Abstract
The assembly of cytosolic and nuclear iron-sulfur (Fe/S) proteins in yeast is dependent on the iron-sulfur cluster assembly and export machineries in mitochondria and three recently identified extramitochondrial proteins, the P-loop NTPases Cfd1 and Nbp35 and the hydrogenase-like Nar1. However, the molecular mechanism of Fe/S protein assembly in the cytosol is far from being understood, and more components are anticipated to take part in this process. Here, we have identified and functionally characterized a novel WD40 repeat protein, designated Cia1, as an essential component required for Fe/S cluster assembly in vivo on cytosolic and nuclear, but not mitochondrial, Fe/S proteins. Surprisingly, Nbp35 and Nar1, themselves Fe/S proteins, could assemble their Fe/S clusters in the absence of Cia1, demonstrating that these components act before Cia1. Consequently, Cia1 is involved in a late step of Fe/S cluster incorporation into target proteins. Coimmunoprecipitation assays demonstrated a specific interaction between Cia1 and Nar1. In contrast to the mostly cytosolic Nar1, Cia1 is preferentially localized to the nucleus, suggesting an additional function of Cia1. Taken together, our results indicate that Cia1 is a new member of the cytosolic Fe/S protein assembly (CIA) machinery participating in a step after Nbp35 and Nar1.
Cells have developed intricate systems to provide metal cofactors for enzymes that rely on metal ions for their function. Many proteins have now been shown to be required for assembly and insertion of the deceivingly simple [2Fe-2S] and cubane [4Fe-4S] clusters into bacterial apoproteins (13). In eukaryotes, the occurrence of Fe/S proteins in mitochondria, the cytosol, and nucleus appears to have necessitated further levels of complexity involving unique biosynthesis enzymes that are just being discovered (for review, see reference 20). The so-called iron-sulfur cluster (ISC) assembly machinery resides in the mitochondrial matrix, and its components display a high degree of sequence similarity to the bacterial ISC counterparts. The ISC assembly machinery includes the cysteine desulfurase Nfs1 for providing sulfide and the scaffold proteins Isu1/Isu2 on which an Fe/S cluster is initially assembled before it is transferred to an apoprotein. Most of the 13 yeast mitochondrial ISC proteins have now been shown to be essential for the maturation of both mitochondrial and extramitochondrial Fe/S proteins. In addition, two mitochondrial proteins have been identified that are required for extramitochondrial Fe/S protein assembly only. These are the ABC transporter Atm1 in the inner membrane and the sulfhydryl oxidase Erv1 in the intermembrane space. Both proteins, together with glutathione, are thought to facilitate the export of an as yet unidentified substance from the mitochondria which is, directly or indirectly, required for Fe/S cluster assembly in the cytosol (15, 18, 28). Recently, the first cytosolic proteins have been characterized that are involved in the maturation of Fe/S proteins in the cytosol and nucleus. These are two similar P-loop NTPases termed Cfd1 and Nbp35, as well as Nar1, a protein with striking similarity to iron-only hydrogenases (3, 9, 24). All three proteins are present throughout the eukaryotic kingdom and contain highly conserved cysteine residues (20). Nbp35 and Nar1 have been shown to coordinate Fe/S clusters. In yeast, down-regulation of Cfd1, Nbp35, or Nar1 leads to similar phenotypical consequences. These include, in addition to the maturation defect in cytosolic and nuclear Fe/S proteins, the accumulation of ribosomal subunits in the nucleus. The latter effect is caused indirectly by the assembly defect in the essential Fe/S protein Rli1, which has a crucial function in rRNA processing and ribosome biogenesis (16, 33).
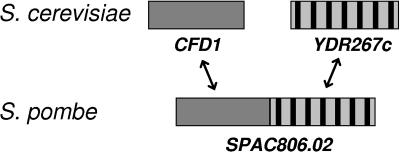
In our search for further components involved in cytosolic Fe/S protein assembly in Saccharomyces cerevisiae, we identified the open reading frame YDR267c as a candidate gene. In the fission yeast Schizosaccharomyces pombe, the homologue of ScCFD1 occurs as a fusion gene with a DNA sequence encoding a WD40 repeat domain at its 3′ end (Fig. 1). The predicted amino acid sequence of the WD40 repeat domain shows striking sequence similarity to ScYDR267c, more so than with any other of the approximately 100 known or predicted WD40 repeat proteins encoded by the S. cerevisiae genome (www.ebi.cam.ac/interpro). The proposed common function of all WD40 repeat proteins is the coordination of multiprotein complex assemblies (19, 29): for example, those functioning in signal transduction (β-transducin), vesicular trafficking (Sec13), transcription regulation (Tup1), or cell cycle control (Cdc4). The crystallization of a number of WD40 repeat domains revealed a seven-bladed β-propeller structure (30-32), where the repeating units form β-sheet “blades” and serve as a rigid scaffold for protein binding.
FIG. 1.
Schematic representation of CFD1 and CIA1 genes in S. cerevisiae and S. pombe. A protein-coding sequence highly similar to S. cerevisiae CFD1 is found in S. pombe as a fusion gene with a C-terminal WD40 repeat domain (systematic name, SPAC806.02).
Here we report the functional characterization of the WD40 repeat protein encoded by the YDR267c gene. Enzyme activity and in vivo 55Fe incorporation studies suggest that it is involved in the assembly of a subset of cytosolic and nuclear, but not mitochondrial, Fe/S proteins. Therefore, we have designated the gene CIA1, for cytosolic iron-sulfur protein assembly 1. The protein coimmunoprecipitated with Nar1, further suggesting that it is a member of the CIA machinery. Depletion of Cia1 also led to the accumulation of the large ribosomal subunit in the nucleus, a phenotype typical for defects in cytosolic/nuclear Fe/S protein assembly. Together, our data suggest that Cia1 is a novel component of cytosolic Fe/S protein assembly acting after Nar1 and Nbp35.
MATERIALS AND METHODS
Yeast strains, cell growth, and plasmids.
