though the previously defined transactivation domains (Neh4 and Neh5) were still intact, Nrf2CTΔ16 failed to activate transcription of either ARE-dependent reporter gene (Fig. 1C). The mutation had no effect on the expression levels of Nrf2 (Fig. 1D).
Because Nrf2CTΔ16 fails to activate gene transcription, we investigated whether its expression might have a dominant-negative effect on wild-type Nrf2-driven gene expression. We cotransfected cells with expression plasmids for both proteins and determined ARE-dependent reporter gene activity. With a fixed amount of full-length Nrf2 plasmid present, increasing amounts of Nrf2CTΔ16 vector in the transfection led to a dose-dependent decrease in reporter gene activity (Fig. 1E). This was true of both the GSTA2 and nqo1 reporter constructs. These results indicate that Nrf2CTΔ16 can effectively compete with wild-type Nrf2 for binding to the ARE.
Nrf2CTΔ16 heterodimerizes with MafK and binds the ARE in vitro.
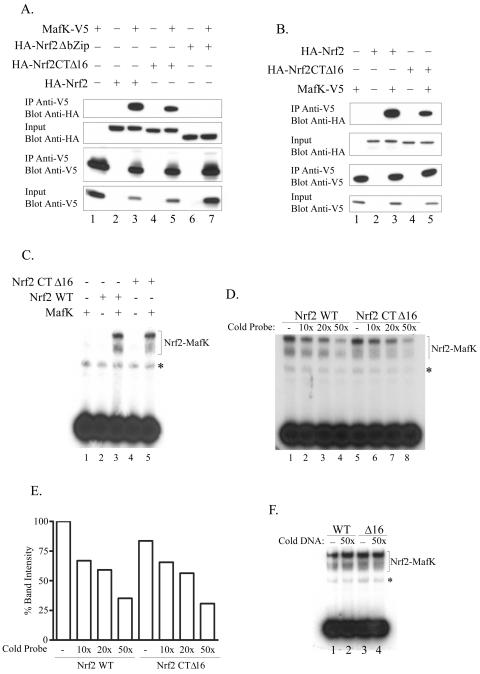
To confirm that Nrf2CTΔ16 retains its ability to bind to DNA, we first assessed whether Nrf2CTΔ16 could dimerize with MafK. We cotransfected cells with plasmids for HA epitope-tagged Nrf2, Nrf2CTΔ16, or Nrf2 with a deleted bZIP region (Nrf2ΔbZIP) and V5-tagged MafK. Following harvesting, whole-cell lysates were incubated with immobilized V5 antibodies, and the IPs were analyzed by Western blotting. The results of this analysis show that full-length Nrf2 and Nrf2CTΔ16 specifically associate with MafK, whereas Nrf2ΔbZIP does not (Fig. 2A). Blotting of input samples with either HA or V5 antibodies revealed that all of the proteins were expressed in the expected cells and at similar levels (Fig. 2A). This experiment was repeated using in vitro-translated proteins with identical results (Fig. 2B).
FIG. 2.
Nrf2CTΔ16 interacts with MafK and binds DNA with affinity similar to that of wild-type Nrf2. (A) Lysates were prepared from Q293 cells expressing HA-tagged wild-type Nrf2, Nrf2CTΔ16, or Nrf2ΔbZip in the presence or absence of V5-tagged MafK, as indicated. Lysates were immunoprecipitated with anti-V5 antibody, and immune complexes were blotted and then probed with either HA or V5 antibodies as shown. Samples of lysates prior to IP were also retained, blotted, and probed with HA or V5 antibodies (input samples). (B) HA-Nrf2, HA-Nrf2CTΔ16, and MafK-V5 were in vitro transcribed/translated and then mixed in the indicated combinations and incubated for 15 min at 30°C. The complexes were then immunoprecipitated with anti-V5 antibodies and blotted and then probed with anti-HA or anti-V5 antibodies. A portion of each sample (10%) was removed prior to IP (input) and was blotted and probed with either anti-HA or anti-V5 antibodies. (C) The indicated combinations of in vitro-transcribed/translated proteins were incubated with radiolabeled ARE probe before being resolved. The Nrf2-MafK and Nrf2CTΔ16-MafK shifted complexes are indicated, and a nonspecific complex is shown with an asterisk. (D) Nrf2-MafK or Nrf2CTΔ16-MafK complexes were preincubated with the indicated molar excess of unlabeled ARE probe prior to addition of radiolabeled DNA. Complexes were resolved, and the specific shifted bands are indicated along with a nonspecific complex shown with an asterisk next to it. (E) Quantitation of the results shown in panel D by densitometry. (F) Nrf2-MafK (wild type [WT]) or Nrf2CTΔ16-MafK (Δ16) complexes were preincubated with the indicated molar excess of unlabeled mutant ARE DNA prior to the addition of radiolabeled probe. Complexes were resolved, and the specific shifted bands are indicated, along with a nonspecific complex highlighted with an asterisk.
