Abstract
Two highly conserved double-strand break (DSB) repair pathways, homologous recombination (HR) and nonhomologous end joining (NHEJ), function in all eukaryotes. How a cell chooses which pathway to utilize is an area of active research and debate. During NHEJ, the DNA-dependent protein kinase (DNA-PK) functions as a “gatekeeper” regulating DNA end access. Here, we provide evidence that DNA-PK regulates DNA end access via its own autophosphorylation. We demonstrated previously that autophosphorylation within a major cluster of sites likely mediates a conformational change that is critical for DNA end processing. Furthermore, blocking autophosphorylation at these sites inhibits a cell's ability to utilize the other major double-strand break repair pathway, HR. Here, we define a second major cluster of DNA-PK catalytic subunit autophosphorylation sites. Whereas blocking phosphorylation at the first cluster inhibits both end processing and HR, blocking phosphorylation at the second cluster enhances both. We conclude that separate DNA-PK autophosphorylation events may function reciprocally by not only regulating DNA end processing but also affecting DSB repair pathway choice.
In response to DNA double-strand breaks (DSBs), the DNA-dependent protein kinase (DNA-PK) initiates the process of nonhomologous DNA end joining (NHEJ) by recognizing and then binding to DNA ends (reviewed in references 23 and 25). DNA-PK's precise function during DNA repair and V(D)J recombination has not been completely elucidated. There is considerable evidence that DNA-PK actually plays at least two roles during DNA repair. Because of the immense size of DNA-PK, it is logical that it might serve as a molecular scaffold for the recruitment of additional repair factors. In fact, DNA-PK is known to interact with XRCC4/ligase IV (3, 18, 22) and with the Artemis nuclease to facilitate coding-end hairpin opening during V(D)J recombination (24). Additionally, since it has been shown that the protein kinase activity of DNA-PK is essential to its function in NHEJ (20, 21), it is likely that DNA-PK also alters the activity of itself and/or other DNA repair factors via serine/threonine phosphorylation (reviewed in references 23 and 25). However, although numerous in vitro and in vivo DNA-PK targets have been defined, the only target to date that has been shown unequivocally to be functionally important for NHEJ is DNA-PK catalytic subunit (DNA-PKcs) itself (4, 9, 33).
Early work demonstrated that when bound to DNA ends, purified DNA-PK undergoes autophosphorylation on its three component polypeptides (the Ku70/86 heterodimer and the catalytic subunit, DNA-PKcs) (5). This autophosphorylation results in loss of activity and disassembly of the kinase complex. From these data, it was proposed that kinase disassembly might be just as important for completing DNA repair as DNA-PK is in initiating repair (5, 11, 26).
Recently, significant efforts from several investigators have focused on defining and characterizing autophosphorylation sites within the large catalytic subunit of DNA-PK (4, 9, 12, 33). We demonstrated that autophosphorylation within a major cluster of sites (between residues 2609 and 2647, referred to hereafter as the ABCDE cluster) (Fig. 1) likely mediates a conformational change in the DNA-PK complex that is critical for DNA end processing (2, 9, 28). V(D)J coding joints mediated by DNA-PKcs that cannot autophosphorylate these sites show markedly decreased DNA end processing. This work is consistent with recent biochemical data showing that ABCDE autophosphorylation is required for efficient joining by XRCC4/ligase IV (2, 28, 36). Surprisingly, cells expressing DNA-PKcs that cannot autophosphorylate ABCDE are more radiosensitive than cells that express no DNA-PKcs at all. We demonstrated that these cells are more radiosensitive because of inhibition of homologous recombination (HR) by the mutant DNA-PKcs (8), suggesting that DNA-PKcs that cannot autophosphorylate ABCDE may block DNA end access to a variety of DNA-modifying enzymes in living cells.
FIG. 1.
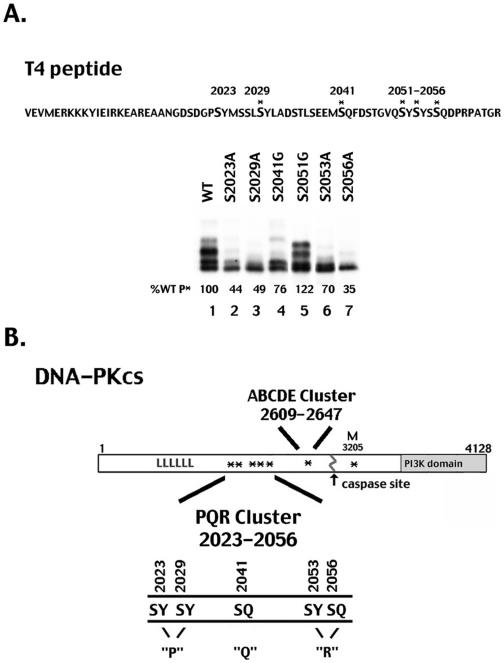
Identification of a second autophosphorylation site cluster in DNA-PKcs. (A) Top panel, sequence of the DNA-PKcs T4 fragment is presented with potential phosphorylation sites (SQ and SY) shown in boldface type. Asterisks indicate serines that are conserved in six or more species. Bottom panel, autoradiogram showing in vitro DNA-PK phosphorylation of equal amounts of His-tagged T4 fragment and mutant T4 fragments as indicated. The percentage of wild-type/T4 peptide phosphorylation (%WT P*) for each mutant is shown. The multiple bands in lanes 1 and 5 represent multiply phosphorylated T4 species consistent with our conclusion that multiple sites within the T4 peptide are targets of DNA-PK's kinase activity. (B) Diagrammatic representation of DNA-PKcs. Autophosphorylation sites are denoted with asterisks. Previously described seven autophosphorylation sites (ABCDE and M) are as follows: T2609, T2620, S2624, T2638, T2647, S2612A, and S3205. Autophosphorylation sites and mutants of those sites described in this report have been denoted as follows: mutant P, S2023 and S2029; mutant Q, S2041; and mutant R, S2056 and S2053.