Saccharomyces cerevisiae strain W303-1A (MATa ura3-1 ade2-1 trp1-1 his3-11,15 leu2-3,112) served as the wild type. The mutant strain Gal-CIA1 was derived from W303-1A by exchange of the endogenous promoter (nucleotides −273 to −1) for the galactose-inducible GAL1-10 promoter by homologous recombination using a PCR product containing the HIS3 marker gene (22). Similarly, the Gal-CFD1 strain was created by replacing nucleotides −242 to −1. Correct insertion of the DNA into the yeast genome was verified by PCR. The Gal-NAR1 and Gal-NBP35 mutant strains have been described previously (3, 9). Cells were grown in rich (yeast extract-peptone [YP]) and minimal (synthetic complete [SC]) media or in minimal medium lacking added iron chloride (“iron poor”), each containing the required carbon sources at a concentration of 2% (wt/vol) unless indicated otherwise (27). The following yeast plasmids were used: pRS416 containing the MET25 promoter and pRS426 with the TDH3 promoter (23). All constructs were verified by DNA sequencing.
Coimmunoprecipitation.
Cells were grown overnight in minimal medium plus galactose, and ∼0.5 g of cells (wet weight) was harvested. The cells were washed with water and resuspended in 0.5 ml of cold extraction buffer (10 mM HEPES-NaOH, pH 7.5, 100 mM sodium acetate, 10% [vol/vol] glycerol, 0.5% [vol/vol] Triton X-100, 2 mM phenylmethylsulfonyl fluoride). All further procedures were carried out at 4°C. The cells were disrupted by vortexing with glass beads, and the lysate was cleared by centrifugation at 13,000 × g for 10 min. Agarose-conjugated hemagglutinin (HA) probe (sc-7392 AC; Santa Cruz Biotechnology, CA) was added to the lysate, and this mixture was incubated for 1 h with gentle agitation. The agarose beads were washed three times with extraction buffer (without phenylmethylsulfonyl fluoride) and resuspended in 1× sodium dodecyl sulfate gel loading buffer (26) to subject the bound proteins to standard protein blot procedures.
Miscellaneous methods.
The following published methods were used: manipulation of DNA and PCR (26); transformation of yeast cells (7); determination of the enzyme activities of alcohol dehydrogenase (EC 1.1.1.1), isopropylmalate isomerase using 2-isopropylmaleate as a substrate (EC 4.2.1.33), aconitase (E.C. 4.2.1.3), and succinate dehydrogenase (E.C. 4.2.1.3) (15, 22); determination of the enzyme activity of sulfite reductase (EC 1.8.1.2) (25); raising of antisera using proteins that were purified after heterologous production in Escherichia coli and protein blot analysis (8); in situ immunofluorescence (17); induction of the iron regulon by monitoring the fluorescence levels of green fluorescent protein (GFP) expressed from a FET3-GFP fusion gene (25); and quantification of iron in mitochondria by the colorimetric chelator ferene (11). Radiolabeling of the Fe/S reporter proteins of interest with 55Fe and cell lysis were performed as previously described (15, 21). All experiments were repeated two to four times. Error bars represent the standard error of the mean value.
RESULTS
The essential protein Cia1 is a soluble protein located in the nucleus and cytosol.
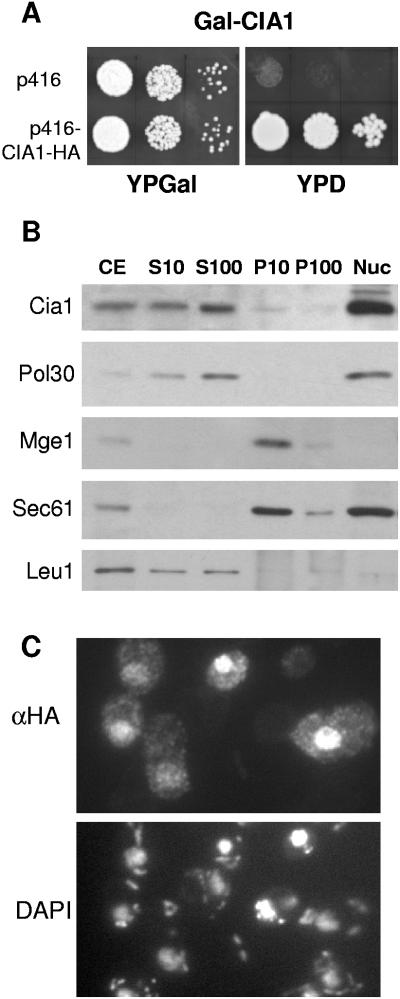
Systematic studies found that deletion of the YDR267c open reading frame was lethal (6). Therefore, a regulatable mutant strain termed Gal-CIA1 was constructed for phenotypic and functional studies. The glucose-repressible GAL1-10 promoter was inserted immediately upstream of the CIA1 gene, using homologous recombination resulting in deletion of 273 nucleotides of the CIA1 promoter. Culturing of Gal-CIA1 cells on glucose-containing rich medium completely inhibited growth (Fig. 2A). This was specifically due to the depletion of Cia1, because transformation with a plasmid expressing the CIA1 gene restored growth on glucose medium.
FIG. 2.
