Abstract
A temporal gradient of the novel nuclear protein LIN-14 specifies the timing and sequence of stage-specific developmental events in Caenorhabditis elegans. The profound effects of lin-14 mutations on worm development suggest that LIN-14 directly or indirectly regulates stage-specific gene expression. We show that LIN-14 can associate with chromatin in vivo and has in vitro DNA binding activity. A bacterially expressed C-terminal domain of LIN-14 was used to select DNA sequences that contain a putative consensus binding site from a pool of randomized double-stranded oligonucleotides. To identify candidates for genes directly regulated by lin-14, we employed DNA microarray hybridization to compare the mRNA abundance of C. elegans genes in wild-type animals to that in mutants with reduced or elevated lin-14 activity. Five of the candidate LIN-14 target genes identified by microarrays, including the insulin/insulin-like growth factor family gene ins-33, contain putative LIN-14 consensus sites in their upstream DNA sequences. Genetic analysis indicates that the developmental regulation of ins-33 mRNA involves the stage-specific repression of ins-33 transcription by LIN-14 via sequence-specific DNA binding. These results reinforce the conclusion that lin-14 encodes a novel class of transcription factor.
The heterochronic genes in Caenorhabditis elegans control the expression of stage-specific developmental events (1). During post-embryonic development, worms go through four larval stages (L1 through L4). Within each stage certain cells express developmental programs specific to their cell type and specific to the larval stage. Early during the first larval stage, the regulatory gene lin-14 specifies L1-specific development, presumably by directly or indirectly promoting the expression of genes involved in the execution of L1-specific developmental programs and/or by repressing L2-specific genes (2). The gene lin-14 encodes a novel nuclear protein (49, 50, 59) whose levels are high at the beginning of L1 and then rapidly decline in the course of the first larval stage (50). This down-regulation of the LIN-14 protein occurs as a result of translational repression by the microRNA product of lin-4. lin-4 RNA inhibits LIN-14 protein synthesis by base pairing with complementary sequences in the lin-14 3′ untranslated region (5, 27, 37, 60). The reduced level of LIN-14 at the start of the L2 stage permits the expression of L2-specific events, presumably through derepression of L2-specific gene expression (2, 3).
LIN-14 protein is localized to the nuclei of most somatic cells in L1 larvae (50, 59), in an anatomical pattern consistent with the cell types affected by lin-14 mutations. LIN-14 is a novel protein that shows no obvious homology to proteins in nonnematode species (20, 41, 59). The C terminus is sufficient for the nuclear localization of the protein and appears to contain a relatively expanded nuclear targeting signal, unlike classical nuclear localization signals (20). The only other predicted structural motifs in LIN-14 are an amphipathic helix in the C-terminal domain of the protein in exon 11 (59) and an alpha helix in exon 10 (P. Olsen, personal communication). These properties of LIN-14 have suggested that LIN-14 regulates stage-specific gene expression in the nucleus. To gain an understanding of the possible mechanism(s) by which lin-14 helps cells establish their temporal identity, we employed a series of assays using the native LIN-14 protein from isolated nuclei and bacterially expressed LIN-14 to characterize LIN-14 biochemical activities. Here we show that native LIN-14 exists in at least two populations in the nucleus, one of which is associated with chromosomal DNA. We show that a bacterially expressed C-terminal fragment of LIN-14 binds double-stranded DNA and can select sequences containing a putative consensus sequence from a pool of randomized double-stranded oligonucleotides. These results support the conclusion that LIN-14 could function as a DNA binding transcriptional regulator.
Based on lin-14 phenotypes, hypothetical lin-14 transcriptional targets can be broadly divided into two classes: L2-specific genes, which would be normally repressed by lin-14 during the L1 stage, and L1-specific genes, which would be normally activated by lin-14 during L1. To identify candidate LIN-14 targets, we employed DNA microarray hybridization. We compared mRNA abundance of 11,899 C. elegans genes in RNA samples from the L1 and L2 stages of wild-type worms, lin-14 loss-of-function (lf) mutants, and lin-4 loss-of-function mutants. We identified 16 genes with characteristics of potential lin-14 targets; these genes display opposite changes in mRNA abundance in response to reduced lin-14 [lin-14(lf)] and elevated lin-14 [lin-4(lf)]. We confirmed that at least one of these genes, ins-33, is a bona fide transcriptional target of LIN-14; ins-33 promoter activity is temporally regulated by lin-14 in vivo, and LIN-14 binds in vitro to ins-33 upstream sequences that are essential for negative developmental regulation by lin-14. These results demonstrate that lin-14 encodes a novel class of transcription factor that controls developmental timing via direct DNA binding.
MATERIALS AND METHODS
Worm strains.
C. elegans (wild type strain N2 var Bristol) was cultured on NGM plates or in liquid culture as previously described (61), at 20°C unless otherwise noted. Synchronized populations of wild-type or mutant larvae were obtained by hypochlorite treatment of gravid adults, allowing embryos to develop to hatching in the absence of food, followed by growth of the L1 larvae on NGM plates with food (bacterial strain OP-50). To obtain lin-14 mutant L1 populations, VT965 [lin-14(n179ts)] starved L1 larvae were placed on NGM plates seeded with bacteria and grown at the nonpermissive temperature (25°C) for 7 to 8 h before harvesting. lin-4 L1 larvae were grown on food for 21 to 22 h at 20°C to obtain L2 larvae. All worm populations were harvested by washing the plates with ice-cold distilled water (dH2O) and rinsed three times with cold dH2O before being snap-frozen in liquid N2.
Nuclear isolation.
Nuclei from L1 larvae were isolated as described (29) except that worms were homogenized in buffer A (250 mM sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1 mM EGTA, 1 mM DTT, 2 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 2 mM Pefabloc). Nuclei were stored at −80°C in 10% glycerol and buffer B (buffer A plus 60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl, pH 7.4, 3 mM MgCl2, 2 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 2 mM Pefabloc) (13). Nuclear DNA concentration was estimated by optical density at 260 nm of an aliquot in 1% sodium dodecyl sulfate (SDS).
MNase assay.
Nuclei were thawed on ice and recovered by centrifugation in a refrigerated microcentrifuge at 11,500 rpm for 10 min. Nuclei were resuspended in micrococcal nuclease (MNase) buffer (10 mM HEPES, 10 mM KCl, 0.5 mM MgCl2, 2 mM CaCl2, 2 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 2 mM Pefabloc) to a DNA concentration of 1 μg/μl (10). MNase (Worthington Biochemical) was added at concentrations from 10 to 150 units/100 μg DNA, and the nuclei were incubated on ice for 35 to 45 min. Digestion was stopped by the addition of an equal volume of MNase buffer supplemented with 10 mM EDTA. After a 10-min spin at 4,000 rpm in a refrigerated microcentrifuge, the supernatant was either loaded onto a sucrose gradient (see below) or subjected to centrifugation at 100,000 × g for 1 h in a Beckman tabletop ultracentrifuge (adapted from Liang and Stillman [28]). For RNase digestion, DNase-free RNase A (Sigma) was used under the same incubation conditions as those for MNase, and the digestion of RNA was verified by agarose gel electrophoresis after RNA isolation using Trizol (GibcoBRL).
For sucrose gradient analysis, 5 ml of 20 to 40% sucrose gradients in MNase stop buffer was centrifuged for 3.25 h at 40,000 rpm in a Beckman ultracentrifuge (SW 55 Ti rotor). Fractions (275 μl each) were collected by hand and split; two-thirds of each fraction was precipitated with trichloroacetic acid and resuspended in SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer, while one-third was phenol extracted and ethanol precipitated to recover DNA.
Nuclear matrix assays. (i) High salt procedure.
Nuclei were thawed on ice and recovered by centrifugation at 4°C and 11,500 rpm for 10 min in a refrigerated microcentrifuge. Nuclear matrix was prepared from the pelleted nuclei according to published protocols (16, 43). As a control, high-salt-extracted pellets were digested with DNase-free RNase A (Sigma).
(ii) Low-salt procedure.
For the low-salt procedure, adapted from Smith et al. (53) and Mirkovitch et al. (35), nuclei were thawed on ice and recovered by centrifugation for 10 min at 4°C, 11,500 rpm, in a microcentrifuge. Nuclei were resuspended in low-salt buffer (5 mM HEPES, pH 7.9, 2 mM EDTA, 2 mM KCl, 0.25 mM spermidine, 2 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 2 mM Pefabloc) at a DNA concentration of 2 μg/μl and incubated at room temperature for 15 min. (As a control, in at least one experiment the nuclei were pelleted after this step to analyze the effect of low-salt buffer alone on nuclear protein release). An equal amount of low-salt buffer supplemented with lithium 3,5-diiodosalicylate to 50 mM was added to the nuclei to a final lithium 3,5-diiodosalicylate concentration of 25 mM, and the nuclei were incubated for another 5 min at 22°C. Extracted nuclei were recovered by centrifugation at 5,000 × g for 3 min at 22°C and washed three times in buffer containing 20 mM Tris, pH 7.4, 20 mM KCl, 70 mM NaCl, 0.5 mM spermine, 0.125 mM spermidine, 2 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 2 mM Pefabloc. Nuclei were resuspended in wash buffer and incubated with RNase-free DNase I (Boehringer Mannheim) at 0.5 U/μg DNA for 15 min at 37°C. After a final 3-min 5,000 × g spin, the nuclear matrix was resuspended in 8 M urea.
LIN-14 fusion protein expression and purification.
LIN-14 coding sequences were amplified from a plasmid containing lin-14 cDNA and were cloned into the glutathione transferase (GST) expression vector pGEX-2T (GE Healthcare) to create a fusion protein with the GST module at the N terminus of the fusion protein. Primers were chosen to amplify sequences coding amino acids 292 to 539 or amino acids 284 to 465 [in the case of GST::LIN-14(292-539) or GST::LIN-14(284-465), respectively] (Fig. 1A). Purification of the GST fusion proteins was carried out as previously described (14), except that cells were lysed by sonication, sarcosyl was omitted from the buffer, and an additional clarifying spin step was carried out at 100,000 × g. Protein bound to glutathione-Sepharose beads (GE Healthcare) was either frozen as a 50% bead slurry in 10% glycerol, 50 mM HEPES, pH 7.9, 150 mM NaCl, and 5 mM dithiothreitol (DTT) or eluted from the beads in the above buffer supplemented with 15 mM glutathione. Elution fractions containing the fusion protein were stored at −80°C. Amino acids 284 to 465 were fused to a six-His tag by PCR amplification with appropriate primers, and the His-tagged fusion protein was purified from Escherichia coli by metal affinity resin. Protein fractions were analyzed using 11% SDS-PAGE gels, and proteins were visualized by Coomassie blue staining.
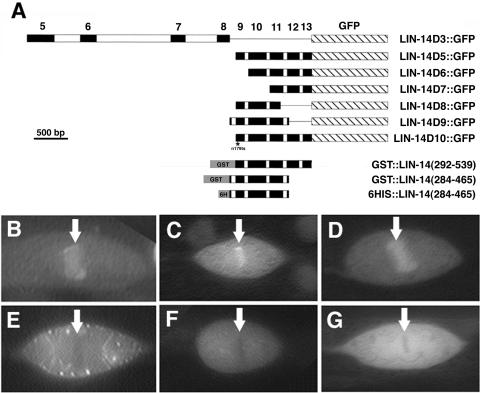
FIG. 1.
