Abstract
The nonhomologous end-joining (NHEJ) pathway of DNA double-strand break repair requires three protein complexes in Saccharomyces cerevisiae: MRX (Mre11-Rad50-Xrs2), Ku (Ku70-Ku80), and DNA ligase IV (Dnl4-Lif1-Nej1). Much is known about the interactions that mediate the formation of each complex, but little is known about how they act together during repair. A comprehensive yeast two-hybrid screen of the NHEJ factors of S. cerevisiae revealed all known interactions within the MRX, Ku, and DNA ligase IV complexes, as well as three additional, weaker interactions between Yku80-Dnl4, Xrs2-Lif1, and Mre11-Yku80. Individual and combined deletions of the Yku80 C terminus and the Xrs2 forkhead-associated (FHA) domain were designed based on the latter two-hybrid results. These deletions synergistically blocked NHEJ but not the telomere and recombination functions of Ku and MRX, confirming that these protein regions are functionally important specifically for NHEJ. Further mutational analysis of Yku80 identified a putative C-terminal amphipathic α-helix that is both required for its NHEJ function and strikingly similar to a DNA-dependent protein kinase interaction motif in human Ku80. These results identify a novel role in yeast NHEJ for the poorly characterized Ku80 C-terminal and Xrs2 FHA domains, and they suggest that redundant binding of DNA ligase IV facilitates completion of this DNA repair event.
The nonhomologous end-joining (NHEJ) pathway of double-strand break (DSB) repair uses proteins to recognize, bridge, process, and ligate the broken DNA strands (20). The first step in eukaryotic NHEJ is end binding by a Ku heterodimer composed of Ku70 and Ku80 subunits (11). In mammalian cells, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) protein facilitates end bridging, whereas in Saccharomyces cerevisiae, which lacks DNA-PKcs, this activity may be provided by the Mre11-Rad50-Xrs2 (MRX) complex (5, 8). Additional processing factors are also used if the DNA termini require resection or polymerization to become ligatable (28, 45). The final step, ligation, is catalyzed by the DNA ligase IV complex (44). With this many proteins, the eukaryotic NHEJ system seems to demand a series of coordinated interactions between its constituent complexes. These interactions must provide an inherent rigidity to maintain association of poorly base paired overhangs while simultaneously providing the flexibility to allow several catalytic domains to access variably spaced DNA termini.
Proper formation of the MRX complex requires three proteins: Mre11, Rad50, and Xrs2 (Nbs1 in mammalian cells). Deletion of any gene results in severe NHEJ deficiency, mild recombination defects, shortened telomeres, and meiotic defects (7, 11). Mre11 harbors a nuclease domain and forms strong protein-protein interactions with Rad50 and Nbs1/Xrs2 (7). Rad50 is similar to structural maintenance of chromosome (SMC) proteins, with a split ABC ATPase domain whose parts are separated by a long coiled-coil region that homodimerizes via a zinc hook (19). Less is known about Nbs1/Xrs2, but it is proposed to harbor forkhead-associated (FHA) and BRCA1 C-terminal (BRCT) domains, which have been shown to mediate phosphopeptide-specific protein-protein interactions (7, 12). Although these domains are important in Nbs1 for binding to γ-H2AX, there were no known binding partners for the corresponding domains of Xrs2 (22). How MRX participates in NHEJ is not well understood.
Like MRX, the Ku heterodimer has multiple cellular functions. In addition to NHEJ, it is required for protection of the telomeres from nucleolytic degradation, epigenetic silencing of genes positioned near telomeres (the telomere position effect [TPE]), and perinuclear anchoring of the telomeres (1, 2, 3, 31, 37). In higher eukaryotes, the Ku heterodimer associates with the DNA-PKcs protein to form the complete DNA-dependent protein kinase (DNA-PK), which is essential for NHEJ (29). The Ku subunits intertwine to form a ring that engages a DSB by sliding onto its end (42), positioning it to guide further events in the repair process.
Unlike those of MRX and Ku, the actions of the DNA ligase IV complex appear restricted to NHEJ. The DNA ligase IV complex consists of DNA ligase IV itself and XRCC4 (Dnl4 and Lif1 in yeast). Dnl4 binds strongly to Lif1, and both proteins are essential for NHEJ (39, 43). Nej1 modulates the NHEJ activity of this complex in yeast by interacting with the globular amino-terminal region of Lif1 (14).
While many studies have elucidated the molecular architecture of MRX, Ku, and DNA ligase IV independently, very little is known about how they interact to execute NHEJ. To systematically identify protein-protein interactions involved in NHEJ, we performed a comprehensive yeast two-hybrid screen on all known Saccharomyces cerevisiae NHEJ proteins and their individual domains. This analysis confirmed all known interactions and, importantly, suggested three additional interactions between specific regions of Dnl4-Yku80, Mre11-Yku80, and Xrs2-Lif1. These results guided mutational analysis of the C terminus of Yku80 and the FHA domain of Xrs2 which showed that these regions are redundantly and specifically required for NHEJ. This Yku80 C-terminal motif mapped to a putative amphipathic helix with intriguing similarities to Ku80 proteins from other species.
MATERIALS AND METHODS
Yeast strains and growth.
All yeast strains were derived from S288C and are isogenic with BY4743 except at the indicated loci. The HO suicide deletion strain YW1276 [MATα-inc ade2::HOSD(+1)::STE3-MET15 his3Δ1 leu2Δ met15Δ ura3Δ] has been described previously (9). The TPE strain YW1452 (MATa his3Δ1 leu2Δ met15Δ ura3Δ ppr1Δ::LEU2 yku80Δ::kanMX4 URA3-TPE) was derived from the yku80Δ strain from the genome deletion array (46). All strains were grown at 30°C (unless otherwise specified) in either a rich medium containing 1% yeast extract, 2% peptone, 2% dextrose, and 40 μg/ml adenine (YPAD) or a synthetic defined (SD) medium with either 2% glucose or galactose.
NHEJ yeast two-hybrid screen.
Yeast two-hybrid constructs were created by gap repair in yeast and tested for interaction by mating as described in the “Supplemental methods” section of the supplemental material. For confirmation, plasmids mediating novel interactions were isolated by electroporation into ElectroMAX DH10B competent cells (Invitrogen). Recovered plasmids were sequenced, and 12 of 12 plasmids tested demonstrated correct recombination, although 3 plasmids contained single-nucleotide changes presumably introduced by PCR. To confirm novel interactions in haploid strains, the corresponding bait and prey plasmids were cotransformed into PJ694-α. Two independent clones were then spotted onto plates lacking histidine or adenine. β-Galactosidase activity was assayed as described elsewhere (34).