However, ABCDE autophosphorylation is clearly not sufficient for kinase inactivation and dissociation in vitro (2, 9, 28), suggesting that additional autophosphorylation sites within DNA-PKcs might be functionally important. Here, we define a second major cluster of DNA-PKcs autophosphorylation sites, and we present evidence that these two autophosphorylation site clusters function reciprocally to regulate DNA end processing.
Finally, it is well established that in addition to NHEJ, HR contributes significantly to DNA double-strand break repair. However, it has been a mystery as to how a cell chooses between these two major repair pathways. The data presented here suggest that the autophosphorylation status of DNA-PKcs may affect DSB repair pathway choice.
MATERIALS AND METHODS
Cloning, expression, and mutagenesis of DNA-PKcs T4 fragment.
cDNA from HeLa cells was used as a template to amplify a fragment of DNA-PKcs (T4), which corresponds to amino acids 1994 to 2065 of human DNA-PKcs. The primers used were as follows (written 5′ to 3′): T4(BamH I), 5′CGGGATCCGTTGAGGTTCCTATGGAAAGAAAG; T4(XmaI), 5′CCTAGACCTGCCACTGGTCGTCCCGGGGGGA; and T4(Hind III), 5′ CCTAGACCTGCCACTGGTCGTAAGCTTGGG.
The PCR product was subcloned into a pQE-30 vector (QIAGEN). The recombinant T4 peptide was expressed in Escherichia coli and purified by nickel-nitrilotriacetic acid affinity chromatography. Serine-to-alanine mutations were generated using a QuikChange site-directed mutagenesis kit (Stratagene) with the following oligonucleotides: S2023A, 5′TCAGATGGTCCTGCCTATATGTCTTC; S2029A, 5′GTCTTCCCTGGCATATTTGGCA; S2041G, 5′GAGTGAGGAAATGGGTCAATTTGATTTCTC; S2051G, 5′CCGGAGTTCAGGGCTATTCATAC; S2053A, 5′GTTCAGAGCTATGCATACAGCTCCC; S2056A, 5′CTATTCATACAGCGCCCAAGACCC; S2034A, 5′ATTTGGCAGACGCTACCCTGAGTG; S2037A, 5′ACAGTACCCTGGCTGAGGAAATGAG; and S2046A, 5′AATTTGATTTCGCAACCGGAGTT.
Phosphorylation condition of DNA-PK T4 mutants by purified DNA-PK.
DNA-PKcs (0.1 μg) and Ku (0.06 μg) (purified from HeLa cells as described previously [17]) were incubated in 25 μl of reaction mixture containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM dithiothreitol, 0.2 mM EGTA, 0.1 mM EDTA, 40 μg/ml sonicated calf thymus DNA, and approximately 1 μg of purified His-tagged DNA-PKcs T4 proteins. Reactions were started by the addition of 0.2 mM ATP containing stabilized [γ-32P]ATP (specific activity, approximately 500 cpm per pmol), and mixtures were incubated for 5 min at 30°C. Reactions were terminated by the addition of sodium dodecyl sulfate (to 1%) and dithiothreitol (to 10 mM). Samples were heated to 100°C for 3 min and were then analyzed by autoradiography after sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 15% polyacrylamide gels.
Oligonucleotides.