Chromatin association of LIN-14::GFP fusion proteins in vivo. (A) Schematic representation of the different truncated versions of LIN-14 used in chromosome association experiments (GFP fusions designated D3 and D5 to D10 [20]) and in DNA binding experiments (GST and six-His fusions). The n179 mutation in exon 9 of D10 is indicated by an asterisk. GST::LIN-14(292-539) contains LIN-14 amino acids 292 to 539, which comprise exons 9 to 13, identical to the fragment included in LIN-14D5::GFP. GST::LIN-14(284-465) and 6HIS::LIN-14(284-465) contain LIN-14 amino acids 284 to 465, identical to the fragment included in LIN-14D9::GFP. Exon numbers are indicated above the constructs. All images are of lin-14(n179) animals carrying transgenic LIN-14 constructs, except for the image in panel C, which is lin-14(ma135). Arrows indicate the mitotic chromosomes. (B to D) Rescuing truncated LIN-14::GFP proteins associate with mitotic chromosomes: LIN-14D5::GFP in lin-14(n179) (B) and lin-14(ma135) backgrounds (C); LIN-14D9::GFP (D). (E to G) Nonrescuing truncated LIN-14::GFP proteins do not associate with mitotic chromosomes: LIN-14D6::GFP (E), LIN-14D7::GFP (F), LIN-14D8::GFP (G). Scale bar, 1.5 μm.
DNA binding of GST::LIN-14(284-465).
DNA cellulose binding was performed using Sigma brand cellulose coupled to double-stranded or single-stranded DNA (D8515 or D8273). Bacterially expressed GST::LIN-14(284-465) fusion protein was diluted in 75 mM NaCl, 50 mM HEPES, 7.5 mM glutathione, 5% glycerol, 2.5 mM, and 0.1 mM EDTA and bound in batch to DNA-cellulose by rocking at 4°C for 1.5 h. The protein-DNA-cellulose complexes were recovered by centrifugation in a clinical centrifuge and washed three to four times with 10 column volumes each of cold wash buffer (150 mM NaCl, 50 mM HEPES, 0.1 mM EDTA, 0.5 mM DTT). Protein was eluted with 3 to 6 column volumes of elution buffer (50 mM HEPES, 0.5 mM DTT, 0.1 mM EDTA, 0.005% Triton X-100) supplemented with increasing concentrations of NaCl (200 mM, 400 mM, 800 mM, and 1.5 M). Equivalent amounts of each fraction were analyzed on SDS-PAGE gels.
Binding site selection assay (SELEX).
DNA sequences for consensus site selection were generated from the following oligonucleotide input sequence: 5′ AGA GGG ATC CGA TTG CAG-N20-GTG TAG GAA TTC GCC GTG 3′ (PO66), where the middle 20 bases (N20) were randomized during synthesis. These sequences have BamHI and EcoRI restriction sites (underlined) in the 5′ and 3′ ends, respectively, for subsequent cloning. Two PCR primers complementary to the 5′ and 3′ constant regions were also synthesized (PO67 and PO68). Double-stranded oligonucleotides were synthesized from the PO66 pool by extension with Klenow (NEB) using primer PO68.
Binding reactions were performed in a 250-μl volume for 30 min at 4°C (adapted from Chittenden et al. [11]). Twenty microliters of frozen glutathione-Sepharose beads complexed with GST::LIN-14(284-465) was diluted in binding buffer (50 mM HEPES, 100 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 5 mM DTT, 10% glycerol, 100 ng/ml) containing double-stranded oligonucleotides. After incubation, the beads were washed three to four times with 1 ml of cold binding buffer to eliminate unbound DNA. Elution of DNA bound to the immobilized protein was carried out as described (7). Approximately 10% of the recovered DNA was amplified via PCR to prepare DNA for the next round of binding.
Amplification conditions were optimized and the following protocol was used: 95°C for 5 min and then 19 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 140 s, with a final step of 72°C for 5 min. After the fourth and fifth rounds of selection, oligonucleotide pools were digested with EcoRI and BamHI and cloned in Bluescript SK vector. Colonies with inserts were selected by blue/white selection and sequenced using a T7 primer. Most colonies contained multiple concatamerized independent inserts. The randomized sequences were compared and aligned manually.
EMSAs.
Probes were end labeled with 32P and T4 polynucleotide kinase, and approximately 20 fmol of labeled probe was added to each binding reaction (2 nM final concentration). Electrophoretic mobility shift assays (EMSAs) were performed essentially as described (7). Binding reactions were performed in binding buffer (50 mM HEPES, pH 7.9, 100 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10% glycerol, dI-dC at 0.02 μg/μl final concentration, bovine serum albumin at a final concentration of 1 μg/μl) for 20 min at room temperature. A fusion protein consisting of a six-His-tagged version of LIN-14 containing amino acids 284 to 465 [6HIS::LIN-14(284-465)] was included in the binding reaction at concentrations of 8 nM to 320 nM.
Binding reactions were fractionated by electrophoresis in 3.5% or 4% (acrylamide/bisacrylamide ratio of 60:1) nondenaturing polyacrylamide gels. All gels were run at 4°C at 30 to 35 mA for 1.5 to 3 h in 0.5× Tris-borate-EDTA. Gels were exposed to a storage phosphor screen and images were captured on a Typhoon 8600 Variable Mode Imager using the ImageQuant Software (Molecular Dynamics). Images were further processed using Adobe Photoshop.
DNase I footprinting.
The binding reaction for the DNase I footprinting analysis was carried out using the same reaction conditions as for EMSA except that protein [6HIS::LIN-14(284-465)] was added in molar excess to probe at quantities sufficient to retard all of the free probe in EMSA experiments. DNase I (Boehringer Mannheim) was then added to the binding reactions at a concentration of 0.01 U/ng of radiolabeled DNA probe, and the reaction mixtures were incubated for 5 min at room temperature. Digestion was stopped by the addition of an EDTA/EGTA cocktail to a final concentration of 2 mM each, and the samples were heated at 65°C for 10 min, dialyzed against dH2O, lyophilized in a Speedvac, and then resuspended in denaturing loading buffer. Samples were then heated and fractionated by electrophoresis through a 6% urea acrylamide gel (SequaGel; National Diagnostics) in 0.5× or 1× Tris-borate-EDTA buffer at 55°C for approximately 1 to 3 h. After drying, gels were exposed to a storage phosphor screen, and images were captured and processed as described above.
Western blots.
Western blot analysis was performed according to standard procedures (7) using Renaissance Enhanced Luminol Chemiluminescence Reagent (New England Nuclear). Proteins were transferred onto Millipore brand Immobilon-P membranes. Primary antibody incubations were performed in 1× phosphate-buffered saline supplemented with 5% nonfat dry milk for 1 to 2 h at room temperature or at 4°C overnight. LIN-14 antisera were generated (Covance Research Products, Inc.) against 6HIS::LIN-14(284-465) (Fig. 1A). Antisera were affinity purified using purified GST::LIN-14(292-539) (Fig. 1A) according to established procedures (18) using cyanogen bromide-activated Sepharose (Amersham Pharmacia Biotech). Whenever reprobing of membranes was necessary, blots were stripped by incubating at 68°C for 30 min in 100 mM Tris, pH 7.4, 100 mM β-mercaptoethanol, and 2% SDS.
Commercially available monoclonal antibodies were used to detect core histones and H1 (Chemicon International). Anti-green fluorescent protein (GFP) antibodies were purchased from Clonetech (Living Colors A.V.). Donkey anti-rabbit horseradish peroxidase-conjugated secondary antibodies were obtained from Amersham Life Sciences. Monoclonal antisera against C. elegans-TATA binding protein were kindly provided by N. Hernandez (48). Polyclonal antisera against C. elegans-lamin (lmn-1) were generously provided by Y. Gruenbaum (26, 45). Polyclonal antisera against LIN-26 were a gift from M. Labouesse (25).
RNA isolation and mRNA purification.
Total worm RNA was isolated using Trizol (GibcoBRL) as follows. Frozen worm pellets were thawed in the presence of 4 volumes of Trizol and vortexed for 10 min; the mixture was spun at 14,000 rpm for 10 min at 4°C, and the supernatant was removed to a fresh tube, to which one-fifth volume CHCl3 was added; the mixture was vortexed and centrifuged for 15 min at 14,000 rpm at 4°C. The aqueous phase was removed to a new tube, and the RNA was precipitated by the addition of an equal volume of isopropanol. RNA samples were stored at −80°C as an ethanol precipitate. mRNA was prepared from total worm RNA using oligo(dT) cellulose (Sigma) and following published procedures (51). mRNA samples were shipped as dried pellets on dry ice to Stuart Kim's laboratory at Stanford University for cDNA synthesis and microarray hybridization (procedures available at http://cmgm.stanford.edu/∼kimlab/wmdirectorybig.html).
Microarray data and statistics.
The data from six experiments [three repeats of N2 L1s versus lin-14(n179ts) L1s and three repeats of N2 L2 larvae versus lin-4(e912) L2 larvae] are stored on the Stanford Microarray Database (http://genome-www4.stanford.edu/cgi-bin/SMD/login.pl; the identifier is “experimenter: MHRISTOVA”). The first three experiments (N2 versus lin-14) were performed using microarrays containing 63% of predicted C. elegans open reading frames (42), while the second three experiments (N2 versus lin-4) were performed using microarrays containing 94% of predicted open reading frames (22). For the purposes of this paper, only the genes that were analyzed in all six experiments will be discussed.
Raw data from each experiment were downloaded from the Stanford Microarray Database into Excel files and processed as follows: (i) sort by Spot Flag and discard any rows where the Spot Flag value was nonzero, indicating a bad PCR; (ii) sort by Failed and discard any rows where the Failed value was nonzero, indicating abnormal hybridization; (iii) import into a common file for each type of experiment (i.e., lin-14 or lin-4) the columns from each raw experimental file [RAT2(R/G), which shows a log base 2 transformed ratio of normalized red/green signal for each spot; name of spot (Wormbase designation); chromosome location and description (www.wormbase.org)]; (iv) calculate an average RAT2(R/G) based on the 2 or 3 values (avg; any rows which had only one good experimental value were discarded); (v) calculate a standard deviation (stdev) for the average value; (vi) calculate a t value for each spot by using the formula t = avg*[sqrt(n −1)]/stdev, where n is the number of experiments for which good data exist, sqrt is square root, and stdev is standard deviation; (vii) sort by absolute t value and discard any rows with a t value below 4.303 (below 95% confidence interval for three experiments) or below 12.706 (below 95% confidence interval for two experiments); (viii) sort by absolute average value and discard any rows with average values below 1.0 (less than twofold change compared to control).
Northern blot hybridization.
Northern blot hybridization using the formaldehyde method of gel electrophoresis was performed on poly(A)-selected RNA following established procedures (7). After exposure to a storage phosphor screen, images were captured on a Typhoon 8600 Variable Mode Imager using the ImageQuant Software (Molecular Dynamics). Images were subsequently processed using Adobe Photoshop.
Reporter fusions.