Prey expression constructs harboring yku80 point mutations were cloned by gap repair into PJ694-α using the oligonucleotide primers OW1646 (5′-CCA CCA AAC CCA AAA AAA GAG ATC GAA TTC CAG CTG GAA TGG ATT CAC CAA CTG) and OW1670 (5′-TCG ACG GAT CCC CGG GAA TTG CCA TGG TGG TCA ATT ATT GCT ATT GTT TGG ACT TCC) and genomic DNA containing the mutated sequence of interest. Strains were then transformed with the Dnl4(1-681) bait. These strains were grown for 2 days in SD medium lacking leucine and tryptophan and plated to nonselective plates and plates lacking histidine to determine the percentage of His+ cells.
Mutational analysis.
All yku80 mutations were constructed in the YKU80 chromosomal gene so that expression was driven from the native promoter. Creation of single and multiple amino acid substitutions involved PCR amplification of tandem DNA regions on either side of the mutation site that contained approximately 20 bp of overlap determined by the two internal PCR primers that encoded the desired mutation (sequences available upon request). These PCR products were then cotransformed into YW1404, a derivative of YW1276 in which the sequence encoding Yku80 amino acids 539 to 629 had been replaced with the URA3 marker. Three-way recombination between the PCR fragments and the endogenous locus results in replacement of the URA3 marker with the amino acid 539-to-629 sequence encoding specific amino acid changes. Three independent 5-fluoroorotic acid (5-FOA)-resistant isolates were subjected to suicide deletion analysis and growth at 37°C. YW1404 displays phenotypes equivalent to the yku80Δ strain; it is severely NHEJ defective and is sensitive to growth at 37°C. Transformants that retained temperature sensitivity thus corresponded to recombination failures and were disregarded. Most mutants were subsequently verified by sequencing. Truncations were created by introduction of a stop codon after residues 618, 604, 590, 576, and 551 in chromosomal YKU80.
xrs2ΔFHA was generated by targeted replacement of amino acids 1 to 125 with the URA3 marker in YW1276. In this allele, URA3 is coupled to the strong ADH1 promoter and a start codon, such that expression of xrs2ΔFHA is driven by the ADH1 promoter beginning at Xrs2 residue 126. As a control, we also generated an amino-terminal deletion of residues 2 to 125 under the control of the endogenous XRS2 promoter. This strain behaved similarly to xrs2ΔFHA (data not shown), and therefore we used the strain containing the URA3 marker in subsequent experiments. The xrs2ΔFHA allele was also introduced by targeted recombination into the YW1276 derivative YW1462, which contained the yku80Δ605-629 allele. The double mutant was mated to MATa wild-type, yku80Δ, and xrs2Δ strains from the genome deletion array (46), followed by sporulation and dissection to generate the strains used in Fig. 4A.
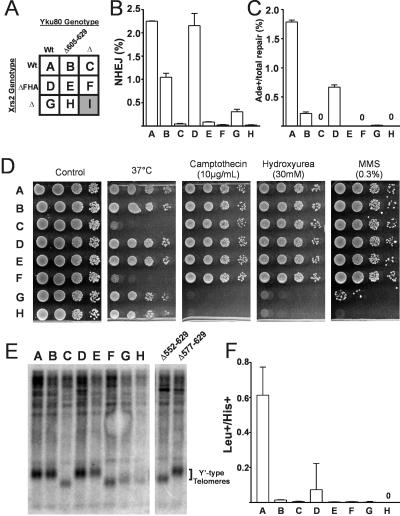
FIG. 4.
The Xrs2 FHA domain and the Yku80 C terminus act synergistically during NHEJ. (A) Genotypes of YW1276 derivative strains used in this figure. Letters from this table are used to designate strains in panels B to F. Strain I (xrs2Δ yku80Δ) displayed rapid senescence and could not be used for further experiments (indicated by shading). (B and C) The yku80Δ605-629 xrs2ΔFHA double mutant (strain E) demonstrates synergistic inhibition of precise NHEJ (B) and +2 imprecise NHEJ (C) compared to the corresponding single mutants (strains B and D) in the HO(+2) suicide deletion assay as described in the legend to Fig. 2. (D) Double-mutant strain E is not temperature sensitive or hypersensitive to camptothecin (10 μg/ml), hydroxyurea (30 mM), or a 30-min exposure to 0.3% MMS. (E) Telomeric Southern blotting showing that the yku80Δ605-629 and xrs2ΔFHA mutants have wild-type telomere length. Y′-type telomeres are indicated. (F) Strains B to H are deficient in recircularization of a plasmid with a DSB in the LEU2 marker following transformation. Error bars indicate standard deviations for at least three replicates.
Suicide deletion.
Our lab has previously described the suicide deletion concept for monitoring precise and imprecise NHEJ (9, 21). Here, all suicide deletion strains are derived from YW1276, which uses the HO mega-endonuclease and its cut sites (see Fig. 2C). The reading frame of the flanking ADE2 marker has been altered so that only imprecise NHEJ events in the +2 register (i.e., with a 2-nucleotide insertion or loss of 1, 4, etc. nucleotides) result in the Ade+ phenotype. The frequency of DSB repair by NHEJ was measured as the ratio of colonies formed on galactose plates compared to glucose plates following growth in glucose liquid culture. The percentage of NHEJ events that are imprecise in the +2 register was measured as the ratio of colonies formed on adenine selective plates compared to nonselective plates following growth in galactose liquid culture. Some Ade− repair events always occur, even in Ku-deficient strains (i.e., residual precise NHEJ and microhomology-mediated end joining), ensuring outgrowth in galactose and therefore a denominator in the +2 ratio (27). However, Ade+ cells are never formed by strains completely lacking Ku. The +2 ratio therefore principally reflects the more extensive loss of the processed HO(+2) NHEJ event.
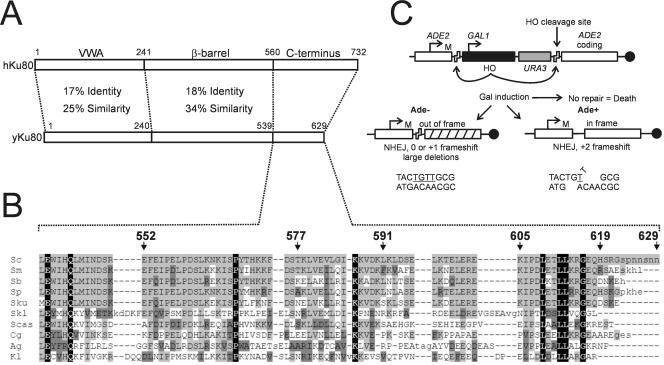
FIG. 2.