Oligonucleotides and their complements (not shown) used to introduce substitutions into the complete human DNA-PKcs cDNA are as follows: P>A, 5′GATTCAGATGGTCCGGCCTATATGTCTTCCCTGGCATATTTGGCAG; P>D, 5′GATTCAGATGGTCCGGATTATATGTCTTCCCTGGATTATTTGGCAG; Q>A, 5′GTACCCTGAGTGAAGAAATGGCTCAATTTGATTTC; Q>D, 5′AGTACCCTGAGTGAAGAAATGGATCAATTTGATTTC; R>A, 5′TCAGAGCTATGCATACAGCGCCCAAGACCCTAGGCCTGCCACTG; and R>D, 5′GTTCAGAGCTATGATTACAGCGACCAAGACCCTAGACCGGCCACTGGTCG. RecA protection oligonucleotides are as follows: 5505, 5′CTGGATGCTTTGAGAGAATTCTTCAGCACAATTGTG; and 6542, 5′ACAATGGAGGAGAAGGAATTCACTACATGGTGGTTG. I-SceI site oligonucleotides are as follows: SalI site, 5′TCGACTATATTACCCTGTTATCCCTAGCGTAACT; and BamHI site, 5′GATCAGTTACGCTAGGGATAACAGGGTAATATAG.
Construction and transfection of expression plasmids.
Construction of the wild-type human DNA-PKcs expression vector was described previously (32). To generate the expression plasmids encoding the “R” phosphorylation site mutant, duplex oligonucleotides encoding the mutation were utilized for QuikChange PCR mutagenesis (Stratagene) of a plasmid containing a fragment of the DNA-PKcs cDNA spanning two EheI restriction sites in the complete DNA-PKcs cDNA. To generate plasmids encoding the “P” and “Q” phosphorylation site mutants, duplex oligonucleotides encoding each mutation were utilized for QuikChange PCR mutagenesis reactions of plasmids spanning an EcoRI fragment of the DNA-PKcs cDNA (nucleotides 5505 to 6542). This restriction fragment was subcloned into the plasmid containing the complete cDNA that had been restricted with EcoRI after EcoRI methylation with protection of the two subcloning sites with RecA as described previously (16).
Cell lines, survival assays, V(D)J recombination assays, plasmid-joining assays, and HR assays.
Derivation of V3 DNA-PKcs transfectants, immunoblot screening, and extrachromosomal recombination assays were performed as described previously (9). Clonal survival assays were done as described previously (8, 9). For plasmid end-joining assays, the RSS in pJH290 were replaced with oligonucleotides containing I-SceI sites. Assays were performed by cotransfecting 1 μg substrate plasmid with 6 μg expression plasmid encoding the restriction endonuclease. Isolation of recombinant plasmids was exactly as with V(D)J assays (9). Details of deletion rates and NHEJ independence of this plasmid end-joining assay will be published elsewhere.
V3 cells harboring the HR substrate (cell strain VD7) were described in detail previously (1). Expression constructs encoding wild-type DNA-PKcs, ABCDE>ala, PQR>ala, and ABCDE+PQR>ala were stably integrated into the VD7 cell strain as described previously (9), except that pCDNA6 was cotransfected to provide blasticidin resistance. An assessment of DSB-induced HR was essentially as described previously (1, 8), except that percent HR is calculated as a percentage of transfection efficiency (measured by puromycin resistance) instead of plating efficiency. Thus, the relative HR percentage is increased compared to that reported in our previous paper, although the actual numbers are completely consistent.
DNA-PK microfractionation, immunoblotting, measurement of protein kinase activity, and DSB-induced nuclear mobilization.
DNA-PK activity was assessed from cell extracts as described previously (9). To visualize mobilization of DNA-PKcs in response to DSBs, V3 transfectants were treated for 1 h with bleomycin (140 μM). Membrane-insoluble fractions were prepared as described previously (13). Antibodies used for immunoblotting include anti-DNA-PKcs (monoclonal 42-2, a generous gift of Tim Carter, St. John's University, New York, NY), anti-β-actin (clone AC-15; Sigma), anti-laminB (goat anti-laminB; Santa Cruz), and anti-γH2AX (clone JBW103; Upstate Biotechnology).
γH2AX focus formation.
To assess γH2AX foci, cells were plated on glass coverslips at ∼60% confluence in six-well plates and were untreated or treated with 7 μM bleomycin in serum-free alpha minimum essential medium. After 1 h, the medium was replaced and cells were cultured for the indicated time periods (0 h to 72 h). At the end of each time point, coverslips were washed twice with phosphate-buffered saline (PBS) and fixed with 3.7% paraformaldehyde for 20 min at room temperature (RT). Coverslips were then incubated for 5 min with 0.2% Triton X-100 in PBS to permeabilize cells. After being rinsed in PBS, coverslips were blocked with 10% fetal bovine serum, 0.2% Triton X-100, and 100 μg/ml RNaseA in PBS for 1 h at RT. Cells were then incubated with anti-γ-H2AX antibody (1:8,000 dilution) overnight at 4°C. After extensive washing, Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G (A21121; Molecular Probes) was applied at a 1:1,000 dilution and incubated for 1 h at RT. DNA was counterstained with 1:500 diluted TO-PRO-3 iodide (T3605; Molecular Probes). Coverslips were then mounted onto glass slides in Permafluor mounting medium (434990; Thermo Electron Corporation). The fluorescently labeled cells were examined with a Zeiss LSM Pascal confocal laser scanning microscope.