The essential Cia1 is a soluble protein in the nucleus and cytosol. (A) Growth of Cia1-depleted cells. Gal-CIA1 cells carrying a galactose-regulatable (Gal) CIA1 gene were transformed with vector p416 without or with a CIA1-HA fusion gene. Cells were grown for 2 × 2 days at 30°C on agar plates containing rich media supplemented with galactose (YPGal) or glucose (YPD). Tenfold serial dilutions are shown. (B) Subcellular fractionation and immunoblot analysis. Wild-type cells were disrupted by removal of the cell wall and homogenization with a Douncer in 0.6 M sorbitol, 20 mM HEPES-KOH, pH 7.4, 1 mM dithiothreitol, and protease inhibitors, followed by differential centrifugation. After discarding a low-speed pellet containing unbroken cells, nuclei, and debris, the cell extract (CE) was centrifuged at 10,000 × g for 10 min to separate soluble proteins (S10) from a fraction enriched in organelles (P10). The S10 fraction was further centrifuged at 100,000 × g for 30 min to remove membranes (P100) from soluble proteins (S100). Nuclei (Nuc) were purified separately using a Ficoll density gradient (1). Equal amounts of protein (20 μg/per lane) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted, and immunostained for Cia1 or marker proteins of the nucleus (DNA polymerase-associated Pol30), mitochondria (Mge1), endoplasmic reticulum (translocon subunit Sec61), and the cytosol (Leu1). (C) In situ localization of Cia1. Wild-type cells were transformed with the high-copy-number expression vector p426 containing CIA1 fused to the HA tag sequence. Log-phase cells were fixed with 2.4% (wt/vol) formaldehyde, permeabilized, and labeled with monoclonal anti-HA (α-HA) antibodies, followed by fluorophore-conjugated secondary antibodies. DNA was counterstained with DAPI (4′,6′-diamidino-2-phenylindole) to show the positions of the nucleus and mitochondria.
To further characterize Cia1, its cellular localization was studied by subcellular fractionation. Wild-type cells were converted into spheroplasts and lysed, and the extracts were separated by centrifugation into supernatant and pellet fractions or loaded onto a Ficoll density gradient for the purification of nuclei. The fractions were analyzed by protein blot analysis. Cia1 was mostly associated with the purified nuclei, but was also found in the soluble fractions (S10 and S100, Fig. 2B). The fractionation pattern of Cia1 was similar to that found for the DNA-associated Pol30. Cia1 was not associated with membranous organelles (P10), since it did not copurify with the mitochondrial protein Mge1 and the endoplasmic reticulum protein Sec61. This suggested that Cia1 is a soluble protein accumulated in the nucleus of yeast cells but is also present in the cytosol.
The cellular distribution of Cia1 was further analyzed by in situ immunofluorescence. Wild-type cells were transformed with a high-copy-number vector encoding an HA-tagged Cia1. Immunolabeling with anti-HA antibodies showed that the Cia1-HA fusion protein was predominantly localized in the nucleus; however, Cia1-HA was also present, at a lower concentration, in the cytoplasm (Fig. 2C). Similar results with low fluorescence intensity were obtained with CIA1-HA inserted into the low-copy-number expression vector p416 (not shown). The microscopy data correspond well to our subcellular fractionation results and are in agreement with systematic localization studies (12).
Cia1 is involved in the assembly of specific cytosolic and nuclear Fe/S proteins.
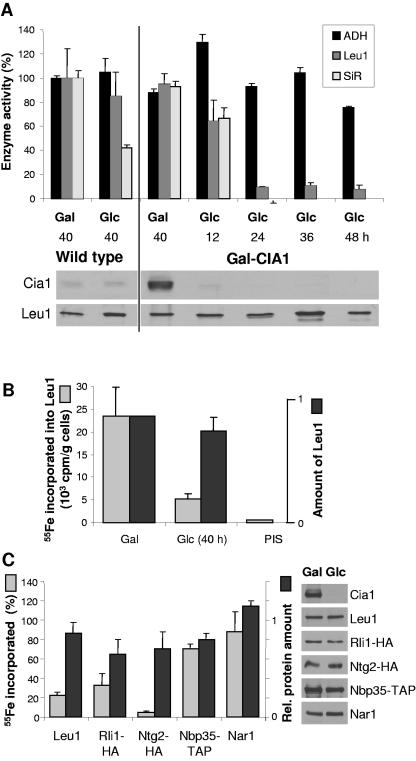
The occurrence of the CFD1-CIA1 gene fusion in S. pombe prompted us to investigate whether the S. cerevisiae Cia1 protein might play a role in cytosolic Fe/S protein assembly like Cfd1 does (24). First, the activities of two cytosolic Fe/S enzymes, isopropylmalate isomerase (Leu1) and sulfite reductase, were measured in cells depleted of Cia1. To this end, Gal-CIA1 cells were grown in minimal medium supplemented with either galactose or glucose to induce or down-regulate, respectively, expression of CIA1. Wild-type cells in equivalent media were used for comparison. After 12 h of culture in glucose, the levels of Cia1 in Gal-CIA1 cells had dropped below those in wild-type cells. Leu1 and sulfite reductase activities in cytosolic extracts were comparable to those in wild-type or CIA1-overexpressing cells (Fig. 3A). At later time points of depletion, the levels of Cia1 decreased to the limit of detection. Sulfite reductase activity could no longer be detected, and only approximately 10% of the Leu1 activity remained, while the levels of Leu1 polypeptide remained unchanged. In contrast, the Zn-dependent alcohol dehydrogenase activity was not affected. Cell growth slowed down considerably after 36 h of Cia1 depletion, but growth continued up to at least 64 h (not shown). The data show that Cia1 was required for maintaining the enzyme activity of two cytosolic Fe/S proteins.
FIG. 3.
Cia1 is required for the assembly of a subset of cytosolic and nuclear Fe/S proteins. Gal-CIA1 cells were grown in minimal medium supplemented with galactose (Gal) or glucose (Glc) for various lengths of time to induce or suppress, respectively, expression of CIA1. (A) Enzyme activities of alcohol dehydrogenase (ADH), isopropylmalate isomerase (Leu1), and sulfite reductase (SiR) in cell extracts are presented relative to the values obtained for wild-type cells grown with galactose. The protein levels of Cia1 and Leu1 were visualized by immunoblot analysis (lower panels). (B) 55Fe incorporation into Leu1. Gal-CIA1 cells were labeled with 55Fe, and a cell lysate was prepared, from which Leu1 was immunoprecipitated with specific antibodies. Coprecipitated 55Fe was quantified by scintillation counting. Immunoprecipitation with preimmune serum (PIS) was used as a control. The amounts of Leu1 protein were quantified by immunoblot analysis using chemiluminescence and densitometry of subsaturated film exposures. (C) 55Fe incorporation into several cytosolic and nuclear Fe/S proteins. Data acquired as in panel B are expressed as the ratio of glucose-grown (40 h) to galactose-grown cells, corrected for background, for each of the reporter proteins. Rli1-HA, Ntg2-HA, Nbp35-TAP, and Nar1 were expressed from high-copy-number vectors (p426) and immunoprecipitated with anti-HA, immnunoglobulin G (IgG), and anti-Nar1 antibodies, respectively. Cells not expressing these constructs were used to determine the background levels of immunoprecipitated 55Fe. The panels on the right show examples of the immunoblot analysis for each of the indicated proteins.