A 2.0-kb fragment of the ins-33 promoter was amplified and cloned into the GFP expression vector pPD95.67 containing a nuclear targeting signal (13a) to generate the ins-33::GFP vector pVT396. Subsequently, a version of this vector containing only 452 bp of the ins-33 upstream region (for sequence, see Fig. S3 available at http://banjo.dartmouth.edu/Hristova_etal_Sup/) was generated (pVT397) by removing intervening sequences of the promoter by digestion using an internal ClaI site and the HindIII pPD95.67 cloning site. To introduce mutations at putative LIN-14 binding sites within the 452-bp fragment, PCR using oligonucleotides with the mutations was carried out, and the new sequence verified by sequencing.
Germ line transformation and analysis of GFP reporter expression.
Worms were transformed by microinjection as previously described (34). Transgenes expressing various regions of LIN-14 protein fused to GFP were previously described (20). The construct carrying the lin-14(ma135) loss-of-function allele was generated by PCR from genomic DNA from mutant animals. The mutation was verified by sequencing. The transgenic strains were generated by microinjection of purified plasmid DNA to a concentration of 20 ng/μl, DraI-digested N2 genomic DNA to a concentration of 100 ng/μl, and pVT301 (col-19::GFP) to a concentration of 15 ng/μl as a coinjection marker (extrachromosomal array maEx177). The following extrachromosomal arrays were generated: maEx171 (plasmid pVT396), maEx172 (plasmid pVT397), and maEx173 (plasmid pVT397mut). Expression of GFP reporters in live transgenic worms was characterized using a Zeiss Axioscop fluorescence microscope. Worms were anesthetized using 1 mM levamisole, and images were captured using an Optronics DEI750 integrating color charge-coupled-device video camera and a Scion CG-7 RGB video capture board on a Power Macintosh G4. Images were further processed using Adobe Photoshop.
RNAi.
RNA-mediated interference (RNAi) was performed as previously described (15, 56) except that the isopropyl-β-d-thiogalactopyranoside concentration in the NGM plates was 5 mM. To make the ins-33 RNAi vector, the coding region of ins-33, including a kilobase of intronic sequence, was amplified and cloned into the feeding vector L4440 (56). Ten L4 or young adult worms were placed onto a plate seeded with the bacterial strain HT115 expressing the RNAi plasmid of interest. After 24 to 30 h, the adults were removed, and progeny were scored 16 to 18 h later for viability and visible phenotypes.
RESULTS
Low-affinity nuclear association and chromosome binding of LIN-14 correlate with lin-14 function in vivo.
The analysis of nuclei from worm extracts reveals three types of interactions between endogenous LIN-14 and worm nuclei: (i) a LIN-14 fraction that elutes easily with the low-salt buffer used to prepare nuclei (Table 1; see Fig. S1 available at http://banjo.dartmouth.edu/Hristova_etal_Sup/); (ii) a LIN-14 fraction that elutes with nuclease treatment, suggesting chromosomal association of this fraction (see Fig. S1 at http://banjo.dartmouth.edu/Hristova_etal_Sup/); and (iii) a tightly bound LIN-14 fraction that appears to be associated with the nuclear matrix (Table 1; see Fig. S2 at http://banjo.dartmouth.edu/Hristova_etal_Sup/). Our results described below suggest that lin-14 function in vivo does not require the tight binding of LIN-14 to the nuclear matrix; rather, lin-14 function seems to be associated with the chromosome binding activity of LIN-14.
TABLE 1.
In vitro nuclear association and in vivo activity of lin-14 deletion constructs
| Genotypea | Exonsa | Nuclear localization in vivob | Nuclear retention in vitroc | Matrix association in vitrod | Functione |
|---|---|---|---|---|---|
| Endogenous LIN-14 | 1-13 | +++ | ∼60% | +++ | +++ |
| lin-14(n179); LIN-14D5::GFP | 9-13 | ++ | ∼0% | NA | ++ |
| lin-14(n179); LIN-14D6::GFP | 10-13 | + | ∼0% | NA |
Transgenic animals were homozygous mutant for lin-14(n179ts) and also contain an extrachromosomal array carrying the indicated LIN-14 deletion construct. (Fig. 1A) (see reference 20 for details).
Native protein localization was determined by indirect immunofluorescence (50) using anti-LIN-14 antibodies (data not shown). Localization of the GFP fusion proteins was determined by microscopy. +++, exclusively nuclear localization; ++, predominantly nuclear localization; +, about two-thirds nuclear localization.
Estimated portion of the LIN-14 protein in the pellet versus supernatant fractions, as determined Western blotting, after low-salt lysis of L1 larvae.
Presence of LIN-14 retained in nuclei after low-salt wash, and extractable with nuclear matrix in lithium 3,5-diiodosalicylate wash. Matrix association assays were not performed, because negligible amounts of LIN-14D5::GFP or LIN-14D6::GFP remained associated with the nuclear fraction after low salt extraction.
See Table 2.
To investigate which domains of LIN-14 mediate the low-affinity and high-affinity nuclear association properties of the protein and which of these properties of LIN-14 best correlates with lin-14 function in vivo, we examined the nuclear association of various LIN-14::GFP fusion proteins. Nuclei were isolated from transgenic worms carrying extrachromosomal arrays expressing N-terminally truncated forms of LIN-14 fused to GFP (Fig. 1A) (20). The behavior of two of these constructs, LIN-14D5::GFP (exons 9 to 13) and LIN-14D6::GFP (exons 10 to 13), in cell fractionation experiments is summarized in Table 1. Previously, the in vivo nuclear localization of both LIN-14D5::GFP and LIN-14D6::GFP was verified by microscopy, and LIN-14D5::GFP was found to be functional in vivo, while LIN-14D6::GFP was not functional (20). Significantly, we found that essentially all LIN-14D5::GFP and LIN-14D6::GFP was easily extracted from nuclei with low salt, indicating that neither fusion protein associates with the nuclear matrix. Since both constructs are nuclear localized in vivo (20) and since LIN-14D5::GFP rescues lin-14 activity, we conclude that nuclear matrix association is not critical for LIN-14 nuclear localization or for lin-14 function in vivo.
Among a set of truncated LIN-14::GFP fusion proteins that we tested in transgenic worms, lin-14 rescuing activity correlated precisely with chromosome association, as revealed during mitotic chromosome condensation (Fig. 1; Table 2). In transgenic worms expressing truncated versions of LIN-14 fused to GFP, fluorescence was diffusely distributed within interphase nuclei, but during mitosis, fluorescence becomes concentrated in association with mitotic chromosomes (20) (Fig. 1B to D). For example, in Fig. 1B and C, LIN-14D5::GFP fluorescence can be seen in association with mitotic chromosomes. LIN-14D5::GFP displayed mitotic chromosome association in lin-14(null) animals (Fig. 1C and Table 2), indicating that endogenous lin-14 activity is not required for the chromatin association of a LIN-14::GFP fusion protein.
TABLE 2.
In vivo chromosome association and genetic activity of lin-14 deletion constructs
| Genotypea | Exonsb | Mitotic chromosome associationc | Functiond |
|---|---|---|---|
| lin-14(n179); LIN-14D3::GFP | 5-8* | − | − |
| lin-14(n179); LIN-14D5::GFP | 9-13 | ++ | ++ |
| lin-14(ma135); LIN-14D5::GFP | 9-13 | ++ | ++ |
| lin-14(n179); LIN-14D6::GFP | 10-13 | − | − |
| lin-14(n179); LIN-14D7::GFP | 11-13 | − | − |
| lin-14(n179); LIN-14D8::GFP | 9-11* | − | − |
| lin-14(n179); LIN-14D9::GFP | 8*-12* | ++ | + |
| lin-14(n179); LIN-14D10::GFP | 9(n179)-13 | − | − |
Transgenic animals were homozygous mutant for either lin-14(n179ts) or for lin-14(ma135) and also contain an extrachromosomal array carrying the indicated LIN-14 deletion construct (Fig 1) (20).
See Hong et al. (20) for details of constructs. Asterisks indicate that the construct does not contain the full-length exon.
++, chromosome association observed; −, no chromosome association evident.
Summary of transgenic rescue results from Hong et al. (20). ++, relatively strong rescue; + relatively weaker rescue; −, no rescue.
The nonrescuing construct, LIN-14D10::GFP, which contains exons 9 to 13 with the n179 point mutation, did not associate with either metaphase or anaphase chromosomes (Table 2), even though the mutant protein's nuclear localization appears to be normal before and after mitosis (data not shown). Other nonrescuing constructs, LIN-14D6::GFP, LIN-14D7::GFP, and LIN-14D8::GFP, also showed no chromosomal association (Fig. 1E to G and Table 2), suggesting that the ability to associate with mitotic chromosomes reflects an activity of LIN-14 that is related to its in vivo function. It should be noted that endogenous LIN-14, assayed by immunohistochemistry, was not detectable in association with mitotic chromosomes (data not shown). Thus, endogenous LIN-14 may associate with mitotic chromosomes in amounts below the sensitivity of the LIN-14 antiserum. Since the truncated LIN-14 fusion proteins (which are produced from extrachromosomal arrays) are expressed at levels that likely exceed that of endogenous LIN-14, their behavior may reflect a natural, low-affinity chromosome binding activity of LIN-14. It is nevertheless possible that the chromosome association exhibited by truncated LIN-14::GFP proteins is chiefly a result of the removal of carboxy-terminal domains of LIN-14.
Bacterially expressed LIN-14 protein can bind DNA.
To explore further the biochemical activity of LIN-14, we generated a GST-tagged LIN-14 fusion protein [GST::LIN-14(244-465)] containing the LIN-14 sequences from the D9-GFP construct (Fig. 1A). D9 had previously been shown by Hong et al. (20) to be sufficient for lin-14 activity in vivo (Table 2). The GST module at the N terminus of the protein was used to purify GST::LIN-14(244-465) using glutathione beads (GE Healthcare) (14).
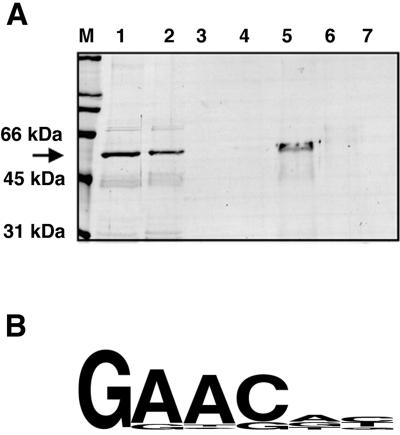
To investigate whether LIN-14 can bind nucleic acids, purified GST::LIN-14(244-465) was applied to a double-stranded DNA cellulose column in low salt (50 to 75 mM NaCl), the column was washed with 150 mM NaCl, and bound protein was eluted step-wise with increasing concentrations of NaCl (Fig. 2A). Eluted fractions were collected and run on an 11% SDS-PAGE gel. The fusion protein bound to DNA and was eluted with 400 mM salt, a concentration typical of other DNA-binding proteins (Fig. 2A, lane 5) (55). The fusion protein can also bind single-stranded DNA and elutes at similar salt concentrations (data not shown). A similar LIN-14 fusion protein tagged with a six-histidine element [Fig. 1A, 6HIS::LIN-14(244-465)] also showed similar affinity to double-stranded DNA (data not shown), indicating that the LIN-14 moiety is responsible for the in vitro DNA-binding activity of these fusion proteins.
FIG. 2.