RESULTS
Identification of a second autophosphorylation site cluster in DNA-PKcs.
Previously, we demonstrated that purified mutant DNA-PKcs containing six autophosphorylation site alanine substitutions was indistinguishable from wild-type DNA-PK in its ability to be stimulated by Ku and to undergo ATP-dependent inactivation in vitro (9). Moreover, this mutant protein still underwent substantial autophosphorylation in vitro (9, 28), and we concluded that DNA-PKcs must contain other autophosphorylation sites besides the ABCDE cluster. Although the ABCDE sites are required for the function of DNA-PKcs in both NHEJ repair and V(D)J recombination, these sites are not responsible for autophosphorylation-induced loss of protein kinase activity.
To identify additional phosphorylation sites, fragments of DNA-PKcs were expressed as His-tagged fusion proteins; several of the soluble polypeptides were purified and assayed for their ability to be phosphorylated in vitro by DNA-PKcs. One of these fragments, identified as T4 and corresponding to amino acids 1994 to 2065, was highly phosphorylated by DNA-PKcs (Fig. 1A, lane 1). A comparison of the amino acid sequences of DNA-PKcs from chicken, frog, dog, horse, human, rat, and mouse indicated that this region contained several highly conserved SQ sites (Fig. 1). This second potential autophosphorylation site cluster is within a region of DNA-PKcs with no previously ascribed function (Fig. 1B).
Like other members of the phosphatidylinositol kinase-related kinase family, DNA-PK phosphorylates many substrates on SQ or TQ motifs but can also phosphorylate some substrates in vitro, such as Ku70, Ku80 (6), and XRCC4 (37), on serines or threonines that are followed by hydrophobic residues. We therefore mutated individual serine residues to alanine (A) or glycine (G) within the T4 fragment and examined the ability of DNA-PKcs to phosphorylate the mutant polypeptides in vitro. As can be seen, mutation of serines 2023, 2029, 2041, 2053, and 2056 to a nonphosphorylatable amino acid significantly reduced the ability of DNA-PK to phosphorylate the T4 fragment, whereas mutation of the other serines did not (Fig. 1A and data not shown). We therefore considered these five serines to be potential DNA-PK phosphorylation sites.
S2056 was recently independently identified by other investigators as a DNA-PK autophosphorylation site; moreover, it was shown to be autophosphorylated in response to ionizing radiation in living cells (7, 35). Thus, just as the ABCDE cluster has been shown to be an authentic in vivo target of DNA-PK's protein kinase activity (4, 7, 12), so is at least one site within the PQR cluster.
To assess the functional relevance of autophosphorylation of this second cluster, several point mutations of the DNA-PKcs cDNA were generated. Initially, three mutant constructs were generated, designated P>ala, QR>ala, and PQR>ala, with two, three, and five mutations, respectively (Fig. 1B).
V3 transfectants expressing DNA-PKcs with multiple alanine substitutions within the PQR cluster are modestly radiosensitive.
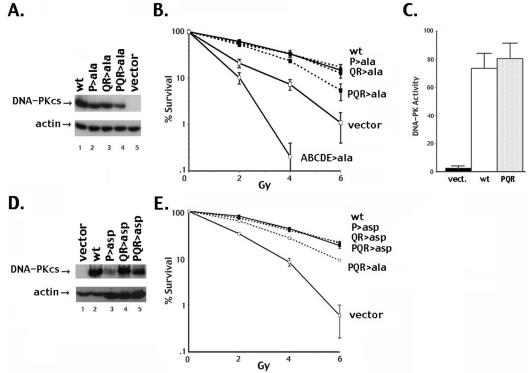
Expression vectors encoding wild-type or alanine-substituted DNA-PKcs were transfected into V3 cells, and stable clones were isolated. The V3 cell line is DNA-PKcs deficient and thus defective in DNA DSB repair. Cell irradiation assays for this panel of transfectants showed that both the P and the QR phosphorylation site mutants reversed the radiosensitivity of V3 cells in a manner similar to that seen with wild-type DNA-PKcs. However, mutant PQR>ala (with five alanine substitutions) leaves the V3 cells partially radiosensitive (Fig. 2B). In this experiment (and all subsequent experiments), although data are presented only for one clone, at least two unique clones with similar levels of DNA-PKcs expression (Fig. 2A) were studied for each mutant, with similar results. These results are similar to our previous study of the ABCDE cluster (9). In that case, five of six phosphorylation sites had to be substituted to observe a dramatic functional difference between the mutant and wild-type proteins, and we concluded that phosphorylation of one or two of the sites within the cluster could suffice functionally. Similarly, with the PQR cluster, although autophosphorylation at any one particular PQR site is not required for DNA-PK's function, autophosphorylation of at least one (or possibly two) of the five sites is needed. It should be noted that cells expressing the PQR>ala mutant are still substantially more radioresistant than cells expressing no DNA-PKcs at all. This is in contrast to cells expressing the ABCDE>ala mutant, which are significantly more radiosensitive than cells expressing no DNA-PKcs at all (Fig. 2B) because of inhibition of HR by the ABCDE>ala mutant (8).