To investigate the de novo biosynthesis of Fe/S clusters, Gal-CIA1 cells were grown in iron-poor minimal medium in the presence of galactose or glucose and labeled with 55Fe for 2 to 3 h. Subsequently, a cell extract was prepared and Leu1 was immunoprecipitated with Leu1-specific antiserum. The 55Fe incorporated into Leu1 was measured by scintillation counting, and the protein levels were assessed by immunoblot analysis and densitometry. When Cia1 was depleted for 40 h by culturing Gal-CIA1 cells in the presence of glucose, the amount of 55Fe incorporated into Leu1 was one-fourth that in galactose-grown cells, whereas the protein levels did not differ significantly (Fig. 3B). Some 55Fe was still associated with Leu1 at this time point, as judged from the background scintillation counts when the experiment was carried out with preimmune serum (PIS). Uptake of 55Fe into the cells was not affected by the choice of carbon source, documenting that the effects were not caused by decreased iron supply (not shown).
We further measured the 55Fe incorporation into a range of cytosolic and nuclear Fe/S proteins as described for Leu1. Our analysis revealed two classes of Fe/S reporters, one dependent on Cia1 function in Fe/S cluster assembly, the other incorporating normal levels of 55Fe in the absence of Cia1. The nuclear reporter protein Ntg2-HA (3) accumulated only ∼5% of its normal levels of 55Fe. Similarly, the cytosolic Rli1-HA incorporated only ∼30% of 55Fe after 40 h and ∼9% after 64 h of Cia1 depletion (Fig. 3C; data not shown). Protein levels were generally slightly decreased (to ∼70 to 90% of the wild-type amounts), indicative of proteolysis which is commonly observed for Fe/S proteins in their apo forms (3). In contrast, the levels of 55Fe incorporation into the predominantly cytosolic Fe/S proteins Nbp35-TAP and Nar1 were ∼70% and ∼90% of wild type, respectively, and did not differ significantly from the respective change in protein levels upon Cia1 depletion (Fig. 3C). Longer periods of Cia1 depletion also did not show decreased 55Fe incorporation into these two reporter proteins (not shown). Earlier results had shown that 55Fe association with Nbp35 and Nar1 is fully dependent on the mitochondrial ISC assembly and export machineries, and that the 55Fe radiolabel associated with these proteins is part of an Fe/S cluster (3, 9). The 55Fe incorporation into Fe/S reporter proteins in galactose-grown Gal-CIA1 cells was, in all cases, similar to that found for wild-type cells. Also, the choice of the carbon source did not influence the 55Fe incorporation into the Fe/S reporter proteins in wild-type cells (not shown). Our results therefore indicated that maturation of Leu1, Ntg2, and Rli1 was strongly dependent on Cia1 function, whereas Fe/S cluster incorporation into Nbp35 and Nar1 did not require Cia1.
Cia1 is not required for mitochondrial Fe/S protein assembly and cellular iron homeostasis.
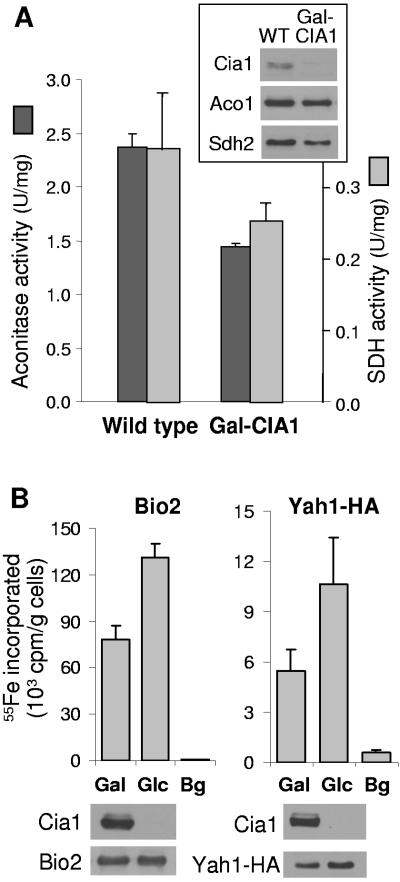
To further investigate the specificity of the Cia1 requirement in the assembly of Fe/S protein targets, we asked whether cofactor assembly on mitochondrial Fe/S proteins was dependent on Cia1. After culturing Gal-CIA1 cells in the presence of glucose for 40 h to deplete Cia1, the activities of the Fe/S-dependent enzymes aconitase and succinate dehydrogenase were decreased by ∼25% compared to those in wild-type cells (Fig. 4A). This decrease correlated with lower protein levels of aconitase (Aco1) and succinate dehydrogenase subunit 2 (Sdh2, Fig. 4A, inset). Next, the de novo Fe/S cluster assembly into two mitochondrial reporters, Bio2 (biotin synthase) and Yah1-HA (ferredoxin), was investigated using the 55Fe incorporation and immunoprecipitation assays described above. After 40 h of Cia1 depletion, the amount of 55Fe incorporated into Bio2 or Yah1-HA was slightly elevated compared to that in Cia1-containing cells (Fig. 4B). The increased 55Fe levels correlated well with the larger amounts of the Fe/S reporter proteins, most notably of Yah1-HA. Even after 64 h of Cia1 depletion, 55Fe labeling of Bio2 was similar to the amount of labeling in galactose-grown cells (not shown). Taken together, our data indicated that Cia1 function was not required for Fe/S protein assembly in mitochondria.
FIG. 4.