Ku80 alignment and suicide deletion assay. (A) Comparison of human and yeast Ku80 showing conserved VWA and β-barrel core domains but little conservation of the C termini. (B) Alignment of the fungal Ku80 C terminus demonstrates a cluster of conserved amino acids between residues 605 and 619. Species shown are as follows: Sc, Saccharomyces cerevisiae; Sm, Saccharomyces mikatae; Sb, Saccharomyces bayanus; Sp, Saccharomyces paradoxus; Sku, Saccharomyces kudriavzevii; Skl, Saccharomyces kluyveri; Scas, Saccharomyces castelli; Cg, Candida glabrata; Ag, Ashbya gossypii; Kl, Kluyveromyces lactis. Numbers indicate S. cerevisiae Yku80 residues. Black shading, residues that are identical in all proteins; light gray shading, residues identical to S. cerevisiae; dark gray shading, conservative substitutions. (C) HO(+2) suicide deletion assay. Galactose-induced expression of the HO endonuclease results in deletion of the HO coding sequence and a DSB in the ADE2 marker. DSB repair by NHEJ allows cell survival, with repair in the +2 frameshift register resulting in Ade+ cells. The principal joint responsible for Ade− and Ade+ colonies is shown.
Telomere position effect assay.
Our telomere position effect allele (URA3-TPE) was similar, but not identical, to published alleles (33). URA3-TPE was generated by PCR amplification of URA3 using oligonucleotide primers OW1720 (5′-ACC ATG TCC TAC TCA CTG TAC TGT TGT TCT ACC CTC CAT ATT GAA CTG TGC GGT ATT TCA CAC CG) and OW1721 (5′-AAC ACC TAT TAT ACT GTT GAC CAA TTC AGT CAC ATT ATT ACG CAT AGA TTG TAC TGA GAT GCA C), which targeted the product to the subtelomeric region of chromosome VII-L (coordinates 5011 to 5356) so that URA3 transcription was toward the centromere. Haploid spores harboring the yku80 mutation, the TPE allele, and the ppr1Δ allele were grown 2 days in SD medium lacking uracil (SD-Ura), diluted 1:1,000, and grown 2 more days in SD complete medium. Strains were then plated or spotted to nonselective, uracil-selective, and 5-FOA plates.
Sensitivity to DNA-damaging agents.
Camptothecin and hydroxyurea sensitivity were determined by spotting serial dilutions of yeast to media containing 10 μg/ml camptothecin or 30 mM hydroxyurea. Methyl methanesulfonate (MMS) sensitivity was determined by incubating exponentially growing cells with 0.3% MMS in 50 mM sodium phosphate buffer (pH 7.5) at 30°C for 30 min, followed by spotting of serial dilutions to SD complete plates.
Plasmid recircularization NHEJ assay.
One hundred nanograms of the LEU2-marked plasmid pRS315 cut with AgeI (Roche) was cotransformed with 10 ng supercoiled pRS413 (4) marked with HIS, as described previously (6). Transformed cells were plated in parallel to a glucose medium lacking either leucine or histidine and were grown at 30°C for 3 days. Relative repair efficiency was measured as the ratio of Leu+ colonies to His+ colonies. Note that because the AgeI-induced DSB is in the LEU2 marker, cotransformation with pRS413 cannot lead to Leu+ colonies by homologous recombination.
Telomere length assessment.
Telomere length was monitored essentially as described elsewhere (3). Briefly, yeast cells were grown for 40 generations in liquid YPAD. Genomic DNA was then prepared by glass bead lysis, digested with RNase A and XhoI, and probed by Southern blotting with radiolabeled poly(GT)20.
RESULTS
Systematic two-hybrid analysis of the MRX, DNA ligase IV, and Ku complexes.
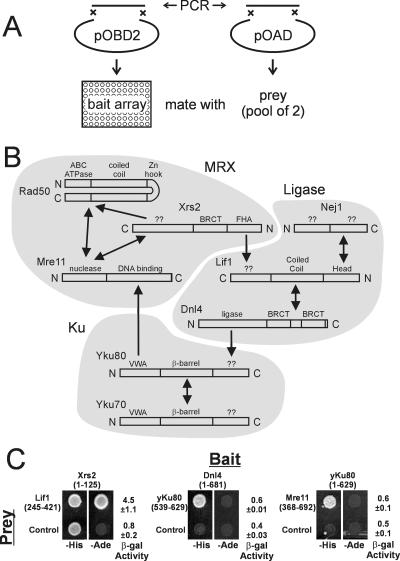
Our two-hybrid analysis generated more than 10,000 data points in a diploid array format (Fig. 1A; see also supplemental material). It included all known NHEJ proteins and their individual domains in both bait and prey constructs to ensure that all potential interactions were queried in a multiply redundant fashion (see Fig. S1 in the supplemental material). This multiplicity also allowed most interactions to be mapped to discrete domains. The screen identified multiple strong positives that included all of the known protein-protein interactions within the MRX, Ku, and DNA ligase IV complexes (Fig. 1B). Specifically, the screen confirmed interactions between the Mre11-Rad50, Mre11-Xrs2, Dnl4-Lif1, Lif1-Nej1, and Ku70-Ku80 protein pairs, which all mapped to the expected domains (see Fig. S2 in the supplemental material) (3, 7, 14, 39, 42). These interactions provide an important validation of our approach. We also observed a weak potential interaction between the C terminus of Xrs2 (residues 185 to 854) and the coiled-coil domain of Rad50 (residues 167 to 1157) that to our knowledge has not been described before (Fig. 1B; see also Fig. S2 and Table S2 in the supplemental material). An additional interaction within MRX could contribute to its overall stability and function, but we note that this was detected only in one bait-prey orientation above substantial autoactivation by the Xrs2(185-854) bait. This potential interaction was not impaired by deletion of MRE11, indicating that it is not due to ternary interactions mediated via Mre11 (see Fig. S3 in the supplemental material).
FIG. 1.
NHEJ yeast two-hybrid screen. (A) All NHEJ gene constructs were cloned by gap repair into both pOBD2 (bait) and pOAD (prey) vectors. Two independent bait isolates were arrayed in a 96-well microtiter dish. Two independent prey isolates were pooled and mated against the bait array to generate arrays of diploids containing paired bait and prey clones. (B) Schematic summarizing the two-hybrid results. Double-headed arrows indicate interactions that were detected by reciprocal bait-prey pairings. Single-headed arrows indicate interactions that were detected in only one orientation, with the arrow pointing to the prey. Arrows point to the domain responsible for the interaction as determined from the pattern of positive clones. Shading is used to indicate the previously known stronger interactions. Note that some proteins are drawn in reverse. (C) Plasmids mediating novel interactions were isolated, sequenced, and transformed into PJ694-α. Two-hybrid interactions were assessed by spotting to −His and −Ade plates and assaying for lacZ expression by measuring β-galactosidase activity.