We next asked whether Nrf2CTΔ16 could bind DNA using gel shift assays. Incubation of in vitro-translated Nrf2, Nrf2CTΔ16, or MafK alone with the radiolabeled ARE probe did not produce a detectable shifted complex. However, heterodimers of Nrf2-MafK or Nrf2CTΔ16-MafK interacted specifically with the ARE, producing two shifted bands, as previously reported (21) (Fig. 2C). Importantly, no gross differences in band intensity could be observed between the wild-type Nrf2-MafK and Nrf2CTΔ16-MafK complexes. To gain some insight into the affinity of the two complexes for the ARE, competition assays were conducted. When a 10-, 20-, or 50-fold molar excess of unlabeled probe was added to the binding reaction, a dose-dependent decrease in band intensity that was comparable between Nrf2-MafK and Nrf2CTΔ16-MafK complexes was observed (Fig. 2D). Quantitative analysis of band intensities by densitometry showed that they were very similar across the range of competitor concentrations used (Fig. 2E). The Nrf2-MafK and Nrf2CTΔ16-MafK complexes were not competed when a probe containing a mutated ARE was used (Fig. 2F). These findings indicate that Nrf2CTΔ16 binds to the ARE with affinity similar to that of wild-type Nrf2.
Nrf2CTΔ16 localizes to the nucleus.
The fact that Nrf2CTΔ16 acts as a dominant-negative mutant implies that, like wild-type Nrf2, the protein is targeted to the nucleus. To confirm this possibility, the localizations of Nrf2CTΔ16 and wild-type Nrf2 were examined by immunocytochemistry and confocal microscopy. Plasmids encoding either HA-tagged wild-type Nrf2 or Nrf2CTΔ16 were transiently transfected into Q293 cells and, following fixation, their distribution was visualized using rat anti-HA primary and goat anti-rat immunoglobulin G Alexa 488 secondary antibodies. Strong nuclear fluorescence was observed in cells overexpressing either wild-type or mutant proteins, indicating that Nrf2 is a nuclear protein, consistent with our previous observation (23), and deleting the last 16 amino acids has no effect on its subcellular localization (Fig. 3).
FIG. 3.
Nrf2 and Nrf2CTΔ16 are localized to the nucleus. Q293 cells were transfected with either HA-Nrf2 or HA-Nrf2CTΔ16, as indicated. Following fixation, the cells were stained with rat anti-HA primary, followed by anti-rat immunoglobulin G Alexa 488 secondary antibodies (A and B). Nuclei were visualized with propidium iodide (P.I.) (C and D), and merged images are shown in panels E and F.
Nrf2CTΔ16 is unable to activate endogenous ARE-regulated genes.
The above data clearly show that although the DNA binding activity of Nrf2CTΔ16 is not compromised, the protein is inactive in reporter gene assays. To put these results in a more physiologically relevant context, we investigated the ability of Nrf2CTΔ16 to activate endogenous ARE-driven gene expression. We established cell lines in which plasmids encoding HA-Nrf2 or HA-Nrf2CTΔ16, under the control of a tetracycline-sensitive promoter, were integrated into single identical genomic loci using FlpIn technology. A time course of Dox treatment showed that expression of HA-Nrf2 and HA-Nrf2CTΔ16 was rapidly induced with identical kinetics, and the maximal expression levels (achieved after only 4 h) were the same for both proteins across the period examined (Fig. 4A). Next, we examined the endogenous expression of two well-characterized ARE-driven Nrf2 target genes (NQO1 and GCLM) in these cell lines, as well as in a control line (293/CAT) in which the chloramphenicol acetyltransferase (CAT) gene was integrated into the FRT site. Taqman analyses demonstrated that Dox treatment resulted in a rapid and sustained increase in NQO1 mRNA expression across the time course in 293/HA-Nrf2 cells (Fig. 4B). By contrast, no induction was observed in 293/HA-Nrf2CTΔ16 or 293/CAT cells (Fig. 4B). Similarly, GCLM expression was also significantly increased by Dox treatment of 293/HA-Nrf2 cells, while no induction was observed in 293/HA-Nrf2CTΔ16 or 293/CAT cells (Fig. 4B). Interestingly, the increase in GCLM mRNA in 293/HA-Nrf2 cells was not sustained across the time course and actually returned to basal levels at the 48-h time point. This is in contrast to the effects observed on NQO1 mRNA, where the level continued to rise throughout the time course. Thus, as yet uncharacterized mechanisms function to limit the ability of Nrf2 to continuously activate certain ARE-driven genes.
FIG. 4.