Cia1 is not required for mitochondrial Fe/S protein assembly. (A) Enzyme activities of aconitase and succinate dehydrogenase (SDH) in wild-type (WT) and Gal-CIA1 cells after growth in minimal medium plus glucose for 40 h. Protein levels of aconitase (Aco1) and succinate dehydrogenase subunit 2 (Sdh2) were visualized by immunoblot analysis (inset), using amounts of mitochondrial protein equivalent to those used for the enzyme assays. (B) 55Fe incorporation into the Fe/S proteins Bio2 and Yah1-HA. Gal-CIA1 cells were grown in iron-poor minimal medium supplemented with galactose (Gal) or glucose (Glc) to induce or suppress, respectively, expression of CIA1. Bio2 and HA-tagged Yah1 were overproduced using the high-copy-number vector p426. Cells were labeled with 55Fe, and a cell lysate was prepared, from which Bio2 and Yah1 were immunoprecipitated with anti-Bio2- or anti-HA-specific antibodies, respectively. Coprecipitated 55Fe was quantified by scintillation counting. The amounts of 55Fe immunoprecipitated from cell extracts that did not contain overproduced Bio2 or Yah1-HA were taken as background (Bg). The levels of the indicated proteins were analyzed by immunoblot analysis.
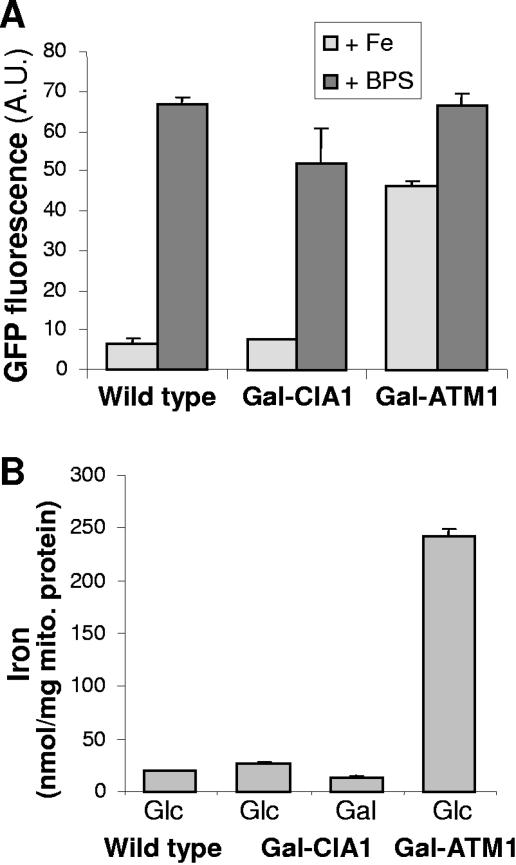
Numerous proteins involved in Fe/S protein maturation affect cellular iron homeostasis, leading to the induction of the Aft1/Aft2 transcription factor-dependent iron regulon, even in the presence of sufficient amounts of iron in the growth medium (25). We therefore analyzed the induction of the Aft1-dependent FET3 gene (encoding a copper-dependent ferroxidase) in Cia1-depleted cells using a fusion construct of the FET3 promoter and the gene coding for GFP as a reporter. A plasmid carrying the FET3-GFP fusion gene was transformed into wild-type, Gal-CIA1, and Gal-ATM1 cells as a control. Cells were analyzed for their GFP fluorescence after growth in the presence of glucose to deplete Cia1 or Atm1, respectively. No significant increase in the fluorescence intensity was observed upon depletion of Cia1 compared to that in wild-type cells under iron-replete conditions, suggesting that depletion of Cia1 did not elicit a disturbance of cellular iron homeostasis as seen upon depletion of Atm1 (Fig. 5A). This finding is in full agreement with the normal 55Fe uptake into Cia1-depleted cells (not shown). Upon addition of the iron chelator bathophenanthroline sulfonate, the iron regulon was induced to a similar extent in Cia1-deficient and in wild-type cells, suggesting that Cia1-deficient cells respond normally to a decreased iron supply (Fig. 5A). We also analyzed the iron content of mitochondria in Cia1-depleted cells, since functional impairment of many proteins involved in Fe/S cluster assembly results in iron accumulation in these organelles (20). However, there was no increase in the iron content of mitochondria purified from Cia1-depleted cells, in contrast to the observation made for Atm1-depleted organelles (Fig. 5B) (15). Taken together, our data document that Cia1 depletion does affect either mitochondrial or cellular iron homeostasis.
FIG. 5.
Normal cellular and mitochondrial iron homeostasis in the absence of Cia1. (A) Wild-type, Gal-CIA1, and Gal-ATM1 cells carrying plasmid p415-FET3-GFP were grown in glucose-containing minimal medium for 24 h, diluted to an optical density at 600 nm of 0.1, and supplemented with either 0.1 mM FeCl3 (Fe) or 0.1 mM of the iron chelator bathophenanthroline sulfonate (BPS). Growth was continued for a further 6 h. The transcriptional activity of the FET3 promoter was determined by recording the fluorescence emission of the cell suspension at 513 nm (excitation at 480 nm). The signal of cells lacking plasmid p415-FET3-GFP was subtracted. A.U., arbitrary units. (B) Wild-type, Gal-CIA1, and Gal-ATM1 cells were grown in minimal medium with either galactose (Gal) or glucose (Glc). Mitochondria were purified by differential centrifugation, and the total iron content was measured using a colorimetric assay based on the iron chelator ferene.
An interaction between Cia1 and Nar1.