Our screen also revealed two putative interactions and refined one known weaker interaction between the DNA ligase IV, Ku, and MRX complexes (Fig. 1B). A Dnl4-Yku80 interaction was detected between the Dnl4 bait corresponding to amino acids 1 to 681 and the Yku80 prey corresponding to the C-terminal 90 amino acids. An interaction between Mre11 and Yku80 corresponded to baits containing the Mre11 C-terminal region (amino acids 368 to 692) and preys containing the Yku80 von Willebrand A (VWA) domain (amino acids 1 to 265). Finally, we mapped the known interaction between Xrs2 and Lif1 to baits containing the FHA domain of Xrs2 (amino acids 1 to 125) and preys containing the C terminus of Lif1 (amino acids 245 to 421). All relevant plasmids were isolated, sequenced, and transformed into haploid strains for retesting. All pairs confirmed their screen phenotype (Fig. 1C). Further testing of mre11, dnl4, and yku80 gene deletion strains demonstrated that the Yku80-Dnl4, Yku80-Mre11, and Xrs2-Lif1 interactions were not dependent on MRX, DNA ligase IV, or Ku, respectively, and are thus likely mediated directly (see Fig. S3 in the supplemental material). Importantly, these weaker positives did not activate both the HIS3 and ADE2 markers, and significant activation of the lacZ marker was observed only for the Xrs2 bait-Lif1 prey combination (Fig. 1C). Consistently, an Xrs2-Lif1 interaction can be observed in vitro (5), but we have so far been unable to demonstrate the Yku80-Dnl4 interaction by coimmunoprecipitation (although such studies are made difficult by the low levels of Dnl4 expression that can be achieved). Thus, although these putative interactions are reproducible in the two-hybrid assay, their weakness clearly demands functional confirmation. We provide this below for the relevant protein regions of Yku80 and Xrs2.
Truncation of the Yku80 C terminus selectively impairs NHEJ.
We chose to focus first on the Yku80 C terminus. This protein region was interesting for reasons described in the Discussion. Moreover, the multiple activities of the Ku heterodimer allow separation of the functional effects of specific mutations. In these studies, NHEJ was assessed primarily by using the suicide deletion assay (Fig. 2C), (9). This assay allows for quantification of the absolute efficiency of DSB repair by NHEJ, as well as of the percentage of total repair that is imprecise in the +2 register (yielding Ade+ yeast). Complete deletion of YKU80 (yku80Δ) results in an approximately 50-fold reduction in total NHEJ and a >1,000-fold defect in imprecise NHEJ (Fig. 3A and B). Three NHEJ-independent Ku telomere phenotypes were also examined. First, yku80Δ strains are unable to grow at 37°C because of telomere deprotection (Fig. 3C) (3). Second, we tested the epigenetic silencing of a subtelomeric marker (2). When our URA3-TPE allele was used, approximately 6% of wild-type but not yku80Δ cells formed colonies on plates containing the URA3 counterselective drug 5-FOA (Fig. 3C). Finally, we performed Southern blotting with a telomere-specific probe, since yku80Δ strains show shortened Y′-type telomeres (Fig. 4E), (3).
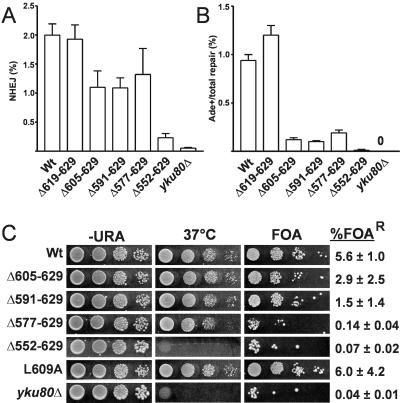
FIG. 3.
Truncation of the Yku80 C terminus results in NHEJ-specific defects. (A) Effects of Yku80 C-terminal truncations on total NHEJ repair by HO suicide deletion. The yku80Δ605-629, Δ591-629, and Δ577-629 mutants showed a modest twofold reduction in total NHEJ. (B) Effects of Yku80 C-terminal truncations on +2 imprecise NHEJ by HO suicide deletion. Data are plotted as the ratio of Ade+ to total colonies. The yku80Δ605-629, Δ591-629, and Δ577-629 yeast strains showed a ninefold reduction in +2 imprecise events. No Ade+ events were recovered with the yku80Δ strain. Error bars indicate standard deviations for at least three replicates. (C) Haploid strains containing the yku80 mutations and the URA3-TPE allele were spotted to SD complete, SD-Ura, and SD 5-FOA plates. Complete plates were incubated at 37°C. Strains were also quantitatively plated to media with and without 5-FOA to determine the percentage of FOA-resistant cells (% FOAR). Unlike the yku80Δ and yku80Δ552-629 yeast strains, the yku80Δ605-629, Δ591-629, and L609A yeast strains did not show compromised growth at 37°C and were also able to epigenetically silence the URA3 marker.
Although the C-terminal regions of human and yeast Ku80 are highly divergent (Fig. 2A), alignments of the fungal Ku80 C terminus do show significant conservation, providing an important tool for guiding mutational analysis of this relatively short region (Fig. 2B). We first constructed a series of truncations of the Yku80 C terminus. Mutation yku80Δ552-629, which deleted almost the entire region present in the Yku80 two-hybrid construct that interacted with Dnl4, resulted in a loss of NHEJ approaching that of the yku80Δ strain (Fig. 3A and B). This mutant also displayed sensitivity to growth at 37°C and shortened Y′-type telomeres (Fig. 3C and 4E), most likely indicating that this truncation destabilizes the Ku heterodimeric ring. Mutations yku80Δ577-629, 591-629, and 605-629 were more informative. Each of these truncations caused a significant twofold reduction in total NHEJ and a ninefold reduction in +2 imprecise NHEJ (Fig. 3A and B). The yku80Δ577-629 strain was only partially temperature sensitive, while strains with the more distal deletions were not at all sensitive to growth at 37°C (Fig. 3C). This suggested that these truncations confer a partial separation of the Yku80 NHEJ and telomeric functions. Consistent with this, yku80Δ605-629 and yku80Δ591-629 yeasts were also able to induce TPE to near-wild-type levels (2.9 and 1.5% 5-FOA-resistant colonies, respectively) and showed wild-type telomere repeat lengths (Fig. 3C and 4E). Truncation of the last 10 Yku80 amino acids (yku80Δ619-629) had no effect on NHEJ efficiency or temperature sensitivity (Fig. 3), consistent with the fact that these residues are not conserved in other fungi (Fig. 2B). Taken together, these results point to amino acids 605 to 619 as an important determinant for Yku80 function in NHEJ, but for none of its telomeric functions. Interestingly, this region of the Yku80 C terminus is the most highly conserved among fungi (Fig. 2).
The Xrs2 FHA domain is functionally redundant with the Yku80 C terminus in NHEJ.