Nrf2CTΔ16 cannot activate endogenous ARE-driven gene expression. (A) 293/HA-Nrf2 or 293/HA-Nrf2CTΔ16 cells were treated with Dox (1 μg/ml) for the indicated periods. Following treatments, lysates were prepared and Western blotting was performed using the indicated antibodies. (B) The indicated cell lines were treated with Dox (1 μg/ml) for 0, 4, 12, 24, or 48 h, and total RNA was subsequently isolated. The RNA was analyzed by TaqMan assay using the ABI Prism 7700 Sequence Detection System. Primers/probes used were specific for NQO1 (left) GCLM (right), and 23hbp (loading control). The values obtained for 23hbp mRNA were used to normalize those obtained for NQO1 and GCLM. Results are mean ± standard deviation of three independent experiments.
These data show that the inability of Nrf2CTΔ16 to activate transcription is not solely a phenomenon observed for reporter genes in transient transfections, but that it is also transcriptionally silent on endogenous ARE-driven genes.
Nrf2CTΔ16 is more stable than wild-type Nrf2 and is regulated normally by Keap1.
Although there were no gross differences in the steady-state levels of wild-type Nrf2 or Nrf2CTΔ16, we also determined the half-lives (t1/2) of these proteins. Cells transiently expressing HA-tagged Nrf2 or Nrf2CTΔ16 were treated with cycloheximide for 0, 15, 30, 45, 60, or 120 min before being harvested. Using immunoblot analysis, we found that the truncated protein appears to be more stable than wild-type Nrf2 (t1/2 of wild-type Nrf2 = 25 min; t1/2 of Nrf2CTΔ16 = 45 min) (Fig. 5A and B). These findings suggest that the inability of Nrf2CTΔ16 to activate transcription is intrinsically related to its transcriptional activity rather than its stability.
FIG. 5.
Nrf2CTΔ16 is more stable than wild-type Nrf2 and is regulated normally by Keap1. (A) Q293 cells were transiently transfected with plasmids encoding HA-Nrf2 or HA-Nrf2CTΔ16, as indicated. Subsequently, cycloheximide (30 μg/ml) was added to the media for the indicated times. Lysates were resolved by SDS-polyacrylamide gel electrophoresis, blotted, and probed with anti-HA antibodies. (B) Semilog graphical depiction of the results shown in panel A. (C) Q293 cells were transfected with 1 μg HA-Nrf2 or HA-Nrf2CTΔ16 plasmid in the presence or absence of 50 ng of Keap1 plasmid. Where indicated, cells were treated with 50 μM t-BHQ or 10 μM MG132 for 4 h prior to being harvested. The blots were probed with either anti-HA (top) or anti-GAPDH (bottom) antibodies.
Since Nrf2 is expressed constitutively and its steady-state level appears to be maintained in part by a Keap1-dependent degradation process (12, 18, 36), we sought to determine whether Nrf2CTΔ16 can still be regulated by Keap1. Cotransfection of cells with plasmids encoding either wild-type Nrf2 or Nrf2CTΔ16 with Keap1 led to a significant decrease in the steady-state level of the Nrf2 proteins (Fig. 5C), which was reflected in a decrease in their half-lives (data not shown). Treatment of cells with either t-BHQ or the proteosome inhibitor MG132 completely reversed the effects of Keap1; in the case of MG132, a ladder of higher-molecular-weight complexes was also observed, most probably representing ubiquitylated forms of Nrf2 and Nrf2CTΔ16 (Fig. 5C).
Identification of residues critical to the activity of Neh3.
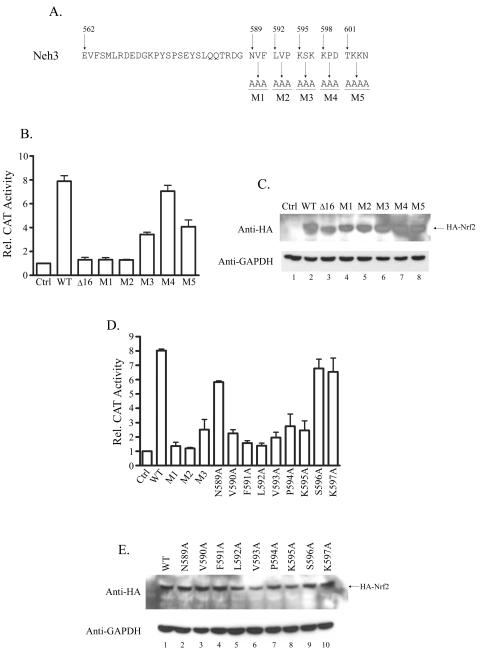
The fact that Nrf2CTΔ16 binds DNA normally and acts as a dominant-negative mutant, effectively blocking the activity of wild-type Nrf2, implies that the final 16 amino acids of Nrf2 must have a defined role in activation of transcription. To clarify the region within the last 16 amino acids that is critical to its transcriptional activity, we constructed expression plasmids for full-length Nrf2 mutants in which sequential blocks of three residues (four in the case of M5) were mutated to alanine (M1 to M5) (Fig. 6A). The functional activities of these mutant Nrf2 proteins were examined by determining the effects of their overexpression on an ARE reporter gene. The results of these experiments indicate that the N-terminal part of this region appears to be the most critical, since the Nrf2 mutants M1 and M2 recapitulated the effects of the 16-amino-acid deletion (Fig. 6B). Mutants M3 and M5 showed partial loss of activity, although they still induced gene expression three- and fourfold, respectively, whereas M4 was as effective as wild-type Nrf2 (Fig. 6B). All mutants were expressed at levels similar to wild-type Nrf2 (Fig. 6C).