FIG. 2.
V3 transfectants expressing DNA-PKcs with multiple alanine but not aspartic acid substitutions within the PQR cluster are modestly radiosensitive. (A) Immunoblot analyses of whole-cell extracts (WCE) (50 μg) from V3 cell transfectants as indicated. (B) Radioresistance of V3 transfectants expressing wild-type DNA-PKcs, vector alone, or mutants P>ala, QR>ala, PQR>ala, or ABCDE>ala was assessed. In this figure and subsequent figures, data are presented as percent survival of unirradiated controls. Error bars indicate standard errors of the means for three independent experiments. (C) WCE prepared from V3 transfectants as indicated were assayed for DNA-PK activity. Each cell extract was tested in duplicate, and three independent extracts were tested. Kinase activity is expressed as [γ-32P]ATP incorporation into the p53 target peptide divided by [γ-32P]ATP incorporation in reactions without peptide, as described in reference 9. (D) Immunoblot analyses of WCE (20 μg) from V3 cells transfected with vector alone or wild-type DNA-PKcs expression vector or (200 μg) WCE from transfectants expressing mutants PQR>asp, P>asp, and QR>asp, as indicated. (E) Radioresistance of V3 transfectants expressing wild-type DNA-PKcs, vector alone, or mutant P>asp, QR>asp, PQR>asp, or PQR>ala was assessed. wt, wild type; vect., vector.
We asked whether these results might be explained by altered enzymatic activity of the PQR>ala mutant. If the PQR sites are involved in autophosphorylation-dependent inactivation of DNA-PK, the protein kinase activity of the alanine mutant might be substantially higher than that of wild-type DNA-PKcs. However, the protein kinase activity of the PQR>ala mutant was indistinguishable from that of the wild-type kinase in standard DNA cellulose “pulldown” experiments (Fig. 2C).
DNA-PKcs mutants with aspartic acid substitutions of the PQR sites completely reverse the NHEJ deficits of V3 cells.
In an attempt to mimic constitutively phosphorylated PQR sites, expression vectors with aspartic acid substitutions for the same combinations of the five autophosphorylation sites as described above for the alanine substitutions were constructed (Fig. 1B). We considered three possibilities. If autophosphorylation of any of the sites within the PQR cluster induces kinase inactivation, then the aspartic acid-substituted PQR DNA-PKcs should lack protein kinase activity and the mutant protein should not complement the NHEJ deficit of the V3 cell line. In this scenario, cells expressing the aspartic acid mutants would be predicted to be more radiosensitive than cells expressing the alanine mutant. Another possibility is that the aspartic acid substitutions would not adequately mimic phosphorylation. In this case, the aspartic acid-substituted mutants might behave like the alanine mutants. Finally, autophosphorylation within the PQR cluster might be important for another reason, perhaps by inducing some conformational change in the repair complex that facilitates subsequent steps in the repair process. In this case, cells expressing the aspartic acid mutant protein would likely display less severe deficits than cells expressing the alanine mutant. Aspartic acid-substituted constructs were transfected into V3 cells, and stable clones were isolated. Though numerous transfections were performed for each of the aspartic acid-substituted constructs, DNA-PKcs expression levels in the resulting transfectants were uniformly lower than in wild-type or alanine mutant-expressing clones (Fig. 2D, note that more extract from the mutants than from the wild-type clones [200 μg versus 20 μg] was loaded to adequately display DNA-PKcs expression). The reason for the low expression with these constructs is under investigation.
Cell irradiation assays for this panel of DNA-PKcs mutants were performed. To more closely match expression levels, we used a wild-type clone expressing a lower level of DNA-PKcs. Still, radioresistance of this wild-type clone was analogous to that of the clone utilized for Fig. 2B. All of the PQR>asp mutants complemented the radiosensitivity of V3 cells similarly to wild-type DNA-PKcs even though the DNA-PKcs expression levels were considerably lower and regardless of whether two, three or five sites were substituted (Fig. 2E). From these data, we surmise that the aspartic acid substitutions are reasonable mimics for phosphorylation. Since the PQR>asp mutant completely reverses the NHEJ deficits of V3 cells whereas alanine mutants do not, it seems clear that (i) the aspartate mutant is an active protein kinase, (ii) autophosphorylation of this cluster of sites does not induce kinase inactivation, and (iii) the aspartic acid mutant can correctly progress through the various steps of NHEJ.
PQR>ala supports only reduced levels of coding-end joining.