We hypothesized that mutation of the Yku80 C terminus caused only partial NHEJ loss because of a potential redundancy of its function. Specifically, the DNA ligase IV complex, composed of Dnl4 and Lif1, appeared to make contacts with both Yku80 and the FHA domain of Xrs2 (Fig. 1B). To examine the role of the Xrs2 FHA domain, we created an amino-terminal deletion of amino acids 1 to 125 (xrs2ΔFHA). This allele and the complete deletion, xrs2Δ, were then introduced into the yku80Δ605-629 and yku80Δ strains in all combinations (Fig. 4A). Myc-tagged derivatives of these strains were also subjected to immunoblotting, which confirmed that yku80Δ605-629 and xrs2ΔFHA were expressed at levels similar to those of their wild-type counterparts (see Fig. S4 in the supplemental material). Complete deletion of XRS2 resulted in approximately 10-fold inhibition of total NHEJ and 150-fold inhibition of +2 imprecise NHEJ (Fig. 4B and C, strain G). This large decrease was expected given the known role of MRX in NHEJ (43) but, interestingly, was less than that observed for yku80Δ (Fig. 4). In contrast, xrs2ΔFHA did not significantly impair total NHEJ and resulted in only a twofold inhibition of +2 imprecise repair (Fig. 4B and C, strain D), even less than the reductions observed for yku80Δ605-629 (strain B). However, the yku80Δ605-629 xrs2ΔFHA double mutant (strain E) showed a 40-fold decrease in total NHEJ and a >1,000-fold decrease in +2 events, very similar to the NHEJ phenotype of yku80Δ yeast and more severe than even xrs2Δ yeast (strains C and G, respectively). This synergism is consistent with the hypothesis that Ku and MRX have partially redundant functions in NHEJ.
To confirm these NHEJ defects by a different method, we tested the ability of our strains to recircularize a linear plasmid. All strains were cotransformed with pRS315 cut in the LEU2 marker with AgeI and a circular plasmid control that contained the HIS3 marker. Repair was measured as the ratio of Leu+ colonies to His+ colonies. The pattern of results (Fig. 4F) was the same as that seen with suicide deletion, with the yku80Δ605-629 xrs2ΔFHA strain (strain E) showing a plasmid joining defect equivalent to that of yku80Δ (strain C). Interestingly, the yku80Δ605-629 and xrs2ΔFHA mutants were more obviously defective in the plasmid assay, with yku80Δ605-629 alone now showing a 44-fold decrease in joining.
To again verify that these double-mutants selectively lost the NHEJ functions of Xrs2 and Yku80, the entire panel of mutants was spotted to plates containing 10 μg/ml camptothecin or 30 mM hydroxyurea. Camptothecin induces topoisomerase-mediated damage during replication, while hydroxyurea produces stalled replication forks. Mutants were also exposed for 30 min to 0.3% MMS, a radiomimetic leading to DSBs requiring homologous recombination for repair (24). All of these lesions require either the recombination or the checkpoint functions of the MRX complex (15, 25, 26). Deletion of XRS2 accordingly resulted in severe drug sensitivity (Fig. 4D, strain G). In contrast, xrs2ΔFHA did not impair MRX-mediated resistance to any of the drugs tested, even in the yku80Δ605-629 background (strain E). The MMS data further supported the role of the Yku80 C terminus in DSB repair, since adding the yku80Δ605-629 mutation eliminated the rare survivors seen in the xrs2Δ background (Fig. 4D).
Both MRX and Ku are required for telomere maintenance, and mutation of either complex leads to reduced numbers of telomere repeats. However, the roles of the two complexes are distinct. While the Ku complex protects the telomeres from excessive nucleolytic degradation, the MRX complex is likely involved in telomerase-based telomere elongation (7). For this reason, combined yku80Δ xrs2Δ mutants display rapid senescence and become nonviable after several generations (Fig. 4, strain I) (30). Indeed, the normal and stable growth rate ofyku80Δ605-629 xrs2ΔFHA yeast already demonstrated preservation of telomeres in this strain, and accordingly, thestrain was also able to grow normally at 37°C (Fig. 4D). Moreover, the xrs2ΔFHA and xrs2ΔFHA yku80Δ605- 629 mutants (strains D and E, respectively) demonstrated telomeres of wild-type length (Fig. 4E). The combined xrs2ΔFHA and yku80Δ605-629 mutations therefore confer marked separation of Ku and MRX functions, with near-complete loss of NHEJ but essentially no impairment of recombination or telomere maintenance.
Identification of Yku80 C-terminal residues important for NHEJ.
Since truncations of the C terminus of Yku80 resulted in NHEJ-specific defects, we next sought to identify the residues important for the function of this undescribed protein region. We initially performed alanine scanning of all hydrophilic amino acids between residues 578 and 618 on the hypothesis that these were most likely to be surface exposed and to mediate a protein-protein interaction. Surprisingly, however, single and even multiple alanine substitutions of these residues had little or no effect on either total or +2 imprecise NHEJ (see Table S1 in the supplemental material). Correspondingly, reexamination of the alignment of the fungal Yku80 C terminus revealed that the residues with the highest degree of conservation are in fact hydrophobic (Fig. 2B). Most notably, there are three highly conserved leucines at residues 609, 612, and 613 in the region most strongly implicated in NHEJ function by deletion analysis (amino acids 605 to 619). Therefore, we also mutated all hydrophobic amino acids in theYku80 C terminus to alanine, alone and in combinations (Fig. 5A and B; see also Table S1 in the supplemental material). Strikingly, the mutations L609A, L612A, and L613A each showed a loss of NHEJ similar to that of yku80Δ605-629, and changing L612 to a proline had an even greater effect (Fig. 5A and B; Table 1). As with yku80Δ605-629, these mutations again did not impair Ku telomere functions, indicating a specific NHEJ defect and implying normal Ku complex formation (Fig. 3C and data not shown).
FIG. 5.
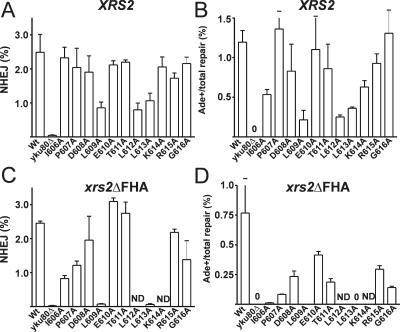
Identification of conserved Yku80 C-terminal residues mediating NHEJ. Residues 606 to 616 were replaced individually with alanine, and the total (A) and +2 imprecise (B) NHEJ efficiencies were determined as for Fig. 2. yku80-L609A, -L612A, and -L613A mutants demonstrate total NHEJ and +2 NHEJ defects similar to those of yku80Δ605-629 in the XRS2 wild-type strain. Deletion of the Xrs2 FHA domain synergistically impaired total (C) and +2 imprecise (D) NHEJ in the leucine mutants and uncovered lesser defects in other residues. ND, not done; mutations were not tested in the xrs2ΔFHA background. Error bars indicate standard deviations for at least three replicates.