DNA binding activity of bacterially expressed LIN-14. (A) Purified bacterially expressed LIN-14::GST was applied to double-stranded DNA-cellulose, and the bound and eluted fractions were analyzed by SDS gel electrophoresis and Coomassie staining. M, molecular weight markers; lane 1, sample of material prior to loading on DNA cellulose; lane 2, flowthrough; lane 3, wash with loading buffer; lanes 4 to 7, elution with 200 mM NaCl, 400 mM NaCl, 800 mM NaCl, and 1.5 M NaCl, respectively. Three elution fractions were collected for each NaCl concentration, pooled, and dialyzed against 150 mM NaCl, and a sample of the dialyzed protein was loaded onto the gel. (B) Sequence logo of SELEX-derived consensus binding specificity of bacterially expressed LIN-14, generated using WebLogo (http://weblogo.berkeley.edu) from 162 sequences (see Tables S3 and S4 at http://banjo.dartmouth.edu/Hristova_etal_Sup/).
Bacterially expressed LIN-14 can select a double-stranded DNA consensus binding site from random double-stranded oligonucleotides.
To determine if LIN-14 prefers a specific DNA sequence, we performed a PCR-based selection assay to screen for consensus binding sites. Starting with double-stranded oligonucleotides that contained random stretches of 20 nucleotides, we performed five rounds of selection and amplification of oligonucleotides bound to GST::LIN-14(244-465). Glutathione beads were used to pellet and wash the protein-DNA complex. The bound DNA was eluted and used as a template for further PCR amplification (see Materials and Methods).
Overall, 188 sequences were recovered from rounds 4 and 5 of the selection process. A total of 163 of these sequences were manually aligned by their core consensus GAAC, which was identified after visual inspection. The results of these sequence alignments (Fig. 2B) (see also Tables S1 and S2 at http://banjo.dartmouth.edu/Hristova_etal_Sup/) suggest a LIN-14 consensus binding sequence GAACRY (complement, RYGTTC).
Genes affected in mRNA abundance in lin-4(lf) and lin-14(lf) mutants.
To identify candidate LIN-14 target genes whose transcription would be repressed or activated by lin-14 in the L1 stage, we performed microarray analysis of C. elegans transcript levels at the L1 and L2 stages in RNA prepared from synchronized populations of wild type, lin-14(lf), and lin-4(lf) larvae. The results from two complementary sets of microarray experiments are summarized in Table 3. Of the 11,899 genes assayed, 463 (classes A to D, G, and H) showed at least a twofold statistically significant change in expression in lin-14(lf) mutants compared to wild-type animals; of those 463 lin-14-responsive genes, 311 genes (classes B, D, and H) were elevated in the absence of lin-14, and 152 genes (classes A, C, and G) were reduced. A total of 442 genes (classes C to H) showed at least a twofold statistically significant change in expression in lin-4(lf) mutants compared to wild-type animals; of the 442 lin-4-responsive genes, 334 genes (classes C, E, and H) were elevated in the absence of lin-4, and 108 genes (classes D, F, and G) were reduced. Twenty-one genes (classes C, D, G, and H) were affected in both lin-14(lf) and lin-4(lf) mutants.
TABLE 3.
Effects of lin-14(lf) and lin-4 (lf) mutations on C. elegans mRNA levels
| Classa | Effect on mRNA level
|
No. of genesb | |
|---|---|---|---|
| lin-14(lf) in L1 | lin-4(lf) in L2 | ||
| A | Down | Nochange | 137 |
| B | Up | Nochange | 305 |
| C | Down | Up | 11 |
| D | Up | Down | 5 |
| E | Nochange | Up | 322 |
| F | Nochange | Down | 99 |
| G | Down | Down | 4 |
| H | Up | Up | 1 |
| I | Nochange | Nochange | 11,015 |
Classes of genes observed by microarray hybridization, based on at least a twofold change in transcript levels in lin-4 and lin-14 mutants compared to the wild-type. Eight different behaviors were observed (corresponding to classes A to H). Genes of class A and B are affected by lin-14(lf) but not lin-4(lf), suggesting that their transcript levels are controlled by lin-14 in the L1 stage only. Genes of class C and D are affected in opposite ways by lin-14(lf) and lin-4(lf), suggesting that their transcription could be regulated by lin-14 in L1, and by lin-4 in L2 via repression of lin-14. Genes of classes E and F are affected by lin-4(lf) but not by lin-14(lf), suggesting these genes are regulated independently of lin-14 in L1 and by lin-4 in L2 via repression of a putative unknown lin-4 target. Genes of class G and H are affected similarly by lin-14(lf) and lin-4(lf) suggesting these genes are regulated independently by lin-14 in the L1 stage and by a lin-4 pathway in the L2 stage.
The total number of genes assayed was 11,899.
Candidate LIN-14 direct targets.
The objective of our expression profiling of lin-14 and lin-4 larval stages was to identify candidate direct targets of lin-14 so that these could be used to test the hypothesis that LIN-14 is a novel transcription factor. The best candidates for lin-14 target genes would be genes whose mRNA levels changed in a complementary fashion in lin-14 and lin-4 mutants. In particular, genes of class D, which were elevated in the lin-14(lf) mutant and reduced in the lin-4(lf) mutant, would be potential L2-specific lin-14 targets that are normally repressed by lin-14 during the L1 stage, while genes of class C, which are reduced in lin-14(lf) mutant worms and elevated in the lin-4(lf) mutant worms, would be potential L1-specific targets that are normally activated by lin-14 during the L1 stage (Table 3). Sixteen genes fit these criteria (Table 3), including 5 genes of the L2-specific category (class D) and 11 genes of the L1-specific category (class C).
Although any of the 16 class C and class D genes could be direct targets of LIN-14, we applied further criteria to narrow the set of candidates. First, we compared the list of 16 candidates to published microarray gene expression data that analyzed mRNA levels in RNA from C. elegans larval stages (19). Four of the class C genes (F41C3.5, C50F2.6, W01F3.3, and C44H4.3) were found to be more abundant in L1 larvae than in L2 larvae in a study by Hill et al. (19) (Table 4). This is consistent with our results that these four genes are reduced in mRNA abundance in lin-14(lf) L1 larvae compared to wild type, and hence could be L1-enriched genes that are normally under positive temporal regulation by LIN-14. One of the class D genes (W09C5.4) was detected at a higher level in L2 larvae than in L1 larvae by Hill et al. (19), which is consistent with our finding that W09C5.4 is elevated in lin-14(lf) L1 larvae compared to the wild type, and hence could be an L2-enriched gene that is normally under negative temporal regulation by LIN-14.
TABLE 4.
Genes whose mRNA levels changed significantly in both lin-4 and lin-14 mutants
| Genea
|
Delta of lin-14(lf)b
|
Delta of lin-4(lf)c
|
Classd | Stagee | Phenotype(s)f | |||
|---|---|---|---|---|---|---|---|---|
| UID | Locus | |||||||
| C15C7.5 | −1.00 | Down | 1.14 | Up | C | U | FATl CLRg GROg UNCg | |
| F01G10.6 | −1.01 | Down | 1.24 | Up | C | U | ||
| F41C3.5 | −1.13 | Down | 1.06 | Up | C | L1 | ||
| F31E3.3 | rfc-4 | −1.22 | Down | 1.17 | Up | C | U | EMBg,k,j GROg STPj |
| D2096.6 | −1.54 | Down | 2.67 | Up | C | L2 | ||
| C50F2.6 | fkb-5 | −1.70 | Down | 2.17 | Up | C | L1 | |
| C02E7.7 | −1.72 | Down | 3.06 | Up | C | U | ||
| F53F1.4 | −1.73 | Down | 2.07 | Up | C | U | ||
| M153.1 | −1.74 | Down | 2.28 | Up | C | U | ||
| W01F3.3 | −1.90 | Down | 2.15 | Up | C | L1 | UNCg,l EMBk LVAi | |
| C44H4.3 | sym-1 | −2.04 | Down | 1.30 | Up | C | L1 | |
| F58E6.4 | 2.98 | Up | −1.57 | Down | D | U | ||
| W09C5.4 | ins-33 | 2.26 | Up | −2.43 | Down | D | L2 | EMBm GROm UNCm |
| F44G3.2 | 1.24 | Up | −3.61 | Down | D | U | ||
| B0412.1a | dac-1 | 1.18 | Up | −1.62 | Down | D | U | TTXo |
| T10B9.7 | cyp-13 | 1.18 | Up | −1.54 | Down | D | U | |
| F59B8.2 | −1.10 | Down | −1.16 | Down | G | U | ||
| C05D9.8/R193.2 | −1.20 | Down | −1.19 | Down | G | U | GROg | |
| F01G10.1 | −1.22 | Down | −1.07 | Down | G | U | CLRg GROg,h EMBg,h,n LVAh | |
| R08E5.2 | −1.39 | Down | −1.39 | Down | G | U | ||
| T02B5.3 | 1.13 | Up | 1.41 | Up | H | U | ||
From www.wormbase.org. The genes shown in bold contain putative LIN-14 recognition sites in their 5′ upstream sequences (see Table S2 at http://banjo.dartmouth.edu/Hristova_etal_Sup/). UID, unique identifier.
Log 2 of the change (n-fold) in lin-14(lf) L1 stage larvae mRNA expression relative to RNA from wild-type L1 larvae.
Log 2 of the change (n-fold) in lin-4(lf) L2 stage larvae mRNA expression relative to RNA from wild-type L2 larvae.
For class, see http://banjo.dartmouth.edu/Hristova_etal_Sup/ (Table S2).
Stage of greater expression in the wild type (considering only the L1 and L2 stages), based on the results of Hill et al. (19). U, less than twofold difference between the L1 and L2 stages.
Abnormalities observed in RNAi experiments of various investigators. EMB, embyronic lethal; GRO, larval growth defects; FAT, defective in fat metabolism; CLR, clear; STP, sterile progeny; UNC, uncoordinated behavior; LVA, larval arrest; TTX, thermosensing defective. For genes showing no phenotype in RNAi, no description is given.
Kamath et al. (23).
Maeda et al. (32).
Simmer et al. (52).
Piano et al. (39).
Sonnichsen et al. (54).
Ashrafi et al. (6).
Fraser et al. (15).
Rual et al. (47).
Mutant analysis of Colosimo et al. (12).
As a final criterion for identifying candidate target genes that LIN-14 could regulate in vivo by DNA binding, we examined the upstream sequences (within 4,000 bp of the start of translation) of the genes in Table 4 for occurrence of sequences that fit the consensus LIN-14 binding sequence generated by SELEX analysis (Fig. 2B; see Table S3 at http://banjo.dartmouth.edu/Hristova_etal_Sup/). For this analysis we used an expanded consensus sequence including a total of 12 bases surrounding the GAACRY core (derived from alignment of the sequences in Table S1 at http://banjo.dartmouth.edu/Hristova_etal_Sup/), followed by processing using the CisOrtho software pipeline (http://wormbase.org/cisortho/) (9). The CisOrtho search of the C. elegans genome yielded five genes (C15C7.5, W09C5.4, T10B9.7, F59B8.2, and F01G10.1) with upstream matches to a LIN-14 consensus (see Table S3 at http://banjo.dartmouth.edu/Hristova_etal_Sup/). For three of these genes (C15C7.5, W09C5.4, and T10B9.7), predicted LIN-14 sites are also found upstream of the Caenorhabditis briggsae ortholog, suggesting that these three genes are the strongest candidates for bona fide LIN-14 targets. By the criteria applied here, W09C5.4 best fits the profile of a gene that could be regulated in vitro by direct binding of LIN-14; W09C5.4 mRNA is (i) L2-specific in the wild type (19), (ii) up-regulated in lin-14(lf) L1 larvae and down-regulated in lin-4(lf) L2 larvae, and (iii) contains conserved LIN-14 consensus binding sites in its upstream sequences. For these reasons we used W09C5.4 to test the hypothesis that LIN-14 can regulate gene expression in vivo by DNA binding.