Because the ABCDE>ala mutant had substantially decreased coding-end joining in our previous report, we similarly tested the PQR mutants. In V(D)J recombination assays of the clonal V3 transfectants, wild-type DNA-PKcs reverses V3's coding-end joining deficit as expected (Table 1). In contrast, V3 transfectants expressing the PQR>ala mutant have modestly reduced coding joint levels (fivefold). For comparison, we also performed V(D)J assays on an ABCDE>ala-expressing clone. Just as in our previous report, a substantial deficit in coding-end joining was observed. V3 cells expressing the PQR>asp mutant join coding ends at a level similar to that of cells expressing wild-type DNA-PKcs.
TABLE 1.
PQR>ala but not PQR>asp mutants reduce the levels of V(D)J recombinationa
| V3 clonal transfectant | PJH290 (coding) Camr/Ampr | % Rb | PJH201 (signal) Camr/Ampr | % R | % Signal fidelityc |
|---|---|---|---|---|---|
| Vector only | |||||
| −RAGd | 0/29,800 | 0 | 0/300 | 0 | |
| 0/8,100 | 0 | 0/9,300 | 0 | ||
| 5/47,900 | 0.01 | 0/15,100 | 0 | ||
| 11/56,700 | 0.02 | 0/5,300 | 0 | ||
| +RAG | 0/9,500 | 0 | 3/12,600 | 0.02 | 79 (23/29) |
| 0/27,500 | 0 | 4/7,900 | 0.05 | ||
| 0/29,100 | 0 | 0/7,000 | 0 | ||
| 1/75,600 | 0.001 | 19/42,300 | 0.045 | ||
| Wild type +RAG | 42/5,000 | 0.84 | 93/4,600 | 2.02 | 87 (26/30) |
| 24/1,800 | 1.33 | 65/2,600 | 2.5 | ||
| 15/1,400 | 1.07 | 27/9,300 | 0.29 | ||
| 37/8,300 | 0.45 | 50/15,000 | 0.33 | ||
| PQR>ala +RAG | 5/16,700 | 0.03 | 65/22,100 | 0.29 | 37 (11/30) |
| 30/10,100 | 0.3 | 146/29,600 | 0.49 | ||
| 11/3,600 | 0.31 | 111/26,700 | 0.42 | ||
| 7/14,300 | 0.05 | 64/23,600 | 0.27 | ||
| PQR>asp +RAG | 210/18,800 | 1.12 | 179/27,900 | 0.64 | 97 (29/30) |
| 47/27,100 | 0.17 | 465/34,500 | 1.35 | ||
| 125/20,800 | 0.6 | 531/57,100 | 0.93 | ||
| ABCDE>ala +RAG | 1/29,000 | 0.003 | 0/8,800 | 0 | 87 (26/30) |
| 4/46,200 | 0.009 | 20/19,800 | 0.1 | ||
| 5/7,500 | 0.07 | 147/71,200 | 0.21 | ||
| ABCDE+PQR>ala +RAG | 5/120,500 | 0.004 | 2/4,300 | 0.047 | 77 (13/17) |
| 1/27,400 | 0.004 | 2/3,000 | 0.067 | ||
| 2/14,300 | 0.014 | 2/4,100 | 0.049 |
RAG expression from plasmid vectors initiates recombination in the V3 cells, as assessed by the plasmid substrate pJH290 that detects coding joints or the pJH201 substrate that detects signal joints, as indicated. Numbers of ampicillin-resistant (Ampr) and chloramphenicol- and ampicillin-resistant (Camr/Ampr) colonies from at least three separate experiments are presented.
%R, recombination rate, calculated as the number of Camr colonies divided by the number of Ampr colonies × 100.
Signal fidelity is equal to the percentage of ApaLI-sensitive signal joints.
Control transfections without RAG expression plasmids (−RAG) were performed for each clonal transfectant with results similar to those presented for the vector only clone. For the sake of brevity, only transfections including RAG plasmids (+RAG) are presented.
Signal end joining is variably depressed in different DNA-PKcs-deficient cell lines and in different animal models of DNA-PKcs deficiency. V3 cells display a fairly significant defect in signal end joining, about 40-fold lower in control V3 transfectants than in cells expressing wild-type DNA-PKcs (Table 1). Although the PQR>ala mutant slightly decreased the level of signal end joining compared to wild-type DNA-PKcs in most experiments, the differences were not significant.
Coding joints mediated by PQR>ala have excessive nucleotide loss from joined coding and signal ends, whereas joints mediated by PQR>asp have minimal nucleotide loss.
In our previous study, V(D)J assays were done by transiently expressing the RAG proteins as well as DNA-PKcs in the parental V3 cells (9). Here, we chose to perform V(D)J assays with clonal transfectants. We compared the characteristics of coding joints isolated by both techniques and found them to be similar (and, of course, dependent on DNA-PKcs expression). Thus, for a more thorough comparison of coding joints mediated by the autophosphorylation mutants, we have included sequences from our previous report in Table 2 and in Fig. S1 in the supplemental material.
TABLE 2.