TABLE 1.
NHEJ defects correlate with loss of the Yku80-Dnl4 two-hybrid interaction
| Yku80 mutation | % His+ cells | Fold defect in NHEJa
|
|||
|---|---|---|---|---|---|
|
XRS2
|
xrs2ΔFHA
|
||||
| Total NHEJ | +2 NHEJ | Total NHEJ | +2 NHEJ | ||
| Vector control | 0.002 | ||||
| Δ::kanMX4 | 50 | ∞ | 100 | ∞ | |
| Δ552-629 | 0.002 | 20 | >100 | ND | ND |
| Δ605-629 | 0.001 | 2 | 8 | 40 | >1,000 |
| L612P | 0.002 | 5 | 21 | 40 | >1,000 |
| L609A | 0.001 | 3 | 5 | 40 | >1,000 |
| L613A | 0.002 | 2 | 3 | 40 | >1,000 |
| I606A | 0.001 | 1 | 2 | 3 | 70 |
| P607A | 0.003 | 1 | 1 | 2 | 20 |
| G616A | 0.005 | 1 | 1 | 2 | 11 |
| R615A | 0.004 | 1 | 1 | 1 | 5 |
| D608A | 5.3 | 1 | 1 | 1 | 7 |
| K578A/K587A/ D589A/K590A | 5.0 | 1 | 2 | 1 | 4 |
| K578/E581A | 3.6 | 1 | 1 | 1 | 3 |
| T611A | 10.0 | 1 | 1 | 1 | 4 |
| S595A | 13.0 | 1 | 1 | 1 | 2 |
| E610A | 76.1 | 1 | 1 | 1 | 2 |
| Wild type | 66.7 | 1 | 1 | 1 | 2 |
∞, no +2 NHEJ events were recovered; ND, not done.
Because deletion of the Xrs2 FHA domain uncovered a more severe NHEJ phenotype for yku80 C-terminal mutants, we next introduced the xrs2ΔFHA allele into most strains with alanine substitutions for residues 606 to 619. The FHA deletion again had a synergistic effect and almost completely eliminated NHEJ when combined with the yku80-L609A and L613A mutations (Fig. 5C and D). In the xrs2ΔFHA background, multiple other residues in the NHEJ critical region now showed intermediate inhibition of NHEJ. The strongest effect was at the hydrophobic residue I606. Hydrophilic residues generally had a milder defect. Thus, the NHEJ-specific function of the Yku80 C terminus requires several highly conserved hydrophobic amino acids between residues 605 and 619.
We finally tested a representative set of yku80 C-terminal mutants in the yeast two-hybrid assay to establish whether their NHEJ phenotype correlated with the observation that stimulated these studies. To obtain a more quantitative assessment, the percentage of total cells that were His+ was determined by plating. The NHEJ competence of the various mutants correlated directly with the strength of the Yku80-Dnl4 two-hybrid interaction, with no outliers (Table 1). Mutants with more severe NHEJ dysfunction, such as yku80-L609A and Δ605-629, consistently showed complete loss of the two-hybrid interaction. Equally revealing were mutants such as yku80-D608A and T611A, which showed weakened two-hybrid interactions and a correspondingly small but measurable NHEJ defect detectable only in the xrs2ΔFHA background. Some mutants, such as I606A and P607A, showed complete loss of the two-hybrid interaction and clear but only partial loss of NHEJ. These and all mutants are consistent with the interpretation that interaction with Dnl4 is an important NHEJ function of the Yku80 C terminus but that the two-hybrid assay is more sensitive to Yku80 mutations than NHEJ. This is expected, because the two-hybrid assay is subject to a detection threshold of His3 expression required for colony formation, and because it used only the 90-amino-acid C-terminal region of Yku80 as opposed to otherwise intact Ku.
DISCUSSION
A crucial aspect of NHEJ must be the ordered and regulated assembly of the ligation machinery at a break, given that the Ku, MRX, and DNA ligase IV complexes are too large to all be coresident at an extreme DSB terminus and that both MRX and Ku have NHEJ-independent functions at DNA ends. What specific protein-protein interactions are required, and how do they facilitate Dnl4-catalyzed ligation of the DSB? To shed light on these questions, we screened a nearly comprehensive set of potential interactions and identified three that could help coordinate NHEJ (Fig. 1B). Functional analysis of the Yku80 and Xrs2 domains, predicted to interact with DNA ligase IV, confirmed their importance to NHEJ in vivo and revealed that they are substantially redundant with respect to NHEJ function. Specifically, clear mutant defects demonstrate that the extreme C terminus of Yku80 and the FHA domain of Xrs2 are required for NHEJ, with a strict separation from the telomere, checkpoint, and recombination functions of these proteins. Below we consider how these protein domains might act to facilitate NHEJ.
Our two-hybrid and mutational analyses of the Yku80 C terminus suggest that it has an important role in binding and perhaps positioning or stimulating Dnl4. All experiments, including our inability to demonstrate such binding in vitro to date, indicate that this interaction is weak, however. Thus, we cannot rule out the possibility that the observed NHEJ function of this Yku80 region is something other than direct Dnl4 binding. Alternatively, the weakness of the interaction may simply reflect its greater relevance in the context of DSB-bound proteins. Indeed, Dnl4 binding by the Yku80 C terminus is consistent with the known structure of DSB-bound Ku. Human Ku, and presumably yeast Ku, forms a ring that slides onto DNA in only one orientation with respect to the end (Fig. 6A) (42). The crystallized Ku protein lacked the C-terminal tail of Ku80, demonstrating that this region is not required for end binding. However, it is clear that the C terminus of Ku80, and not Ku70, will be oriented to face the DSB, putting it in position to promote ligation.
FIG. 6.
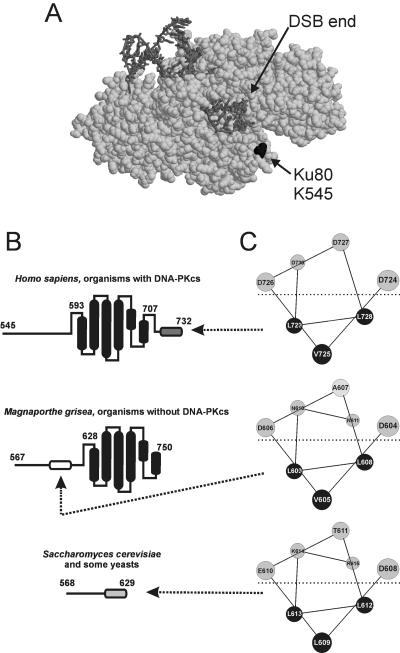
The NHEJ critical region of theYku80 C terminus. (A) The crystal structure of the human Ku70/80 heterodimer (light gray) bound to DNA (dark gray) reveals that the C terminus of Ku80 is oriented toward the DSB terminus. The C-terminal residue of the partial Ku80 protein is shown in black. (B) Schematic representation of three classes of Ku80 C termini. The α-helical bundle seen in human Ku80 NMR structures is schematized in black. The DNA-PKcs-interacting region is shown in dark gray. The putative Dnl4-interacting region is shown in light gray. A possible homologous region in M. grisea is shown in white. Numbers indicate amino acid positions. (C) Helical wheel predictions for the three regions indicated in panel B.