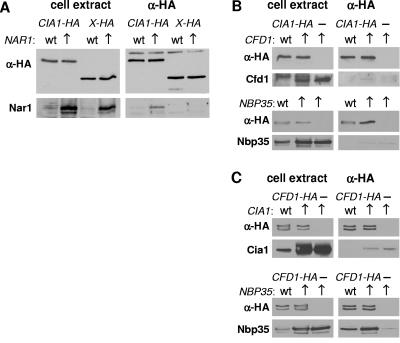
The S. pombe CFD1-CIA1 fusion gene and our functional data for Cia1 in S. cerevisiae (see above) hinted at the possible existence of direct protein interactions between Cia1 and Cfd1 or the other known CIA components, Nbp35 and Nar1. We performed coimmunoprecipitation assays employing yeast cell extracts, followed by protein blot analyses to assess the amounts of candidate partners binding to HA-tagged Cia1 (Fig. 6). Because of the extremely low endogenous levels of Cfd1, Nbp35, and Nar1 in yeast (3, 5, 9), these proteins were overproduced. When Cia1-HA was coexpressed in cells with enhanced levels of Nar1, a significant amount of Nar1 was coimmunoprecipitated with Cia1-HA using anti-HA agarose (Fig. 6A). This protein interaction was specific for Nar1 and Cia1 because, first, a weaker yet significant immunostaining signal for Nar1 was observed in the coimmunoprecipitation reaction from cells with wild-type levels of Nar1. Second, Nar1 could not be detected in precipitation assays using another HA-tagged protein, Ymr134-HA (2), in cell extracts with either induced or wild-type levels of Nar1. Our data are further supported by a yeast two-hybrid interaction reported for Cia1 and Nar1 (10).
FIG. 6.
Cia1 undergoes a protein interaction with Nar1. Cells producing the HA-tagged proteins as indicated were grown in minimal medium and harvested, and a cell extract was prepared. HA-agarose was added for 1 h of incubation at 4°C and washed several times with buffer. Both the cell extract and the washed HA-agarose were subjected to protein blot analysis to detect proteins of interest. (A) Coimmunoprecipitation of HA-tagged Cia1 with Nar1 in extracts from cells with wild-type (wt) or overproduced (↑) levels of Nar1. Another HA-tagged protein was used as a control (X-HA, where X represents Ymr134w). The indicated proteins were detected by protein blot analysis in total cell extracts (left panels) and after immunoprecipitation (anti-HA [α-HA], right panels). (B and C) As in panel A, using cells overproducing Cfd1, Nbp35, or Cia1 as indicated. Cia1-HA (B) or Cfd1-HA (C) was immunoprecipitated using anti-HA antibodies before protein blot analyses.
Next, similar coimmunoprecipitation assays were carried out with extracts from cells that overexpressed either CFD1 or NBP35 (Fig. 6B). In both cases, no specific coimmunoprecipitation of either Cfd1 or Nbp35 with Cia1-HA was seen. To independently confirm these results, the immunoprecipitation reactions were carried out vice versa by immunoprecipitating HA-tagged Cfd1 and analyzing the amount of coprecipitated Cia1 (Fig. 6C). In this case, the protein interaction between Cfd1 and Nbp35 (D. J. Aguilar Netz et al., unpublished) served as a positive control (Fig. 6C, lower panels). No endogenous or overproduced Cia1 was precipitated with Cfd1-HA in a specific fashion. In summary, our data demonstrate a specific protein interaction between Cia1 and Nar1 but do not identify a stable interaction between Cia1 and Cfd1 or Cia1 and Nbp35.
Depletion of Cia1 impairs the export of the large ribosomal subunit from the nucleus.
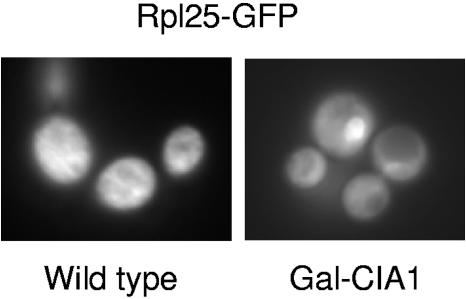
The requirement of Cia1 for Fe/S cluster assembly on Rli1 predicts that cells lacking Cia1 display phenotypic features similar to those of Rli1-depleted cells. Rli1 performs a crucial function in the export of ribosomal subunits from the nucleus to the cytosol. We therefore expected that Cia1-depleted cells accumulate ribosomal precursors in the nucleus. Such a defect can be visualized using GFP-tagged ribosomal proteins (16, 33). Gal-CIA1 cells were transformed with a plasmid encoding a polypeptide of the large ribosomal subunit, Rpl25, fused to GFP. The expression of CIA1 was down-regulated by culturing Gal-CIA1 cells in minimal medium with glucose, and the localization of Rpl25-GFP was observed by fluorescence microscopy at various time points. After approximately 40 h of Cia1 depletion, nuclear accumulation of Rpl25-GFP became evident (Fig. 7). At this time point, 55Fe incorporation into Rli1-HA was diminished to ∼30% of its normal levels (Fig. 3C). A similar nuclear export defect of ribosomal proteins was observed in cells depleted of Cfd1, Nar1, and Nbp35 (16, 33). Together, this can be taken as an additional argument that Cia1 is a member of the CIA machinery.
FIG. 7.
Cia1 is required for export of the large ribosomal subunit from the nucleus. Wild-type and Gal-CIA1 cells expressing an RPL25-GFP fusion gene were cultured in minimal medium supplemented with glucose for 40 h. The cellular distribution of GFP was monitored by fluorescence microscopy. The spot accumulating GFP in the Gal-CIA1 cells is the nucleus, as judged by phase-contrast microscopy and DAPI (4′,6′-diamidino-2-phenylindole) staining (not shown; see reference 16).