W09C5.4 encodes a developmentally regulated insulin/insulin-like growth factor (IGF) homolog, ins-33.
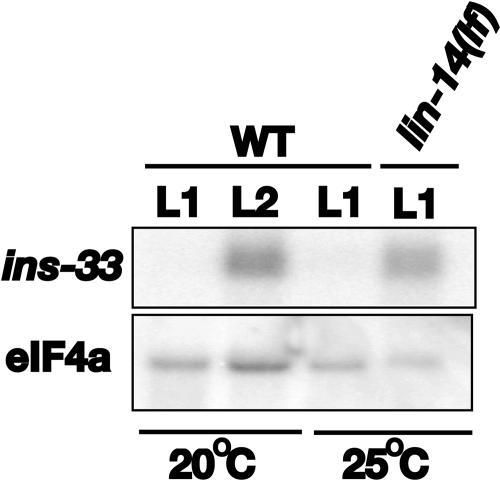
W09C5.4 encodes a predicted protein of 124 amino acids that is homologous to members of the C. elegans-specific alpha group of insulin/IGF-like proteins (40). In our microarray experiments, W09C5.4 transcripts were at least four times more abundant in lin-14(lf) L1 larvae compared to wild-type L1 larvae and at least three times lower in lin-4(lf) L2 larvae compared to wild-type L2 larvae. These data suggest that W09C5.4 may be an L2-specific gene that is repressed by lin-14 in the L1 stage of the wild type. To confirm this supposition, the developmental profile of ins-33 was examined using Northern blot analysis of RNA from wild-type larval stages. ins-33 transcripts are absent in the L1 and then accumulate in the L2 stage (Fig. 3, compare first and second lanes). In lin-14(lf) L1 animals, ins-33 mRNA accumulates precociously (Fig. 3, compare third and fourth lanes). In lin-4(lf) L2 larvae, where LIN-14 levels are abnormally high, ins-33 message is reduced compared to wild type (data not shown). These results indicate that ins-33 gene expression is developmentally regulated and that lin-14 negatively regulates ins-33 during the first larval stage.
FIG. 3.
Northern blot analysis of ins-33 mRNA levels in poly(A)-selected RNA from wild-type or lin-14(lf) mutant animals at the indicated stages that had been grown at the indicated temperatures. mRNA samples from either wild-type (lanes 1 to 3) or lin-14(lf) (lane 4) animals, which were grown at either 20°C (lanes 1 and 2) or 25°C (lanes 3 and 4), were run on a formaldehyde-agarose gel and transferred onto a nylon membrane. The membrane was probed for ins-33 message (upper panel) and eIF4a (lower panel) as a loading control (46). ins-33 mRNA displays stage-specific expression in wild-type animals: low or absent message in L1 animals and high transcript levels in L2 animals. The ins-33 mRNA level is elevated in the L1 stage in the absence of lin-14 (compare lane 4 to lane 3).
The ins-33 5′ upstream regulatory region confers stage-specific reporter expression.
To determine whether the observed developmental regulation of ins-33 mRNA is controlled at the level of transcription, we generated transcriptional reporter plasmids containing approximately 2 kb of 5′ upstream sequence from the ins-33 gene (pVT396) or 452 bp of ins-33 upstream sequence (pVT397) fused to GFP (see Fig. S3 at http://banjo.dartmouth.edu/Hristova_etal_Sup/). Each plasmid was injected into wild-type worms, and stable transgenic lines were established. Both pVT396 and pVT397 conferred identical stage-specific and cell type-specific GFP expression. The transgene carrying pVT397 (maEx172) was used as the basis for all subsequent experiments.
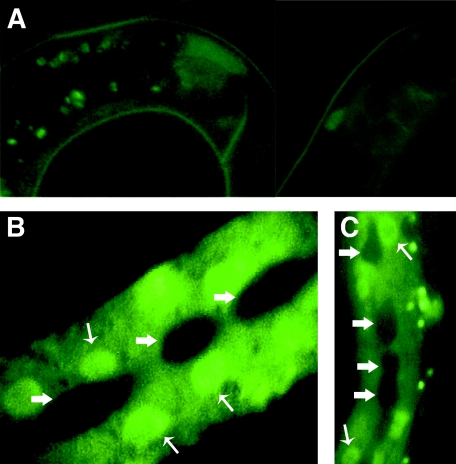
Faint expression of the maEx172 reporter was observed in wild-type L1 animals in the head and in one or two cells in the posterior intestine (Fig. 4A). In contrast to this relative inactivity of the ins-33 promoter in L1 larvae, beginning at the end of the L1 and continuing during the L2 and later stages, expression of ins-33::GFP was dramatically up-regulated in the head and tail hypodermis, the ventral hypodermis, and the main body hypodermis, hyp-7 (although not in the hypodermal seam cells) (Fig. 4B). These observations indicate that the 452 bp of the ins-33 upstream region contained in maEx172 can mediate transcriptional upregulation of ins-33 expression in hyp-7 after the L1 stage of the wild type.
FIG. 4.
ins-33::GFP expression in the hypodermis is stage specific and is controlled by lin-14. (A) Wild-type L1 larvae. GFP expression is relatively low throughout the animal. (B) Wild-type L2 larva. The most striking expression of the GFP reporter is observed in the hypodermis after the L1 stage. Expression is excluded from the seam cells (thick arrows), which can be seen as dark areas. Expression can be observed in hypodermal cells in the head and tail (not shown), as well as in the main body hypodermal synctium, hyp-7, shown here (thin arrows indicate hyp-7 nuclei). (C) lin-14(lf) L1 larva. Precocious expression of ins-33::GFP is observed throughout the hypodermis, in a pattern similar to that of wild-type L2 larvae (compare with panel B).
Stage-specific expression of ins-33::GFP is regulated by lin-14.
The stage-specific expression of GFP under the control of ins-33 upstream regulatory sequences is consistent with the observed ins-33 mRNA levels (Fig. 3), which start accumulating in RNA samples of wild-type L2 larvae. Northern analysis had revealed that in a lin-14(lf) mutant, endogenous ins-33 mRNA appears precociously during the L1 stage (Fig. 3). To determine if the stage-specific regulation of ins-33 mRNA detected in Fig. 3 reflects regulation of ins-33 promoter activity, the ins-33::GFP promoter fusion transgene (maEx172) was crossed into lin-14(n179ts). These maEx172 lin-14(n179) animals were raised at the permissive (15°C) or nonpermissive (25°C) temperature for lin-14(n179) function. L1 larvae raised at the permissive temperature showed the usual faint L1 expression of maEx172 typical of wild-type larvae (see above). By contrast, L1 larvae raised at 25°C exhibited GFP expression in the hypodermis (Fig. 4C) at levels similar to wild-type L2 larvae. Thus, based on Northern analysis of endogenous ins-33 mRNA (Fig. 3) and the expression of ins-33::GFP (Fig. 4), we conclude that lin-14 functions via ins-33 upstream sequences to repress transcription of ins-33 in the hypodermis of L1 larvae.
Bacterially expressed LIN-14 binds specifically to two sites in the ins-33 upstream sequences.
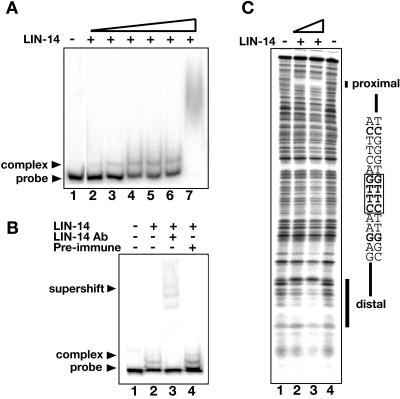
The above results indicate that lin-14 regulates ins-33 expression at the level of transcription. This regulation could occur directly, through direct binding of the LIN-14 protein to ins-33 regulatory sequences in the DNA, or indirectly, by the binding of LIN-14 to another protein or gene that in turn regulates the ins-33 promoter. LIN-14 is a novel protein, and hence its structure does not easily allow prediction of biochemical function. However, evidence in favor of a hypothetical direct DNA binding activity for LIN-14 comes from its nuclear localization (50) and our findings (described herein) that bacterially expressed LIN-14 can bind double-stranded DNA and can select a specific binding site via SELEX. To test if LIN-14 protein can directly bind to ins-33 upstream sequences, we performed EMSA using bacterially expressed LIN-14 and ins-33 upstream DNA. The fusion protein 6HIS::LIN-14(284-465), which contains a fragment of LIN-14 that is sufficient to rescue lin-14 function (20) (Fig. 1A), was incubated with an end-labeled 452-bp ins-33 DNA fragment (see Fig. S3 at http://banjo.dartmouth.edu/Hristova_etal_Sup/) corresponding to the sequence that we found to confer lin-14-dependent expression of a GFP reporter in transgenic worms (Fig. 4). LIN-14 protein bound to the 452-bp DNA fragment, resulting in a species of altered electrophoretic mobility (Fig. 5A, lanes 2 to 6). At higher protein concentrations, an apparent aggregate between the protein and DNA is evident (Fig. 5A, lane 7). To further delineate the location of LIN-14 binding, a shorter fragment (see Fig. S3 at http://banjo.dartmouth.edu/Hristova_etal_Sup/) of 365 bp was tested. This fragment also shifted in the presence of LIN-14 protein (Fig. 5B, lane 2). The binding was due specifically to LIN-14 protein, as the addition of anti-LIN-14 antibody to the binding reaction caused a supershift of the probe (Fig. 5B, lane 3), while addition of preimmune serum had no effect (Fig. 5B, lane 4).
FIG. 5.
LIN-14 directly binds ins-33 upstream sequences in vitro. (A and B) EMSAs. Bacterially expressed 6HIS::LIN-14(284-465) (Fig. 1A) was bound to end-labeled fragments of the ins-33 promoter (see Fig. S3 at http://banjo.dartmouth.edu/Hristova_etal_Sup/). (A) Protein-DNA complexes formed between 6HIS::LIN-14(284-465) (8 to 320 nM) and a labeled 452-bp fragment of ins-33 upstream sequences. The super-retarded smeared signal formed at 320 nM 6HIS::LIN-14(284-465) (lane 7) may represent an aggregate too large to be resolved properly on the gel. (B) Protein-DNA complexes formed between 6HIS::LIN-14(284-465) (40 nM) and labeled 365-bp fragment of the ins-33 upstream sequence. Addition of anti-LIN-14 antibody (lane 3) further retards the mobility of the bound material; mobility is unaffected by the addition of preimmune serum (lane 4). (C) LIN-14 protects two sites in the ins-33 promoter from DNase I digestion. The 365-bp fragment of the ins-33 promoter was end labeled at the 3′ end and incubated with a molar excess of 6HIS::LIN-14(284-465). Lanes 1 and 4 are samples where no LIN-14 was added, while lanes 2 and 3 are samples with increasing concentrations of excess 6HIS::LIN-14(284-465). The DNA was partially digested with DNase I and fragments were separated on a sequencing gel. Two sites are protected from DNase I digestion in the presence of 6HIS::LIN-14(284-465); these sites are referred to as proximal and distal, indicating their relationship to the predicted translation start site. An alignment of the two protected sequences shows a core of four nucleotides shared by the two sequences and identical to the core consensus binding site determined for LIN-14 in vitro (Fig. 2B).