Coding joints and rejoined I-SceI breaks mediated by the PQR>ala mutant have excessive nucleotide loss, whereas those mediated by the PQR>asp mutant have minimal nucleotide lossa
| Transfectant | V(D)J coding joint result
|
I-SceI joint result
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of sequences | bp loss/joint | % Complete ends | % Sequences with SSH | % P segments/complete end | % N+ joints | N segmented length | No. of sequences | bp loss/joint | % Complete ends | |
| Wild type | 74 | 4.8 | 31 (46/148) | 50 (37/74) | 24 (11/46) | 63 (24/38) | 2.3 | 30 | 11.7 | 10 (6/60) |
| Wild type + Tdt | 38 | 5.6 | 28 (21/76) | 36 (5/14) | 38 (8/21) | |||||
| PQR>ala | 67 | 14.1 | 9 (12/134) | 66 (44/67) | 8 (1/12) | 48 (13/27) | 1.8 | 33 | 31.4 | 1.5 (1/66) |
| PQR>ala + Tdt | 27 | 10.5 | 6 (3/54) | 71 (10/14) | 0 (0/3) | |||||
| PQR>asp | 40 | 2.2 | 56 (45/80) | 20 (6/30) | 16 (7/45) | |||||
| ABCDE>ala | 29 | 1.4 | 71 (41/58) | 0 (0/29) | 32 (13/41) | 38 (6/16) | 1.2 | 33 | 6 | 19.7 (13/66) |
| ABCDE>ala +Tdt | 16 | 2.6 | 66 (21/32) | 20 (2/10) | 62 (13/21) | |||||
| ABCDE>asp | 25 | 3.1 | 38 (19/50) | 52 (13/25) | 26 (5/19) | |||||
| ABCDE+PQR>ala | 39 | 6.7 | 46 (36/78) | 28 (11/39) | 36 (13/36) | 33 | 25.4 | 6.1 (4/66) | ||
| RAGS/I-SceI only | 16 | 14.7 | 41 (13/32) | 31 (5/16) | 85 (11/13) | 33 | 19.8 | 6.1 (4/66) | ||
Some of the coding joint sequences presented (61/74 of the wild type without Tdt, 28/29 of the ABCDE>ala without Tdt, and 16/16 of the vector) are from reference 9. Average nucleotide loss/joint, % intact coding ends, % sequences with SSH (calculated only from joints lacking N segments), % P segments/complete end, % N segment containing (% N+) joints, and N segmented length were calculated from sequences presented in Fig. S1 in the supplemental material. Although only mean nucleotide loss is presented, both the median and the average were calculated and did not differ substantially from one another. The one ABCDE+PQR>ala sequence with a 200-bp deletion was not included in calculating the mean nucleotide loss.
Coding joints isolated from cells expressing wild-type DNA-PKcs display diverse joined termini. Nucleotide loss from coding ends ranges from 0 to 14 per end, with an average nucleotide loss from each joint of 4.8 (summarized in Table 2). In contrast, coding joints recovered from cells expressing the PQR>ala mutant had lost significantly more nucleotides. In these joints, nucleotide loss from coding ends ranged from 0 to 23 per end, with an average of 14.1 nucleotides lost from each joint. Although at first glance, these joints might seem similar to joints isolated from cells lacking DNA-PKcs (where the average nucleotide loss is 14.7), they are not. First, the rate of recombination in cells expressing the PQR>ala mutant is three logs higher than the rate of recombination in V3 cells expressing no DNA-PKcs (Table 1). As with classic SCID joints, the rare joints isolated from the V3 vector control cells have a wide range of deletions, from 0 to 70 bp for any single end. However, complete ends are relatively common in joints recovered from vector-alone cells (13 of 32 ends). Furthermore, when a complete end is present, a P segment (often abnormally long) is almost always apparent (11 out of 13 joints). In contrast, joints mediated by the PQR>ala mutant rarely maintain complete ends (only 12 of 134 ends were complete), and only one P segment was observed. Thus, classic SCID joints either maintain a complete end with a P segment present or have large deletions. Joints mediated by PQR>ala uniformly display increased nucleotide loss, but with no drastic deletions, few complete unmodified ends, and a high incidence of short sequence homology (SSH) at the junctures. To our knowledge, this is the first description of an NHEJ defect that affects end processing without dramatically affecting joining rates.
Surprisingly, the characteristics of coding joints mediated by PQR>asp are exactly opposite of those of the joints isolated from cells expressing the PQR>ala mutant. In these joints, nucleotide loss from any single end ranged from 0 to only 7, with an average nucleotide loss from each joint of just 2.2. Fifty-six percent of the ends analyzed in these joints were complete (45/80); of the complete ends, 18% (8/45) contained a P segment. SSH at the junctions is absent. Joints mediated by the PQR>asp mutant are remarkably similar to joints mediated by the ABCDE>ala mutant (Table 2; see also Fig. S1 in the supplemental material). Joints mediated by the ABCDE>ala mutant also display minimal end processing, losing up to just 7 nucleotides from any end, with an average nucleotide loss from each joint of only 1.4 bp. The ABCDE>asp mutant that poorly mimics phosphorylation of ABCDE (9) partially reverses this pattern.