FIG. 6.
Characterization of the last 16 amino acids of Nrf2. (A) Sequence of the Neh3 domain of Nrf2. The numbers shown represent the positions in the rat protein sequence, and the sequence separated into blocks highlights the final 16 amino acids of Nrf2. The mutations shown (M1 to M5) were introduced into the full-length protein by site-directed mutagenesis of an expression vector encoding wild-type Nrf2. (B) Q293 cells were cotransfected with an ARE-CAT reporter construct and the indicated HA-tagged Nrf2 expression construct: wild-type Nrf2 (WT), Nrf2CTΔ16 (Δ16), or Nrf2M1 to -M5. CAT assays were performed using standard procedures. Activity is shown as activation (n-fold) over the level obtained when reporter constructs were transfected alone (Ctrl), and results are the mean plus standard deviation of three independent experiments. (C) CAT lysates from panel B were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), blotted, and probed with anti-HA or anti-GAPDH. (D) Experiments were conducted exactly as described for panel B using the indicated HA-tagged Nrf2 expression constructs. (E) CAT lysates from panel D were resolved by SDS-PAGE, blotted, and probed with anti-HA or anti-GAPDH.
We further defined the critical region by individually mutating each amino acid encompassed by mutants M1, M2, and M3 (N589 to K597) to alanine. Again, the transcriptional capacity of each mutant was tested by overexpressing them in the presence of an ARE-CAT reporter construct. This analysis showed that mutating any of the amino acids in the sequence V590FLVPK595 resulted in a large decrease in the transcriptional activity of Nrf2 (Fig. 6D). Most notably, Nrf2F591A and Nrf2L592A appeared essentially unable to drive transcription of the ARE reporter gene. The VFLVPK motif is highly conserved across species, suggesting it may have a critical role for Nrf2 activity (Fig. 1B). In contrast, Nrf2N589A, Nrf2S596A, and Nrf2K597A activated transcription to levels comparable with wild-type Nrf2 (Fig. 6D). All proteins were expressed at similar levels (Fig. 6E).
CHD6 interacts with the Neh3 domain of Nrf2, and the VFLVPK motif is critical for this association.
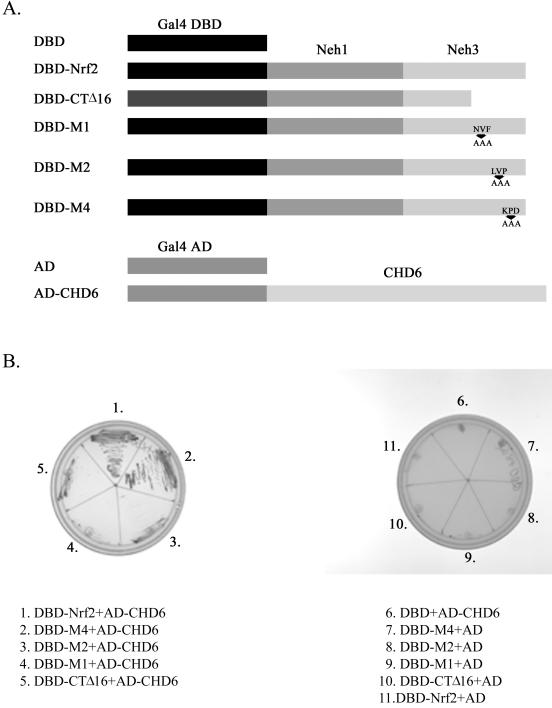
Since our results indicate that the Neh3 domain is not required for DNA binding activity, we explored whether it may be involved in interacting with components of the transcriptional machinery. To assess this possibility, we used DBD-Nrf2, which contains the Gal4 DNA binding domain fused in frame to the Neh1 and -3 domains of Nrf2 (Fig. 7A), as bait in a yeast two-hybrid screen of a human liver cDNA library. We obtained multiple clones, many of which contain sequence encoding a protein identified as CHD6. CHD6 is a member of the chromo-ATPase/helicase DNA binding protein family and contains two tandem chromodomains and a DNA-helicase domain (28), making it a plausible candidate for an Neh3-interacting protein. To further assess this possibility, we created mutants of the DBD-Nrf2 construct in which the final 16 amino acids were deleted (DBD-CTΔ16) or the M1, M2, and M4 mutations depicted in Fig. 6 were introduced (Fig. 7A). Subsequently yeast cells bearing the mutated plasmids were mated with yeast containing the Gal4-activation domain-CHD6 (AD-CHD6 [Fig. 7A]) construct. Strong growth was observed only for DBD-Nrf2/AD-CHD6 and DBD-M4/AD-CHD6 diploid yeast (Fig. 7B), which is consistent with the observation that the M4 mutant remains as transcriptionally active as wild-type Nrf2 (Fig. 6B). All other mutants failed to interact with AD-CHD6, which correlated perfectly with the loss of transcriptional activity associated with these deletions/substitutions (Fig. 7B and 6B). Thus, the loss of transcriptional activity associated with mutation of the VFLVPK motif correlates with loss of interaction with AD-CHD6.