To more precisely map the LIN-14 binding site(s), we performed DNase I footprinting. In the presence of 6HIS::LIN-14-(284-465), two sites within either the 452-bp fragment (not shown) or the 365-bp fragment (Fig. 5C) were protected from digestion. The specific regions protected were mapped more precisely using shorter fragments of DNA (not shown), and the sequences of the approximately 15-bp regions protected is shown in Fig. 5C. The two regions are referred to as “proximal” and “distal” in reference to their location with respect to the ins-33 translation start site. Alignment of the two sequences revealed the common core G(TT/AA)C, which is identical to the core consensus sequence identified through PCR-assisted binding site selection (Fig. 2B).
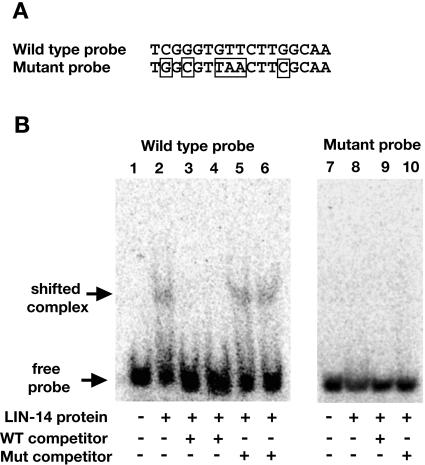
These results indicate that the LIN-14 protein can directly bind to at least two sites in the ins-33 upstream regulatory region. Moreover, the presence of identical nucleotides found at the core of each protected sequence strongly suggests that these nucleotides are required for binding of LIN-14 to the DNA. To test this hypothesis, we introduced mutations at six of the positions in the proximal protected element (Fig. 6A). EMSA assays using 50-bp oligonucleotides with sequences containing the wild-type or mutated proximal protected element were performed to test binding specificity. The 6HIS::LIN-14(284-465) protein bound the oligonucleotides containing the wild-type but not the mutated sequence (Fig. 6B, compare lanes 2 and 8). Moreover, the mutated sequences did not compete away the binding between 6HIS::LIN-14(284-465) and the wild-type sequences (Fig. 6B, lanes 5 and 6), whereas the wild-type sequence did (Fig. 6B, lanes 3 and 4). These results support the conclusion that LIN-14 can bind specifically to two sequences in the ins-33 upstream regulatory region and that residues in the core sequence G(TT/AA)C, possibly in conjunction with residues outside the core, are necessary for this binding.
FIG. 6.
Bacterially expressed LIN-14 binds a fragment of the ins-33 promoter containing the wild-type LIN-14 consensus binding site but does not bind a fragment containing a mutated binding site. (A) The wild-type proximal LIN-14 binding site or the indicated mutant sequence (mutated sequence boxed) was included in 50-bp double-stranded DNA oligonucleotides used in gel shift assays with purified 6HIS::LIN-14(284-465) protein. (B) 6HIS::LIN-14(284-465) binds the wild-type fragment of ins-33 promoter (compare lanes 1 and 2). This binding requires the presence of a wild-type core consensus and surrounding sequences, as the mutated fragment did not compete for binding to LIN-14 (compare lanes 2 and 3 with lanes 4 and 5). Lanes 3 and 5 have a 50× molar excess of unlabeled competitor; lanes 4 and 6 have a 200× molar excess of unlabeled competitor. LIN-14 did not bind a fragment of the ins-33 promoter containing the mutated sequences shown in panel A (compare lanes 7 and 8 with lanes 1 and 2). The wild-type and mutant competitors in lanes 9 and 10 were present at a 50× molar excess.
Stage-specific regulation of ins-33::GFP by lin-14 requires wild-type sequences in the two LIN-14 binding sites.
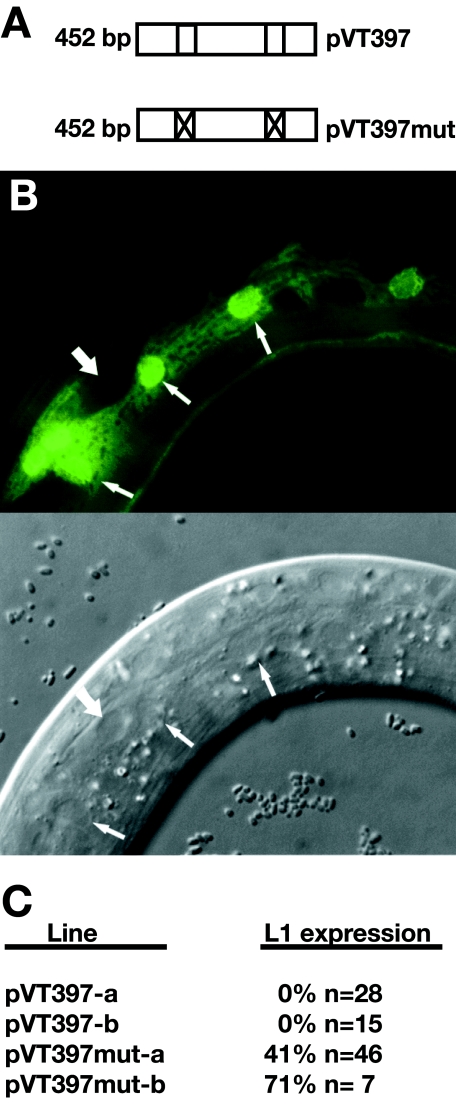
The 6HIS::LIN-14(284-465) fusion protein was found to bind two sites within 452 bp of the ins-33 upstream regulatory sequence in vitro. To test if the binding sites are important for the lin-14 repression of ins-33::GFP hypodermal expression in the L1, we engineered the mutations described above (Fig. 6A) into both putative LIN-14 binding sites in the 452-bp upstream region in pVT997, creating pVT997mut (Fig. 7A). Wild-type and mutant constructs were injected into lin-14(n179ts) animals, and stable transgenic lines were established containing either the wild-type or the mutant transgene. At 15°C, which is the permissive temperature for lin-14(n179ts), two independent lines carrying the mutant upstream sequences showed precocious expression of GFP during the L1 stage (Fig. 7B and C). This result indicates that the mutant promoter is substantially resistant to repression by lin-14. In contrast, the wild-type transgene was not expressed in lin-14(n179ts) L1 larvae at 15°C (Fig. 7C) but was expressed when worms were grown at the nonpermissive temperature for lin-14 (25°C) (data not shown).
FIG. 7.
An ins-33::GFP transgene carrying mutations in the LIN-14 binding sites is expressed precociously in the hypodermis of L1 larvae. (A) Wild-type and mutant versions of the 452-bp ins-33 upstream sequence. The mutant sequence is altered at both the proximal and distal binding sites (Fig. 4A). (B) An L1 animal expressing ins-33::GFP from the pVT397mut reporter transgene (top panel). Arrows indicate hyp-7 nuclei, while the arrowhead points to a nonexpressing seam cell (also indicated in the differential interference contrast image, lower panel). (C) Frequency of precocious expression during the L1 stage of the mutated transgene (for two independent transgenic worm lines, pVT397mut-a and pVT397mut-b) compared to the wild-type transgene (for two independent lines, pVT397-a and pVT397-b). All four lines are of genotype lin-14(n179ts), and so the animals were grown at the permissive temperature of 15°C. No animals carrying the wild-type binding site reporter transgene showed expression of the ins-33::GFP reporter precociously, while 41% and 71% of animals carrying the mutant binding site showed precocious ins-33::GFP expression. At the restrictive temperature (25°C) for lin-14(n179ts), the line carrying the wild-type promoter showed precocious expression of the reporter in 71% of animals (not shown). n, number of larvae scored.
These results indicate that the repression of ins-33::GFP expression in the L1 stage requires lin-14 activity and also requires the presence of functional LIN-14 binding sites in the ins-33 upstream regulatory region. We therefore conclude that, at least in the case of ins-33, LIN-14 appears to repress transcription by directly binding to a specific recognition sequence within 5′ regulatory sequences.
DISCUSSION
DNA binding activity of LIN-14.
Several findings suggest that chromatin association and DNA binding, in particular, are important for LIN-14 function in vivo. First, although LIN-14 is a novel protein, a carboxy-terminal domain of LIN-14 that is necessary and sufficient for in vivo function of LIN-14 (20) contains basic residues and a predicted helical structure consistent with nucleic acid binding (59). Second, we find that this LIN-14 carboxy-terminal domain, when expressed in bacteria, can bind double-stranded DNA in vitro, indicating that LIN-14 could indeed function in vivo by direct DNA binding. This LIN-14 carboxy-terminal domain also bound single-stranded DNA. Other nuclear proteins, such as RPA and p53, have been shown to bind both double- and single-stranded DNA (55, 57, 58). LIN-14 might possess both double-stranded binding and single-stranded DNA binding activities in vivo, or the single-stranded binding activity of the carboxy-terminal domain that we tested may represent a relaxed substrate specificity caused by the absence of amino terminal sequences.
Further evidence in support of a DNA binding activity for LIN-14 in vivo is our finding that LIN-14 exhibits sequence specificity of DNA binding in vitro. We performed a PCR-based binding site selection assay (SELEX) and recovered the putative consensus GAACRY (complement, RYGTTC). This consensus sequence does not appear to match previously known transcription factor binding sites, suggesting that our results identify LIN-14 as a transcriptional regulatory factor of novel sequence specificity.
Also in support of in vivo DNA binding by LIN-14 is the apparent chromatin association of endogenous full-length LIN-14 and LIN-14::GFP fusion protein. We found that a small fraction of endogenous LIN-14 is released from the bulk of chromatin by digestion of worm nuclei with MNase (see Fig. S1 available at http://banjo.dartmouth.edu/Hristova_etal_Sup/). For LIN-14::GFP fusion proteins, the evidence for functional interaction with chromatin is from a correlation between the in vivo rescuing activity of the transgenes and the association of the corresponding fusion proteins with mitotic chromosomes (Fig. 1). Other proteins known to interact directly with DNA and that also interact with condensed mitotic chromatin include ATRX (8) and the C. elegans SMAD, daf-3 (38). By contrast, some DNA binding proteins such as Sp1 and HSF1 appear to be excluded from mitotic chromatin (reference 33 and references therein). Although we find that mitotic chromosome association correlates with function for truncated, overexpressed forms of LIN-14::GFP, the endogenous LIN-14 protein was not detected in association with metaphase chromosomes (data not shown). Accordingly, we interpret the mitotic chromatin binding behavior of the LIN-14 carboxy-terminal domain to represent a low-affinity activity of the native LIN-14, which is not detected unless the protein is present at abnormally abundant amount.