To explore this in more detail, we compared multiple Ku80 sequences, available solution structures of the human Ku80 C terminus (18, 47), and secondary structural predictions using PHD/PROF (32). This analysis revealed three Ku80 C-terminal types in eukaryotic cells (Fig. 6B). The first type, exemplified by human Ku80, shows an extended region terminating in an α-helical bundle, as seen by nuclear magnetic resonance (NMR), and a 12-amino-acid C-terminal region that interacts with DNA-PKcs (16). The second type, exemplified by the fungus Magnaporthe grisea, retains the same apparent helical-bundle structure but lacks the last DNA-PKcs-interacting region. Indeed, this helical bundle is conserved in most eukaryotes (18), but only organisms with DNA-PKcs homologues contain the last region (10). Intriguingly, the third class, including Saccharomyces cerevisiae and its closest relatives, does not even harbor the helical bundle.
Given this divergence of the Ku80 C terminus, it is not immediately clear if it functions analogously in all organisms. However, the Yku80 C terminus does contain a single putative α-helix that corresponds precisely to the NHEJ critical region (amino acids 608 to 615). Strikingly, this helix may be conserved just N-terminal to the M. grisea helical bundle (Fig. 6B and C). Moreover, the DNA-PKcs-interacting region at the extreme C terminus of human Ku80 is also predicted to fold as a very similar amphipathic α-helix (Fig. 6C) (18, 47), although, intriguingly, this was unstructured insolution NMR analyses in the absence of DNA-PKcs. Thus,despite their differences, all three Ku80 configurations present a similar potential protein interaction motif on the DSB side of Ku that could act by partner-induced formation and sequestration of the hydrophobic face of an amphipathic helix. This would explain our finding that both yeast NHEJ and the Yku80-Dnl4 two-hybrid interaction are dependent on three highly conserved leucines, since these would all cluster on one helical face (Fig. 6C). The especially severe defect of L612P mutation is also consistent with this model (see Table S1 in the supplemental material). It is not clear whether this pattern of putative Ku80 helices represents convergent or divergent evolution, but it is intriguing that a Ku80-DNA-PKcs-type interaction motif is also present in Nbs1 and ATRIP, where it mediates interactions with ATM and ATR, respectively (13).
Previous work identified an in vitro interaction between Xrs2 and Lif1 (5). Our studies indicate that the region encompassing the FHA domain of Xrs2 and the C terminus of Lif1 mediates this interaction. We further show its functional importance for NHEJ in vivo. It is most striking that severe NHEJ inhibition requires disruption of both this interaction between Xrs2 and Lif1 and the proposed interaction between Yku80 and Dnl4. Why might two interactions with DNA ligase IV be required? It is possible that multiple interactions are needed to accommodate the variety of DSB end configurations presented to the NHEJ machinery. Indeed, the fact that imprecise NHEJ is more severely affected by the corresponding Yku80 and Xrs2 mutations than simple religation NHEJ implies that these protein regions are especially necessary for more-complex repair events. Another possibility is that redundant binding of Dnl4 increases the kinetics and efficiency of NHEJ, which would be especially important if DNA ligase IV is the rate-limiting step. This two-contact model is also consistent with published data characterizing an inefficient microhomology-mediated end-joining (MMEJ) mechanism, which involves MRX and DNA ligase IV but not Ku (27). The Xrs2-Lif1 interaction can account for this Ku-independent role of Dnl4, while the absence of the Ku interaction could explain at least in part the inefficiency of MMEJ.
Several recent studies have addressed the role of the Xrs2/Nbs1 FHA domain in the cell. In mammalian cells, the Nbs1 FHA domain is important for nuclear focus formation and checkpoint signaling, but its role in DNA repair is unclear (38). In yeast, no definitive role for the Xrs2 FHA domain in DSB repair has been identified, and indeed, several recent studies have revealed no phenotype for xrs2 mutants lacking this domain (35, 41). Our work identifies an important NHEJ role for the Xrs2 region containing its FHA domain, but this role is robustly apparent only in the Yku80 mutant background. FHA domains bind with high specificity to phosphothreonine residues (12). We do not yet know if a similar function of the Xrs2 amino terminus mediates its interaction with Lif1, but it should be noted that the FHA domain of polynucleotide kinase 3′ phosphatase interacts with the C terminus of XRCC4 (mammalian homologue of Lif1) in a manner dependent on phosphorylation of XRCC4 by CK2 (23).
Importantly, the Yku80 C terminus and Xrs2 FHA domains cannot account for the full role of Ku and MRX in NHEJ, because both the yku80Δ605-629 and xrs2ΔFHA mutants retain substantial NHEJ activity, unlike the complete gene deletion mutants. This may indicate that MRX and Ku both serve other essential functions in NHEJ. Specifically, Ku may be required for protecting ends from degradation, and MRX may be required to tether the DNA ends together (11). A nonexclusive possibility is that the putative Mre11-Yku80 interaction detected in our screen, and perhaps other interactions, may help drive formation of an NHEJ repairosome that contains both MRX and Ku. Indeed, although we have identified many protein-protein interactions involved in NHEJ, there are certainly additional interactions we did not detect. Some may be realized only during NHEJ, either because they are DNA dependent or because they require context-dependent modification of proteins. Moreover, the two-hybrid method has a sensitivity threshold that is biased toward stronger interactions. In this regard, Tseng and Tomkinson described an interaction between Dnl4/Lif1 and Pol4 in vitro (40) that was not recovered in our screen. Further biochemical work will be required to fully understand the extent of interactions between the various NHEJ proteins.
Supplementary Material
Acknowledgments
We thank the Wilson lab for support and critical readings of the manuscript and Aidan Doherty for reading and evaluation of the manuscript. We also thank Peter Uetz and Stan Fields, who kindly provided pOAD, pOBD2, PJ69-4a, and PJ694-α.