FIG. 7.
Nrf2 interacts with CHD6 in a yeast two-hybrid assay, and this is dependent upon the VFLVPK motif. (A) Depiction of constructs used in the yeast two-hybrid assay. (B) Yeasts transformed with the indicated plasmids were mated, and diploids were selected on SD-Leu-Trp plates and subsequently streaked onto SD-Leu-Trp-His-Ade-X-α-Gal plates as shown.
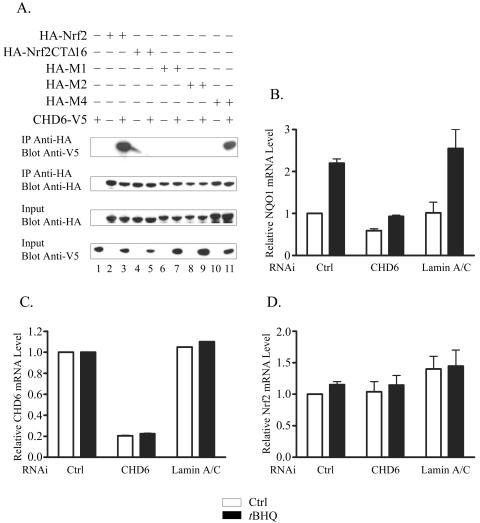
To confirm these data in mammalian cells, we cotransfected an expression construct encoding a V5-tagged portion of the CHD6 protein with one of the plasmids encoding HA-Nrf2, HA-Nrf2CTΔ16, HA-Nrf2M1, HA-Nrf2M2, or HA-Nrf2M4. Following harvesting, lysates were incubated with immobilized HA antibodies, and the IPs were analyzed by Western blotting. The results of this experiment demonstrate that full-length Nrf2 and Nrf2M4 associate with CHD6, whereas Nrf2CTΔ16, Nrf2M1, and Nrf2M2 interact much more weakly with the helicase (Fig. 8A), supporting the findings of our yeast two-hybrid experiment.
FIG. 8.
CHD6 interacts with Nrf2 in mammalian cells and is required for expression of NQO1. (A) Lysates were prepared from Q293 cells expressing HA-tagged Nrf2, Nrf2CTΔ16, Nrf2M1, Nrf2M2, or Nrf2M4 in the presence or absence of a V5-tagged CHD6 protein (amino acids 124 to 324), as indicated. Following IP with immobilized HA antibodies, immune complexes were blotted and probed with V5 or HA antibodies, as specified. A portion of each sample (10%) was removed prior to IP (input) and was blotted and probed with either anti-HA or anti-V5 antibodies. (B to D) HeLa cells were transfected with nonspecific siRNA (Ctrl) or siRNA specific for CHD6 or lamin A/C, as indicated. Subsequently, cells were treated for 24 h with either 0.1% (vol/vol) DMSO (white bars) or 50 μM t-BHQ (black bars). Total RNA was isolated and analyzed by TaqMan with a primer-probe set specific for NQO1 mRNA (B), CHD6 mRNA (C), or Nrf2 mRNA (D). The error bars indicate standard deviations.