Evidence supports the idea that the association of LIN-14 with DNA and/or chromatin may not represent the only in vivo activity of LIN-14. In particular, we found that a significant fraction of LIN-14 in worms is tightly bound to nuclei, perhaps in association with the nuclear matrix (see Fig. S2 at http://banjo.dartmouth.edu/Hristova_etal_Sup/), and at least some of the LIN-14 released from nuclei by MNase digestion appeared to sediment apart from polynucleosomes (see Fig. S1 at http://banjo.dartmouth.edu/Hristova_etal_Sup/). Perhaps LIN-14 exists in distinct pools in normal nuclei. We propose that one nuclear pool of LIN-14 functions to regulate developmental timing and is in equilibrium between chromatin-bound and chromatin-unbound fractions and that another pool is tightly bound (likely matrix associated). Our results suggest that nuclear matrix association may be secondary to the primary function of LIN-14. In particular, we observed that a truncated LIN-14::GFP fusion protein that can rescue lin-14 heterochronic mutant phenotypes in the hypodermis nevertheless is quantitatively eluted from nuclei with low-salt incubation. Thus, this LIN-14 variant with virtually no detectable nuclear matrix binding still retains at least partial function. We therefore conclude that the pool of LIN-14 strongly associated with the nuclear matrix in vivo does not participate directly in the regulation of gene expression, at least not in hypodermal cells, and may serve as a reservoir of LIN-14 that could be recruited for function in some physiological or developmental circumstances. It remains to be determined whether the loosely bound and tightly bound nuclear pools of LIN-14 are normally both found within the same individual nucleus and, if so, whether there is dynamic interchange among them.
LIN-14 is a transcription factor.
The assignment of transcription factor activity to LIN-14 is based on (i) its apparent chromosome association in vivo, (ii) its in vitro DNA biding activity and, in particular, (iii) its functional and biochemical relationship to a canonical LIN-14 target, ins-33. Several characteristics of ins-33 suggest that ins-33 is a direct transcriptional regulatory target of LIN-14 during worm development. First, the ins-33 mRNA is developmentally up-regulated at the L2 stage of wild-type development (Fig. 3) (19), consistent with repression by LIN-14, which is high in the L1 stage and reduced in L2. Second, the level of ins-33 mRNA is dramatically elevated in the L1 stage of animals lacking lin-14 (Fig. 3 and Table 4) and is reduced in the L2 of animals with elevated LIN-14 [lin-4(lf)] (Table 4). Third, the stage-specific expression of ins-33 mRNA in the wild type and its behavior in lin-14(lf) animals are recapitulated by a reporter transgene containing 452 bp of the ins-33 upstream region (Fig. 4). Since this 452-bp region contains no ins-33 coding sequence, these results indicate that ins-33 stage-specific expression and the regulation of ins-33 by lin-14 result from transcriptional regulation via the 452-bp upstream sequence. Finally, two specific sequences within this 452-bp sequence are protected from DNase I digestion in the presence of purified LIN-14 protein (Fig. 5); these sequences are necessary for LIN-14 binding to the ins-33 452-bp region in vitro (Fig. 6) and for lin-14-dependent transcriptional repression in vivo (Fig. 7). Taken together, these results provide strong evidence that LIN-14 is a direct repressor of ins-33 transcription in wild-type worms.
Sixteen putative lin-14 targets.
Because our objective has been to test the hypothesis that LIN-14 is a transcription factor, we applied strict criteria to identify a high-confidence candidate target to use as a test case. Therefore, we have not attempted to comprehensively catalog LIN-14 targets. The genes we identified by microarray hybridization as lin-14-responsive undoubtedly include direct targets as well as indirect targets. It is also quite likely that we may have missed numerous targets in our microarray screens for lin-14-responsive genes. One reason could be that a target might be broadly expressed in the animal but regulated by lin-14 in only a subset of cells. An example is cki-1 (T05A6.1), which has been shown to be regulated by lin-14 in the vulva precursor cells (VPCs) (21). By our microarray assays, cki-1 mRNA was essentially unchanged in lin-14(lf) and lin-4(lf). This is most likely because cki-1 is expressed in diverse cell types besides the VPCs, while the VPCs are the only cell type were cki-1 expression is affected by lin-14 mutations (21).
A key criterion for classifying genes as potential LIN-14 direct targets was the occurrence of a sequence fitting the consensus LIN-14 site derived by SELEX. Five of the 16 genes contain a good match to a 12-nucleotide consensus derived from the SELEX data. However, the sequence specificity of LIN-14 binding has not been explored in detail, and so the stringent consensus that we used to search for sites may not encompass all bona fide LIN-14 sites. Many of the 16 predicted target genes contain core G(AA/TT)C sequences in their upstream sequences and, hence, could be targets with sites that more loosely fit the search consensus (for sites see Table S2 at http://banjo.dartmouth.edu/Hristova_etal_Sup).
If the genes listed in Table 4 are bona fide lin-14 targets, how might they be involved in cell fate decisions controlled by lin-14 in L1 versus L2? The lin-14 mutations affect a diverse set of cells and cellular behaviors, including the hypodermal cell division pattern, intestinal cell cycle behavior, neuroblast identity (1), neuronal polarity (17), dauer larva arrest (30), and VPC cell cycle (21). The precise nature of the cellular behaviors controlled by lin-14 depends on cell type, and so it is reasonable to expect that LIN-14 may repress or activate target genes in a cell type-specific manner. Among the 16 high-probability LIN-14 targets listed in Table 4, only one, B0412.1, encodes a predicted transcription factor, suggesting that lin-14 does not chiefly function by regulating the expression of other transcription factors. None of the putative lin-14 targets in Table 4 has been shown to have a loss-of-function heterochronic phenotype. Several of these candidate targets have RNAi phenotypes that include general growth problems, suggesting that these genes function in one or more essential cellular processes. But since their phenotypes are not simply developmental timing defects as with lin-14(lf) or lin-4(lf), it would appear that the products of these genes may function as effectors of the cellular behaviors whose timing is controlled by lin-14.
The ins-33 gene and the function of lin-14 in developmental timing.
The evidence presented here strongly suggests that stage-specific expression of ins-33 in hypodermal cells of wild-type worms is controlled by LIN-14 on the level of transcription, suggesting a role for lin-14 as a transcription factor controlling the timing of an L2-specific insulin/IGF signal. One developmental event that is known to be regulated by both lin-14 and by insulin signaling in C. elegans is the expression of the optional third larval stage, the dauer larva. Dauer larvae are developmentally arrested and specialized larvae formed in crowded or starved cultures. In wild-type worms, dauer larva formation can occur only at the L2 molt. The lin-14 loss-of-function mutations affect the timing of dauer formation, including the execution of the dauer remodeling program by the hypodermis (30). lin-14(lf) animals can become dauer larvae precociously, at the end of the L1 stage; moreover, they form partial dauer larvae, where the hypodermis does not remodel properly in patches of the animal. The animal's decision to arrest development as a dauer larva is controlled by at least two environmental signaling pathways: a transforming growth factor β cascade, which detects a worm-specific pheromone and thus responds to crowding, and an insulin/IGF cascade, which likely reflects the metabolic state of the animal and thus responds to nutrient availability (44). Since dauer larva development is in part controlled by insulin signaling, ins-33 could function to couple the dauer larva program to temporal signals. Thirty-seven insulin-related ligands have been predicted in the worm, compared to three in vertebrates (40). DAF-2 is the C. elegans homolog of the insulin and IGF-1 receptors from vertebrates (24). It appears that daf-2 can mediate different insulin signals for different life history events; for instance, overexpression of ins-1 affects dauer formation, larval growth, and longevity, while overexpression of ins-9 and ins-31/19 affects larval growth but not dauer formation or longevity (40). It is possible that daf-2 also mediates different insulin signals in different cell types. These signals would then collaborate to influence the dauer arrest decision. This collaboration hypothesis is consistent with the finding that, for dauer development, daf-2 functions cell nonautonomously in a number of tissues, including neurons and hypodermis (4). The precise expression pattern of daf-2 has not been reported. Perhaps ins-33 is the insulin signal or one of the insulin signals from the hypodermis, which contribute(s) to the final dauer larva initiation decision.
A phenotype was previously reported for ins-33 loss-of-function by RNAi (15). The phenotype included embryonic lethality and slow-growing and uncoordinated larvae. The authors did not report heterochronic or dauer-related phenotypes. However, the original RNAi bacterial strain used in the study by Fraser et al. (15) has been lost (J. Ahringer, personal communication), and we did not observe an ins-33 phenotype in our own RNAi experiments (data not shown). Since a negative result in RNAi experiments could reflect factors other than the actual requirement for the gene, we cannot say at present whether ins-33 is required for the normal timing or execution of dauer larva development. It is possible that lin-14 controls the timing of dauer larva development through the regulation of ins-33 transcription in concert with another, redundant factor.
Complexity of regulatory pathways involving lin-14 and lin-4.
If lin-14 were the only target of the microRNA product of lin-4, then one simple expectation would be that all the genes affected by lin-14(lf) should be reciprocally affected by lin-4(lf). Genetic analysis has shown that mutations in lin-14 are epistatic to lin-4(lf) (1), which would suggest that most, and perhaps all, of the lin-4(lf) phenotype depends on lin-14 activity. However, our results described here suggest that the lin-4/lin-14 pathway is more complex than the simplifying expectation, and may diverge significantly. For example, we observed that numerous genes were significantly altered in lin-4(lf) that were unaffected in lin-14(lf) mutants (Table 3). These genes could include bona fide lin-14 targets that respond to LIN-14 levels in the L2 stage but are independent of lin-14 in the L1 stage. However, lin-4 microRNA is known to have at least one other target, lin-28 (36), which encodes an RNA binding protein that could regulate mRNA levels posttranscriptionally. Many of the genes affected by lin-4(lf) and not by lin-14(lf) in our microarray experiments, indeed, may not be targets of LIN-14 but, rather, could be regulated by lin-4 via lin-28 or other unidentified targets of lin-4.
Some bona fide LIN-14 target genes could be among the genes that did not respond reciprocally to lin-14(lf) and lin-4(lf). In particular, genes of classes A and B (Table 3) changed in the lin-14(lf) microarray experiments but were not affected by lin-4(lf). For many of these genes, expression might have been altered as an indirect consequence of the physiology of the lin-14(lf) mutant. Others could be direct LIN-14 targets whose expression is activated by LIN-14 in the L1 stage but becomes governed by other factors in the L2 stage (rendering them insensitive to removal of lin-4).
The results reported here establish that LIN-14 likely regulates the timing of developmental events through the direct binding to specific recognition sequences of diverse target genes. Down-regulation of LIN-14 levels from the L1 to the L2 stage likely results in the derepression of genes, such as ins-33, that are negatively regulated by LIN-14. It remains to be determined whether most LIN-14 target genes are repressed by LIN-14 binding or whether LIN-14 can also activate transcription in some situations. LIN-14 presumably acts in combination with cell type-specific transcriptional regulators to affect repression or activation of target genes in conjunction with regulating L1 and L2 developmental programs. A complete understanding of how the lin-4/lin-14 circuit regulates diverse developmental events in C. elegans larvae will require a comprehensive catalog of all the genes targeted by LIN-14 in vivo; knowledge of where, when, and how they are affected by lin-14 activity; and genetic analysis of their functional relationships to lin-14.