DISCUSSION
In this paper, we describe the identification and initial characterization of the essential protein Cia1 as a novel component of the CIA machinery. Cia1 is required for cytosolic and nuclear Fe/S protein assembly and interacts with Nar1, another recently identified member of the CIA machinery. Depletion of Cia1 resulted in the loss of the enzyme activities of the cytosolic Fe/S proteins isopropylmalate isomerase (Leu1) and sulfite reductase. The decrease in the enzyme activities is explained by a requirement of Cia1 in the de novo assembly of the Fe/S cofactor in Leu1 as shown by in vivo 55Fe labeling studies. The same experimental approach revealed that Fe/S cluster assembly was strongly decreased in the mostly cytosolic Rli1 and the nuclear Ntg2 Fe/S proteins. As expected from the extramitochondrial location of Cia1, both the enzyme activities and 55Fe incorporation of mitochondrial Fe/S proteins were not negatively affected. Surprisingly, 55Fe incorporation into the predominantly cytosolic Fe/S proteins Nar1 and Nbp35 was also not decreased following depletion of Cia1. How may this latter observation be explained? Nar1 and Nbp35 were previously shown to be required for Fe/S protein assembly outside mitochondria. In particular, Nar1 depletion resulted in diminished 55Fe incorporation into Nbp35 (4), and depletion of Nbp35 abolished 55Fe incorporation into Nar1 (9). Thus, Cia1 behaves differently from these proteins in that it is required only for “true” Fe/S protein targets such as Leu1 and Rli1 (16), but not for the Fe/S protein members of the CIA machinery. These in vivo iron-labeling data place the function of Cia1 clearly after that of Nar1 and Nbp35. The fact that Nar1 and Nbp35 can assemble their Fe/S clusters independently of Cia1 suggests that these cofactors may bind transiently to Nbp35 and Nar1, and the function of Cia1 may be, e.g., in the delivery of the clusters to the apoproteins. It is therefore tempting to speculate that these proteins serve as scaffolds for Fe/S cluster assembly in the cytosol. In support of these ideas, the Fe/S clusters bound to Nbp35 are unstable in vivo compared to those of target Fe/S proteins such as Leu1 (P. Smith and R. Lill, unpublished). Rigorous testing of these ideas will require successful in vitro reconstitution of the assembly process of Fe/S clusters in the eukaryotic cytosol.
We demonstrate a specific and stable protein interaction of Cia1 with Nar1, but not with Cfd1 or Nbp35, suggesting a close collaboration of the former components in Fe/S protein biogenesis. While Cfd1, Nbp35, and Nar1 are predominately located in the cytosol with only a minor fraction in the nucleus (3, 9, 24), a large proportion of Cia1 is localized in the nucleus. Further, Cia1 appears to be at least 10-fold higher in its cellular concentration than Cfd1, Nbp35, and Nar1 (5). These localization and expression differences between Cfd1/Nbp35/Nar1 and Cia1 may indicate a second function of the latter protein in the nucleus. Interestingly, the human protein with the highest sequence similarity to Cia1 was found to interact with a zinc finger transcription factor, WT1, the Wilms' tumor suppressor protein (14). This human WD40 repeat protein named Ciao1 (Chinese for “bridge”) modulates the mobility of the WT1-DNA complex in gel shift assays and decreases the transcriptional activity of WT1. Although Cia1 and Ciao1 exhibit 64% amino acid similarity, no obvious sequence homologue of WT1 can be found in the yeast genome. Nevertheless, other yeast zinc-finger protein or proteins may interact with Cia1. For instance, Cia1 was found to interact with the zinc-dependent transcription factor Hms1 using systematic yeast two-hybrid studies (10).
Cia1 did not show a stable protein interaction with Cfd1 (or Nbp35) under our experimental conditions. This was somewhat surprising, since in Schizosaccharomyces pombe the CFD1 and CIA1 sequence homologues are linked as a fusion gene. Now that more genome sequences have become available, it appears that this gene arrangement is unique for S. pombe. It is important to note that there is no experimental evidence at this point for the existence of a fusion protein in S. pombe, since posttranslational cleavage may occur. Nevertheless, the presence of a fusion gene in S. pombe points to a common function of both encoded proteins.
The function of Cia1 in the incorporation of the essential Fe/S clusters at the N terminus of Rli1 explains why Cia1 is indispensable for cell growth. Rli1 plays a crucial role in ribosome biogenesis, and its depletion results in the accumulation of ribosomal subunits in the cell nucleus (16, 33). The function of Cia1 in Fe/S cluster assembly on Rli1 also satisfactorily explains why Cia1 was needed for ribosome biogenesis, as demonstrated by the requirement of Cia1 for export of the large ribosomal subunit from the nucleus, visualized using an Rpl25-GFP fusion protein. Even though the function of Cia1 in nuclear export of ribosomes appears to be indirect, this phenotype has also been observed after down-regulation of Cfd1, Nbp35, and Nar1 (16, 33) and thus may be a general, yet secondary, consequence of defects in cytosolic Fe/S protein biogenesis. These data support the previous notion of an intimate link between cellular Fe/S protein biogenesis and protein synthesis, two evolutionarily ancient and essential processes (16).
The WD40 repeat domain is a common structural module in eukaryotes and is found as part of many protein complexes exhibiting diverse functions. The WD40 repeat proteins form characteristic β-propeller structures. Specificity of each of the WD40 repeat proteins for its binding target and cellular process is thought to be defined by the protein loops protruding from one side of the propeller blades (31). Crystallographic studies are currently being undertaken to structurally define these loops in Cia1. This information can then aid in mutagenesis studies to unravel the precise molecular function of Cia1 and its interaction with Nar1. Furthermore, modeling of the human Ciao1 protein on the crystal structure of Cia1 could give an indication of whether these two proteins may be functional homologues.
Depletion of Cia1 did not lead to the stimulation of iron uptake into the cell nor to hyperaccumulation of iron in mitochondria, unlike defects in the mitochondrial ISC components involved in Fe/S protein assembly (15, 20). It appears that none of the known cytosolic Fe/S protein assembly components (Cfd1, Nbp35, Nar1, and Cia1) is critically involved in regulating cellular iron homeostasis via the Aft1/Aft2 transcription factors (25). Moreover, decreased iron uptake under iron-limiting conditions does not necessarily lead to a (cytosolic) Fe/S cluster assembly defect, as growth in iron-depleted medium did not affect cytosolic Fe/S protein assembly (J. Balk, unpublished). Hence, regulation of cellular and particularly mitochondrial iron homeostasis in yeast does not require active Fe/S protein assembly in the cytosol. Together, these data underline the importance of yeast mitochondria and the ISC system for cellular iron homeostasis and distinguish them from the cytosolic Fe/S protein assembly apparatus.