We next asked whether reducing the level of endogenous CHD6 using RNAi would affect the expression of ARE-driven genes. Our initial attempts focused on the Q293 cell line used throughout this study; however, small interfering RNA (siRNA) directed against CHD6 had no effect on basal or inducible expression of NQO1 in these cells. This was most likely due to inefficient transfection of this cell line, since we could not detect a decrease in the level of mRNA corresponding to CHD6. Therefore, we decided to use the HeLa cell line, because with our optimized protocol we can achieve transfection efficiencies in excess of 95% in these cells. Cells were transfected with siRNA corresponding to CHD6 mRNA, Lamin A/C mRNA, or a nonspecific control, and 48 h later, they were treated with either DMSO or t-BHQ and incubated for a further 24 h; total RNA was then isolated and analyzed using TaqMan chemistry. Both basal and inducible levels of mRNA corresponding to NQO1 were significantly reduced by RNAi against CHD6 compared with levels observed in cells transfected with control or lamin A/C siRNA (Fig. 8B). We confirmed that levels of CHD6 mRNA were efficiently and specifically reduced by RNAi (Fig. 8C). We also found that Nrf2 mRNA levels were unaffected by any of the double-stranded RNAs, demonstrating that the observed effects are not explained by a decrease in Nrf2 transcription (Fig. 8D).
These data indicate that the specific interaction between CHD6 and the Neh3 domain appears to be important for Nrf2 activity and suggest that Neh3 may be involved in recruiting accessory proteins or serve to bridge the transcription factor to the active transcription apparatus. The precise mechanisms whereby the interaction between Neh3 and CHD6 leads to gene transcription are not understood and remain to be explored.
DISCUSSION
Nrf2 is a key activator of genes encoding antioxidant and phase II drug-metabolizing enzymes through the ARE. The molecular mechanisms governing the activity of Nrf2 are starting to become apparent through analysis of conserved regions within the protein, including the Neh2 domain for interaction with the repressor protein Keap1 (11), the transactivation domain within the Neh4-Neh5 region (13), the basic DNA binding and dimerization domain of Neh1, and the Neh6 domain serving as a redox-insensitive degron (19).
In this report, we present evidence demonstrating that the Neh3 domain at the C terminus of Nrf2 harbors a critical activity required for activation of transcription and may function as a TAD, along with the Neh4 and Neh5 domains. This conclusion is supported by our finding that deletion of the last 16 amino acids from Nrf2 results in a protein that is transcriptionally inactive. Although the truncated protein failed to activate ARE-dependent gene expression, its nuclear localization, as well as its ability to bind to the ARE with high affinity, are indistinguishable from those of wild-type Nrf2, suggesting that its inability to activate gene transcription is due to a defect in the transactivational activity. This is consistent with the observation that it can block wild-type Nrf2 from activating gene transcription and thus effectively act as a dominant-negative form of the transcription factor. Furthermore, although the truncated protein appears to be slightly more stable than wild-type Nrf2, it is efficiently targeted for degradation by Keap1 and is stabilized by either t-BHQ or MG132. These findings are consistent with the fact that Nrf2 is a modular protein comprising multiple functional domains that act independently of each other. These data provide strong evidence suggesting that the Neh3 domain may act as a TAD that is distinct and in addition to the TAD found in the Neh4-Neh5 domains; furthermore, it is absolutely required for Nrf2 to activate expression of its target genes.
Thus, the transcriptional activity of Nrf2 appears to be dependent on three separate domains that act in concert to recruit the full complement of proteins required to form an active transcriptional complex. This is supported by the fact that deletion of the TADs within the Neh4 and Neh5 domains also inactivates Nrf2, despite the presence of the Neh3 domain (13). We also noted that a Gal4-Neh3 fusion is unable to activate Gal4-dependent transcription in our yeast two-hybrid assay. The Neh3 domain and its interacting factors may not be capable of activating transcription in isolation but rather represent essential components of a larger multiprotein transcription complex that is formed on ARE-driven genes. These findings are reminiscent of p53, where deleting its N-terminal activation domain renders the protein unable to activate several of its target genes, even though the protein can still bind to the p68 coactivator protein (2, 4, 35).
Although the Neh4 and Neh5 domains have not been fully characterized, their activities are thought to be due in part to their cooperative binding of CBP (13). Using purified proteins, we have found that Nrf2CTΔ16 interacts with p300 as competently as wild-type Nrf2 (data not shown). This finding suggests first that the structures of Neh4 and Neh5 are not perturbed by the deletion and secondly that Neh3 functions through a mechanism distinct from those of the other TADs. Furthermore, if CBP/p300 is still recruited to DNA by Nrf2CTΔ16, then this would indicate that CBP/p300 alone is insufficient for transactivation of at least some Nrf2-dependent genes, supporting the hypothesis that the recruitment of multiple factors is required.
The activity of the Neh3 domain is reliant upon the VFLVPK motif, which is predominantly hydrophobic in nature and could be a site of interaction with a novel partner molecule. The critical F591 and L592 residues would most likely form essential interactions within such a protein complex. Notably, the VFLVPK motif is highly conserved between a number of CNC family members, including the drosophila CNC protein (Fig. 9), and likely represents a fundamental means of activating transcription that has been conserved throughout the evolution of the CNC-bZIP family. Sequences N terminal to the VFLVPK motif are also very highly conserved (Fig. 9), and this is consistent with our unpublished observations indicating that these regions are also critical for the activity of Nrf2.