Acknowledgments
We are grateful to members of the Ambros laboratory, especially Rosalind Lee, Philip Olsen, and Richard Roy, for technical help and discussion; to Cecilia Henricksson-Spencer for technical assistance at Stockholm University; to Stuart Kim and the Stanford Microarrray Facility for help with the microarray assays; and to N. Hernandez, Y. Gruenbaum, and M. Labouesse for antisera.
This work was supported by PHS grant GM34028 to V.A.
REFERENCES
- 1.Ambros, V., and H. R. Horvitz. 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 266:409-416. [DOI] [PubMed] [Google Scholar]
- 2.Ambros, V., and H. R Horvitz. 1987. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1:398-414. [DOI] [PubMed] [Google Scholar]
- 3.Ambros, V. 2000. Control of developmental timing in Caenorhabditis elegans. Current Opin. Genet. Devel. 10:428-433. [DOI] [PubMed] [Google Scholar]
- 4.Apfeld, J., and C. Kenyon 1998. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell 95:199-210. [DOI] [PubMed] [Google Scholar]
- 5.Arasu, P., B. Wightman, and G. Ruvkun. 1991. Temporal regulation of lin-14 by the antagonistic actin of two other heterochronic genes, lin-4 and lin-28. Genes Dev. 5:1825-1833. [DOI] [PubMed] [Google Scholar]
- 6.Ashrafi, K., F. Y. Chang, J. L. Watts, A. G. Fraser, R. S. Kamath, J. Ahringer, and G. Ruvkun. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421:268-272. [DOI] [PubMed] [Google Scholar]
- 7.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 8.Bérubé, N. G., C. A. Smeenk, and D. J. Picketts. 2000. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum. Mol. Genet. 9:539-547. [DOI] [PubMed] [Google Scholar]
- 9.Bigelow, H. R., A. S. Wenick, A. Wong, and O. Hobert. 2004. CisOrtho: a program pipeline for genome-wide identification of transcription factor target genes using phylogenetic footprinting. BMC Bioinformatics 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burtelow, M. A., S. H. Kaufmann, and L. M. Karnitz. 2000. Retention of the human Rad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J. Biol. Chem. 275:26343-26348. [DOI] [PubMed] [Google Scholar]
- 11.Chittenden, T., D. M. Livingston, et al. 1991. The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell 65:1073-1082. [DOI] [PubMed] [Google Scholar]
- 12.Colosimo, M. E., A. Brown, S. Mukhopadhyay, C. Gabel, A. E. Lanjuin, D. T. S. Aravinthan, and P. Sengupta. 2004. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr. Biol. 14:2245-2251. [DOI] [PubMed] [Google Scholar]
- 13.Dixon, D. K., D. Jones, and E. P. Candido. 1990. The differentially expressed 16-kD heat shock genes of Caenorhabditis elegans exhibit differential changes in chromatin structure during heat shock. DNA Cell Biol. 9:177-191. [DOI] [PubMed] [Google Scholar]
- 13a.Fire, A., S. W. Harrison, and D. Dixon 1990. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene 93:189-198. [DOI] [PubMed] [Google Scholar]
- 14.Frangioni, J. V., and B. G. Neel. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210:179-187. [DOI] [PubMed] [Google Scholar]
- 15.Fraser, A. G., R. S. Kamath, P. Zipperten, M. Martinez-Campos, M. Sohrmann, and J. Ahringer. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408:325-333. [DOI] [PubMed] [Google Scholar]
- 16.Grondin, B., M. Bazinet, and M. Aubry. 1996. The KRAB zinc finger gene ZNF74 encodes an RNA-binding protein tightly associated with the nuclear matrix. J. Biol. Chem. 271:15458-15467. [DOI] [PubMed] [Google Scholar]
- 17.Hallam, S. J., and Y. Jin. 1998. lin-14 regulates the timing of synaptic remodeling in Caenorhabditis elegans. Nature 395:78-82. [DOI] [PubMed] [Google Scholar]
- 18.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Hill, A. A., C. P. Hunter, B. T. Tsung, G. Tucker-Kellogg, and E. L. Brown. 2000. Genomic analysis of gene expression in C. elegans. Science 290:809-812. [DOI] [PubMed] [Google Scholar]
- 20.Hong, Y., R. Lee, and V. Ambros. 2000. Structure and function analysis of LIN-14, a temporal regulator of postembryonic developmental events in Caenorhabditis elegans. Mol. Cell. Biol. 20:2285-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong, Y., R. Roy, and V. Ambros. 1998. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development 125:3585-3597. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, M., J. Ryu, M. Kiraly, K. Duke, V. Rinke, and S. K. Kim. 2001. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 98:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 2003. 421:231-237. [DOI] [PubMed] [Google Scholar]
- 24.Kimura, K. D., H. A. Tissenbaum, Y. Liu, and G. Ruvkun. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277:942-945. [DOI] [PubMed] [Google Scholar]
- 25.Labouesse, M., E. Hartwieg, and H. R. Horvitz. 1996. The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development 122:2579-2588. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K. K., Y. Gruenbaum, P. Spann, J. Liu, and K. L. Wilson. 2000. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol. Biol. Cell 11:3089-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, R., R. Feinbaum, and V. Ambros. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843-854. [DOI] [PubMed] [Google Scholar]
- 28.Liang, C., and B. Stillman. 1997. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11:3375-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichtsteiner, S., and R. Tjian. 1995. Synergistic activation of transcription by UNC-86 and MEC-3 in Caenorhabditis elegans embryo extracts. EMBO J. 14:3937-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, Z., and Ambros, V. 1989. Heterochronic genes control the stage-specific initiation and expression of the dauer larva developmental program in Caenorhabditis elegans. Genes Dev. 3:2039-2049. [DOI] [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.Maeda, I., Y. Kohara, M. Yamamoto, and A. Sugimoto. 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11:171-176. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Balbas, M. A., A. Dey, S. K. Rabindran, K. Ozato, and C. Wu. 1995. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83:29-38. [DOI] [PubMed] [Google Scholar]
- 34.Mello, C. C., J. M. Kramer, D. Stinchcomb, and V. Ambros. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirkovitch, J., M.-E. Mirault, and U. K. Laemmli. 1984. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell 39:223-232. [DOI] [PubMed] [Google Scholar]
- 36.Moss, E., R. Lee, and V. Ambros. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88:637-646. [DOI] [PubMed] [Google Scholar]
- 37.Olsen, P., and V. Ambros. 1999. The lin-4 regulatory RNA controls developmental timing in C. elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216:671-680. [DOI] [PubMed] [Google Scholar]
- 38.Patterson, G. I., A. Koweek, A. Wong, Y. Liu, and G. Ruvkun. 1997. The DAF-3 Smad protein antagonizes TGF-β-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes Dev. 11:2679-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piano, F., A. J. Schetter, D. G. Morton, K. C. Gunsalus, V. Reinke, S. K. Kim, and K. J. Kemphues. 2002. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12:1959-1964. [DOI] [PubMed] [Google Scholar]
- 40.Pierce, S. B., M. Costa, et al. 2001. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 15:672-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhart, B. J., and G. Ruvkun. 2001. Isoform-specific mutations in the Caenorhabditis elegans heterochronic gene lin-14 affect stage-specific patterning. Genetics 157:199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinke, V., H. E. Smith, J. Nance, J. Wang, C. Van Doren, R. Begley, S. J. Jones, E. B. Davis, S. Scherer, S. Ward, and S. K. Kim. 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6:605-616. [DOI] [PubMed] [Google Scholar]
- 43.Reyes, J. C., C. Muchardt, and M. Yaniv. 1997. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137:263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riddle, D. L., and P. S. Albert. 1997. Genetic and environmental regulation of dauer larva development, p. 739-769. In D. L. Riddle, T. Blumenthal, B. Meyer, and J. Priess (ed.), C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 45.Riemer, D., H. Dodemont, and K. Weber. 1993. A nuclear lamin of the nematode Caenorhabditis elegans with unusual structural features: cDNA cloning and gene organization. Eur. J. Cell Biol. 62:214-223. [PubMed] [Google Scholar]
- 46.Roussell, D. L., and K. L. Bennett. 1992. Caenorhabditis cDNA encodes an eIF-4A-like protein. Nucleic Acids Res. 20:3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rual, J. F., J. Ceron, J. Koreth, T. Hao, A. S. Nicot, T. Hirozane-Kishikawa, J. Vandenhaute, S. H. Orkin, D. E. Hill, S. van den Heuvel, and M. Vidal. 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14:2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruppert, S. M., V. McCulloch, M. Meyer, C. Bautista, M. Falkowski, H. G. Stunnenberg, and N. Hernandez. 1996. Monoclonal antibodies directed against the amino-terminal domain of human TBP cross-react with TBP from other species. Hybridoma 15:55-68. [DOI] [PubMed] [Google Scholar]
- 49.Ruvkun, G., V. Ambros, A. Coulson, R. Waterston, J. Sulston, and H. R. Horvitz. 1989. Molecular genetics of the Caenorhabditis elegans heterochronic gene in lin-14. Genetics 121:501-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruvkun, G., and J. Giusto. 1989. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature 338:313-319. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Simmer, F., C. Moorman, A. M. van der Linden, E. Kuijk, P. V. van den Berghe, R. S. Kamath, A. G. Fraser, J. Ahringer, and R. H. Plasterk. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1:E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, H. C., R. L. Ochs, D. Lin, and A. C. Chinault. 1987. Ultrastructural and biochemical comparisons of nuclear matrices prepared by high salt or LIS extraction. Mol. Cell. Biochem. 77:49-61. [DOI] [PubMed] [Google Scholar]
- 54.Sonnichsen, B., L. B. Koski, A. Walsh, P. Marschall, B. Neumann, M. Brehm, A. M. Alleaume, J. Artelt, P. Bettencourt E. Cassin, M. Hewitson, C. Holz, M. Khan, S. Lazik, C. Martin, B. Nitzsche, M. Ruer, J. Stamford, M. Winzi, R. Heinkel, M. Roder, J. Finell, H. Hantsch, S. J. Jones, M. Jones, F. Piano, K. C. Gunsalus, K. Oegema, P. Gonczy, A. Coulson, A. Hyman, and C. J. Echeverri. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434:462-469. [DOI] [PubMed] [Google Scholar]
- 55.Steinmeyer, K., and W. Deppert. 1988. DNA binding properties of murine p53. Oncogene 3:501-507. [PubMed] [Google Scholar]
- 56.Timmons, L., and A. Fire. 1998. Specific interference by ingested dsRNA. Nature 395:854. [DOI] [PubMed] [Google Scholar]
- 57.Treuner, K., C. Eckerich, and R. Knippers. 1998. Chromatin association of replication protein A. J. Biol. Chem. 273:31744-31750. [DOI] [PubMed] [Google Scholar]
- 58.Vousden, K. 2000. p53: death star. Cell 103:691-694. [DOI] [PubMed] [Google Scholar]
- 59.Wightman, B., T. R. Bürglin, J. Gatto, P. Arasu, and G. Ruvkun. 1991. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans. Genes Dev. 5:1813-1824. [DOI] [PubMed] [Google Scholar]
- 60.Wightman, B., I. Ha, and G. Ruvkun. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formulation in C. elegans. Cell 75:1-20. [DOI] [PubMed] [Google Scholar]
- 61.Wood, W. B., and the community of C. elegans researchers (ed.). 1988. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.