The past 2 years have led to the discovery of four extramitochondrial components that are functionally involved in Fe/S protein assembly in cytosolic and nuclear apoproteins in yeast. Currently, the mechanism by which these CIA components assist the assembly process is unknown, apart from the finding that Cia1 acts late in biogenesis. Further progress can now be made by a combination of in vivo and in vitro approaches to unravel the precise mode of action of the individual proteins and by identifying even more members of the CIA system. Yeast is an ideal model system for developing a more and more complete picture of Fe/S protein assembly in the eukaryotic cell.
Acknowledgments
We thank H. Beinert for critical reading of the manuscript, E. Hurt for plasmid p316-RPL25-eGFP, H. Ulrich for affinity-purified anti-Pol30p antibodies, and D. Darley for chemical synthesis of 2-isopropylmaleate.
Our work was supported by grants of the Sonderforschungsbereich 593, Deutsche Forschungsgemeinschaft (Gottfried-Wilhelm Leibniz program), the European Commission (Marie Curie European Fellowship HPMF-CT-2002-01750 to J.B.), and Fonds der Chemischen Industrie.
REFERENCES
- 1.Aris, J. P., and G. Blobel. 1991. Isolation of yeast nuclei. Methods Enzymol. 194:735-749. [DOI] [PubMed] [Google Scholar]
- 2.Babcock, M., D. de Silva, R. Oaks, S. Davis-Kaplan, S. Jiralerspong, L. Montermini, M. Pandolfo, and J. Kaplan. 1997. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276:1709-1712. [DOI] [PubMed] [Google Scholar]
- 3.Balk, J., A. J. Pierik, D. J. Aguilar Netz, U. Mühlenhoff, and R. Lill. 2004. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23:2105-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balk, J., A. J. Pierik, D. J. Aguilar Netz, U. Mühlenhoff, and R. Lill. 2005. Nar1p, a conserved eukaryotic protein with similarity to Fe-only hydrogenases, functions in cytosolic iron-sulphur protein biogenesis. Biochem. Soc. Trans. 33:86-89. [DOI] [PubMed] [Google Scholar]
- 5.Ghaemmaghami, S., W.-K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 6.Giaever, G., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 7.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harlow, E., and D. Lane. 1998. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Hausmann, A., D. J. Aguilar Netz, J. Balk, A. J. Pierik, U. Mühlenhoff, and R. Lill. 2005. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. USA 102:3266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazbun, T. R., L. Malmström, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. Muller, S. Fields, D. Baker, J. R. Yates, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 11.Hennessy, D. J., G. R. Reid, F. E. Smith, and S. L. Thompson. 1984. Ferene—a new spectrophotometric reagent for iron. Can. J. Chem. 62:721-724. [Google Scholar]
- 12.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, D. C., D. R. Dean, A. D. Smith, and M. K. Johnson. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247-281. [DOI] [PubMed] [Google Scholar]
- 14.Johnstone, R. W., J. Wang, N. Tommerup, H. Vissing, T. Roberts, and Y. Shi. 1998. Ciao1 is a novel WD40 protein that interacts with the tumor suppressor protein WT1. J. Biol. Chem. 273:10880-10887. [DOI] [PubMed] [Google Scholar]
- 15.Kispal, G., P. Csere, C. Prohl, and R. Lill. 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kispal, G., K. Sipos, H. Lange, Z. Fekete, T. Bedekovics, T. Janáky, J. Bassler, D. J. Aguilar Netz, J. Balk, C. Rotte, and R. Lill. 2005. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 24:589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krebber, H., T. Taura, M. S. Lee, and P. A. Silver. 1999. Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes Dev. 13:1994-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange, H., T. Lisowsky, J. Gerber, U. Mühlenhoff, G. Kispal, and R. Lill. 2001. An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep. 2:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, D., and R. Roberts. 2001. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol. Life Sci. 58:2085-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lill, R., and U. Mühlenhoff. 2005. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 30:133-141. [DOI] [PubMed] [Google Scholar]
- 21.Mühlenhoff, U., J. Gerber, N. Richhardt, and R. Lill. 2003. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 22:4815-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mühlenhoff, U., N. Richhardt, M. Ristow, G. Kispal, and R. Lill. 2002. The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum. Mol. Genet. 11:2025-2036. [DOI] [PubMed] [Google Scholar]
- 23.Mumberg, D., R. Müller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 24.Roy, A., N. Solodovnikova, T. Nicholson, W. Antholine, and W. E. Walden. 2003. A novel eukaryotic factor for cytosolic Fe/S cluster assembly. EMBO J. 22:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rutherford, J. C., L. Ojeda, J. Balk, U. Mühlenhoff, R. Lill, and D. R. Winge. 2005. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial, but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 280:10135-10140. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 28.Sipos, K., H. Lange, Z. Fekete, P. Ullmann, R. Lill, and G. Kispal. 2002. Maturation of cytosolic iron-sulfur proteins requires glutathione. J. Biol. Chem. 277:26944-26949. [DOI] [PubMed] [Google Scholar]
- 29.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 30.Sondek, J., A. Bohm, D. G. Lambright, H. E. Hamm, and P. B. Sigler. 1996. Crystal structure of a Gα protein dimer at 2.1 Å resolution. Nature 379:369-374. [DOI] [PubMed] [Google Scholar]
- 31.Sprague, E. R., M. J. Redd, A. D. Johnson, and C. Wolberger. 2000. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 19:3016-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wall, M. A., D. E. Coleman, E. Lee, J. A. Iñiguez-Lluhi, B. A. Posner, A. G. Gilman, and S. R. Sprang. 1995. The structure of the G protein heterotrimer Giα1β1γ2. Cell 83:1047-1058. [DOI] [PubMed] [Google Scholar]
- 33.Yarunin, A., V. G. Panse, E. Petfalski, C. Dez, D. Tollervey, and E. C. Hurt. 2005. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 24:580-588. [DOI] [PMC free article] [PubMed] [Google Scholar]