FIG. 9.
Alignment of Neh3 domains from members of the CNC transcription factor family. The VFLVPK motif is highlighted by asterisks.
The notion that the VFLVPK motif may serve as a protein-protein interaction module is confirmed by our yeast two-hybrid screen, which identified CHD6 as an Neh3-interacting protein. This association is highly specific and dependent upon the integrity of the VFLVPK motif; mutations that reduce the transactivity of Nrf2 (i.e., within the VFLVPK motif) also decrease the strength of interaction with CHD6, whereas mutations that have no effect on transactivity also have no impact on the interaction with CHD6. Furthermore, reducing the level of endogenous CHD6 by RNAi decreases both the basal and t-BHQ-inducible expression of NQO1, a prototypical Nrf2-regulated gene. Our findings clearly show that loss of binding to CHD6 is sufficient to explain the impaired transcriptional activity of Nrf2CTΔ16. All of the available evidence demonstrates that Nrf2CTΔ16 binds DNA normally; however, this could not be unequivocally confirmed in vivo.
CHD6 is particularly interesting, since it is characterized by the presence of two tandem chromodomains and a DNA helicase domain, but its function is not well understood (28). However, the founding member of the CHD family, CHD1, is important for the expression of certain genes and associates with actively transcribed chromatin in both yeast and mammalian cells (29, 30, 31). CHD1 has been implicated in several processes, which may explain its importance in gene expression. First, CHD1 may be involved in transcription elongation, since it has been shown to associate with several factors involved in this process and can be localized to the coding regions of certain genes (29). Secondly, CHD1 can alter nucleosome structure to produce regions of DNase I hypersensitivity in a test substrate, possibly providing access to DNA for additional DNA-interacting factors (31). Finally, one of the chromodomains of CHD1 specifically interacts with methylated lysine 4 in histone H3, a modification that is associated with transcriptional activity, and by doing so recruits enzymes with histone acetyltransferase activity to chromatin (25). Importantly, the activity of CHD1 is reliant upon both its helicase domain and chromodomains, both of which are highly conserved in CHD6 (14, 25, 29). Therefore, recruitment of CHD6 to Nrf2-dependent genes by the Neh3 domain has the potential to nucleate multiple activities associated with transcriptional activation. These data suggest that the specific interaction between CHD6 and the Neh3 domain may be important for Nrf2 activity. The precise role of CHD6 in ARE-driven gene expression remains to be explored.
In summary, as a modular protein, Nrf2 possesses a number of distinct functional domains the concerted actions of which are critical to the maintenance of cellular redox homeostasis. We have established that the Neh3 domain harbors a transactivation-like activity and functions in parallel with the Neh4 and Neh5 domains to activate transcription of Nrf2 target genes. The function of the Neh3 domain is reliant upon the VFLVPK motif, which is conserved across species and among other members of the CNC-bZIP family. The Neh3 domain may function as a protein-protein interaction module to facilitate the recruitment of putative coactivator proteins, such as CHD6, to Nrf2-dependent genes.
Acknowledgments
We thank Ahmed Samatar for assistance with RNAi experiments.