This work was supported by the Pew Scholars Program in the Biomedical Sciences of the Pew Charitable Trusts, Public Health Service grants R01CA90911 and R01CA102563 (T.E.W.), and the Loeb Predoctoral Fellowship (P.L.P.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bertuch, A. A., and V. Lundblad. 2003. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol. Cell. Biol. 23:8202-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulton, S. J., and S. P. Jackson. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24:4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 5.Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8:1105-1115. [DOI] [PubMed] [Google Scholar]
- 6.Daley, J. M., and T. E. Wilson. 2005. Rejoining of DNA double-strand breaks as a function of overhang length. Mol. Cell. Biol. 25:896-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Amours, D., and S. P. Jackson. 2002. The mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 8.DeFazio, L. G., R. M. Stansel, J. D. Griffith, and G. Chu. 2002. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 21:3192-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Della, M., P. L. Palmbos, H. M. Tseng, L. M. Tonkin, J. M. Daley, L. M. Topper, R. S. Pitcher, A. E. Tomkinson, T. E. Wilson, and A. J. Doherty. 2004. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 306:683-685. [DOI] [PubMed] [Google Scholar]
- 10.Dore, A. S., A. C. Drake, S. C. Brewerton, T. L. Blundell, and A. M. Robert. 2004. Identification of DNA-PK in the arthropods. Evidence for the ancient ancestry of vertebrate non-homologous end-joining. DNA Repair 3:33-41. [DOI] [PubMed] [Google Scholar]
- 11.Dudasova, Z., A. Dudas, and M. Chovanec. 2004. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 28:581-601. [DOI] [PubMed] [Google Scholar]
- 12.Durocher, D., I. A. Taylor, D. Sarbassova, L. F. Haire, S. L. Westcott, S. P. Jackson, S. J. Smerdon, and M. B. Yaffe. 2000. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6:1169-1182. [DOI] [PubMed] [Google Scholar]
- 13.Falck, J., J. Coates, and S. P. Jackson. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434:605-611. [DOI] [PubMed] [Google Scholar]
- 14.Frank-Vaillant, M., and S. Marcand. 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 15:3005-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuta, T., H. Takemura, Z. Y. Liao, G. J. Aune, C. Redon, O. A. Sedelnikova, D. R. Pilch, E. P. Rogakou, A. Celeste, H. T. Chen, A. Nussenzweig, M. I. Aladjem, W. M. Bonner, and Y. Pommier. 2003. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 278:20303-20312. [DOI] [PubMed] [Google Scholar]
- 16.Gell, D., and S. P. Jackson. 1999. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 27:3494-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Harris, R., D. Esposito, A. Sankar, J. D. Maman, J. A. Hinks, L. H. Pearl, and P. C. Driscoll. 2004. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR). J. Mol. Biol. 335:573-582. [DOI] [PubMed] [Google Scholar]
- 19.Hopfner, K. P., C. D. Putnam, and J. A. Tainer. 2002. DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 12:115-122. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, S. P. 2002. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23:687-696. [DOI] [PubMed] [Google Scholar]
- 21.Karathanasis, E., and T. E. Wilson. 2002. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161:1015-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi, J., H. Tauchi, S. Sakamoto, A. Nakamura, K. Morishima, S. Matsuura, T. Kobayashi, K. Tamai, K. Tanimoto, and K. Komatsu. 2002. NBS1 localizes to γ-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 12:1846-1851. [DOI] [PubMed] [Google Scholar]
- 23.Koch, C. A., R. Agyei, S. Galicia, P. Metalnikov, P. O'Donnell, A. Starostine, M. Weinfeld, and D. Durocher. 2004. XRCC4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 23:3874-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kupiec, M. 2000. Damage-induced recombination in the yeast Saccharomyces cerevisiae. Curr. Genet. 38:23-32. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, L. K., F. Storici, S. Van Komen, S. Calero, P. Sung, and M. A. Resnick. 2004. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics 166:1701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, C., J. J. Pouliot, and H. A. Nash. 2002. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. USA 99:14970-14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, J. L., E. M. Kim, J. E. Haber, and S. E. Lee. 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23:8820-8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 29.Meek, K., S. Gupta, D. A. Ramsden, and S. P. Lees-Miller. 2004. The DNA-dependent protein kinase: the director at the end. Immunol. Rev. 200:132-141. [DOI] [PubMed] [Google Scholar]
- 30.Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger, J. K. Moore, J. E. Haber, and V. Lundblad. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8:657-660. [DOI] [PubMed] [Google Scholar]
- 31.Polotnianka, R. M., J. Li, and A. J. Lustig. 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8:831-834. [DOI] [PubMed] [Google Scholar]
- 32.Rost, B. 2004. The PredictProtein server. Nucleic Acids Res. 32:W517-W521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy, R., B. Meier, A. D. McAinsh, H. M. Feldmann, and S. P. Jackson. 2004. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J. Biol. Chem. 279:86-94. [DOI] [PubMed] [Google Scholar]
- 34.Rupp, S. 2002. LacZ assays in yeast. Methods Enzymol. 350:112-131. [DOI] [PubMed] [Google Scholar]
- 35.Shima, H., M. Suzuki, and M. Shinohara. 2005. Isolation and characterization of novel xrs2 mutations in Saccharomyces cerevisiae. Genetics 170:71-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reference deleted.
- 37.Taddei, A., F. Hediger, F. R. Neumann, C. Bauer, and S. M. Gasser. 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23:1301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tauchi, H., J. Kobayashi, K. Morishima, S. Matsuura, A. Nakamura, T. Shiraishi, E. Ito, D. Masnada, D. Delia, and K. Komatsu. 2001. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50-hMRE11-NBS1 complex DNA repair activity. J. Biol. Chem. 276:12-15. [DOI] [PubMed] [Google Scholar]
- 39.Teo, S. H., and S. P. Jackson. 2000. Lif1p targets the DNA ligase Lig4p to sites of DNA double-strand breaks. Curr. Biol. 10:165-168. [DOI] [PubMed] [Google Scholar]
- 40.Tseng, H. M., and A. E. Tomkinson. 2002. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J. Biol. Chem. 277:45630-45637. [DOI] [PubMed] [Google Scholar]
- 41.Tsukamoto, Y., C. Mitsuoka, M. Terasawa, H. Ogawa, and T. Ogawa. 2005. Xrs2p regulates mre11p translocation to the nucleus and plays a role in telomere elongation and meiotic recombination. Mol. Biol. Cell 16:597-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker, J. R., R. A. Corpina, and J. Goldberg. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412:607-614. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, T. E. 2002. A genomics-based screen for yeast mutants with an altered recombination/end-joining repair ratio. Genetics 162:677-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, T. E., U. Grawunder, and M. R. Lieber. 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388:495-498. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, T. E., and M. R. Lieber. 1999. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase β (Pol4)-dependent pathway. J. Biol. Chem. 274:23599-23609. [DOI] [PubMed] [Google Scholar]
- 46.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C.Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Z., W. Hu, L. Cano, T. D. Lee, D. J. Chen, and Y. Chen. 2004. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions. Structure (Cambridge) 12:495-502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.