REFERENCES
- 1.Alam, J., D. Stewart, C. Touchard, S. Boinapally, A. M. Choi, and J. L. Cook. 1999. Nrf2, a Capcn'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274:26071-26078. [DOI] [PubMed] [Google Scholar]
- 2.Bates, G. J., S. M. Nicol, B. J. Wilson, A. M. Jacobs, J. C. Bourdon, J. Wardrop, D. J. Gregory, D. P. Lane, N. D. Perkins, and F. V. Fuller-Pace. 2005. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 24:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho, H. Y., A. E. Jedlicka, S. P. Reddy, T. W. Kensler, M. Yamamoto, L. Y. Zhang, and S. R. Kleeberger. 2002. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell. Mol. Biol. 26:175-182. [DOI] [PubMed] [Google Scholar]
- 4.Courtois, S., G. Verhaegh, S. North, M. G. Luciani, P. Lassus, U. Hibner, M. Oren, and P. Hainaut. 2002. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 21:6722-6728. [DOI] [PubMed] [Google Scholar]
- 5.Cullinan, S. B., J. D. Gordon, J. Jin, J. W. Harper, and J. A. Diehl. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24:8477-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enomoto, A., K. Itoh, E. Nagayoshi, J. Haruta, T. Kimura, T. O'Conner, T. Harada, and M. Yamamoto. 2001. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 59:169-177. [DOI] [PubMed] [Google Scholar]
- 7.Favreau, L. V., and C. B. Pickett. 1991. Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic compounds and phenolic antioxidants. J. Biol. Chem. 266:4556-4561. [PubMed] [Google Scholar]
- 8.Furukawa, M., and Y. Xiong. 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitylation by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 25:162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, H. C., T. Nguyen, and C. B. Pickett. 2002. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 277:42769-42774. [DOI] [PubMed] [Google Scholar]
- 10.Itoh, K., T. Chiba, S. Takahashi, T. Ishii, K. Igarashi, Y. Katoh, T. Oyake, N. Hayashi, K. Satoh, I. Hatayama, M. Yamamoto, and Y. I. Nabeshima. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236:313-322. [DOI] [PubMed] [Google Scholar]
- 11.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, T. O'Conner, and M. Yamamoto. 2003. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8:379-391. [DOI] [PubMed] [Google Scholar]
- 13.Katoh, Y., K. Itoh, E. Yoshida, M. Miyagishi, A. Fukamizu, and M. Yamamoto. 2001. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 6:857-868. [DOI] [PubMed] [Google Scholar]
- 14.Kelley, D. E., D. G. Stokes, and R. P. Perry. 1999. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma 108:10-25. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, A., M. I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J. M., K. Chan, Y. W. Kan, and J. A. Johnson. 2004. Targeted disruption of Nrf2 causes regenerative immune-mediated hemolytic anemia. Proc. Natl. Acad. Sci. USA 101:9751-9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon, M., K. Itoh, M. Yamamoto, S. A. Chanas, C. J. Henderson, L. I. McLellan, C. R. Wolf, C. Cavin, and J. D. Hayes. 2001. The Cap′n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 61:3299-3307. [PubMed] [Google Scholar]
- 18.McMahon, M., K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 278:21592-21600. [DOI] [PubMed] [Google Scholar]
- 19.McMahon, M., N. Thomas, K. Itoh, M. Yamamoto, and J. D. Hayes. 2004. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 279:31556-31567. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen, T., T. H. Rushmore, and C. B. Pickett. 1994. Transcriptional regulation of a rat liver glutathione S-transferase Ya subunit gene. Analysis of the antioxidant response element and its activation by the phorbol ester 12-O-tetradecanoylphorbol-13-acetate. J. Biol. Chem. 269:13656-13662. [PubMed] [Google Scholar]
- 21.Nguyen, T., H. C. Huang, and C. B. Pickett. 2000. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J. Biol. Chem. 275:15466-15473. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, T., P. J. Sherratt, H.-C. Huang, C. S. Yang, and C. B. Pickett. 2003. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 278:4536-4541. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen, T., P. J. Sherratt, P. Nioi, C. S. Yang, and C. B. Pickett. 2005. NRF2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by keap1. J. Biol. Chem. 280:32485-32492. [DOI] [PubMed] [Google Scholar]
- 24.Nioi, P., M. McMahon, K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 374:337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433:434-438. [DOI] [PubMed] [Google Scholar]
- 26.Ramos-Gomez, M., M. K. Kwak, P. M. Dolan, K. Itoh, M. Yamamoto, P. Talalay, and T. W. Kensler. 2001. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. USA 98:3410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rushmore, T. H., M. R. Morton, and C. B. Pickett. 1991. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 266:11632-11639. [PubMed] [Google Scholar]
- 28.Schuster, E. F., and R. Stoger. 2002. CHD5 defines a new subfamily of chromodomain-SWI2/SNF2-like helicases. Mamm. Genome 13:117-119. [DOI] [PubMed] [Google Scholar]
- 29.Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa, A. D. Johnson, G. A. Hartzog, and K. M. Arndt. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22:1846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stokes, D. G., and R. P. Perry. 1995. DNA-binding and chromatin localization properties of CHD1. Mol. Cell. Biol. 15:2745-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran, H. G., D. J. Steger, V. R. Iyer, and A. D. Johnson. 2000. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 19:2323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venugopal, R., and A. K. Jaiswal. 1996. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA 93:14960-14965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakabayashi, N., A. T. Dinkova-Kostova, W. D. Holtzclaw, M. I. Kang, A. Kobayashi, M. Yamamoto, T. W. Kensler, and P. Talalay. 2004. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 101:2040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wild, A. C., H. R. Moinova, and R. T. Mulcahy. 1999. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 274:33627-33636. [DOI] [PubMed] [Google Scholar]
- 35.Yin, Y., C. W. Stephen, M. G. Luciani, and R. Fahraeus. 2002. p53 stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat. Cell Biol. 4:462-467. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, D. D., and M. Hannink. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23:8137-8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, D. D., S. C. Lo, J. V. Cross, D. J. Templeton, and M. Hannink. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24:10941-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]