The fidelity of signal end joining was also affected by PQR mutations. In contrast to the diversity of coding joints, joining of signal ends is usually precise, directly ligating heptamer to heptamer, generating an ApaLI site. In V3 transfectants expressing wild-type DNA-PKcs, joining was precise in 87% of signal joints (Table 1). In cells with no DNA-PKcs, 79% of the signal joints were precise. In contrast, signal joints isolated from cells expressing the PQR>ala mutant were much more abundant than those from the vector-alone control cells but notably less precise (37%). The loss of fidelity is completely reversed in joints recovered from cells expressing PQR>asp, where 97% of recovered signal joints are precise.
It seemed possible that terminal deoxynucleotidyl transferase (Tdt) might affect coding joint structure differently in the presence of DNA-PKcs autophosphorylation mutants. However, recombination rates were not altered by the presence or absence of Tdt (data not shown), and the number of nucleotides lost in joints mediated by the wild-type DNA-PKcs or the ABCDE>ala and PQR>ala mutants was largely unchanged by Tdt expression (Table 2; see also Fig. S1 in the supplemental material). Some effects on N segment addition were seen. The ABCDE>ala mutant modestly inhibits N segment addition by Tdt; a smaller fraction of the joints have N segments, and N segments present are shorter. This effect of the ABCDE>ala mutant is less dramatic than its impact on nucleotide loss. No significant change in N nucleotide addition was seen with joints mediated by the PQR>ala mutant compared to wild-type DNA-PKcs.
To determine whether these autophosphorylation events affect end processing during NHEJ not associated with V(D)J recombination, a plasmid end-joining assay was performed. Briefly, the RSS in the V(D)J substrate plasmids were replaced with two restriction sites for the endonuclease I-SceI. This plasmid was cotransfected with an expression construct for the I-SceI endonuclease, and plasmids that delete the small oop sequence between the restriction sites were recovered by chloramphenicol selection. Similarly to other plasmid end-joining assays of mammalian cells, plasmid rejoining does not depend on NHEJ, but end processing is highly dependent on intact NHEJ (19, 36). Just as with V(D)J joining, the PQR>ala mutant promotes excessive nucleotide loss when joining I-SceI breaks (31.4 nucleotides lost per end compared to 11.7 with wild-type DNA-PKcs), whereas the ABCDE>ala mutant reduces end processing (6 nucleotides lost per end compared to 11.7 with wild-type DNA-PKcs) (Table 2). We conclude that autophosphorylation within the ABCDE and PQR clusters functions reciprocally to regulate DNA end processing during both V(D)J recombination and non-V(D)J-associated DSB.
PQR phosphorylation exacerbates radiosensitivity of cells expressing the ABCDE>ala mutant.
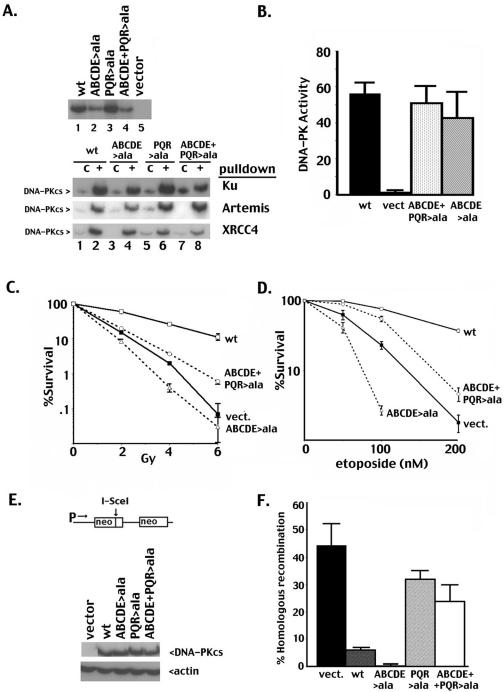
Since autophosphorylation of these two clusters has opposing effects on DNA end processing (and thus probably end access), we predicted that blocking phosphorylation at both clusters would partially relieve the severe phenotype observed with the ABCDE>ala mutant. To test this model, an expression plasmid that encodes alanine substitutions at both clusters (ABCDE+PQR>ala) was constructed; the paired mutant is stably expressed in V3 cells. “Pull-down” experiments demonstrate that the interactions of all of these mutant proteins with His-tagged XRCC4, Ku, and Artemis are similar to those of wild-type DNA-PKcs (Fig. 3A, bottom panel), and robust DNA-PK activity is present in cell extracts (Fig. 3B). We conclude that the alanine substitutions at ABCDE and PQR do not significantly disrupt the basic structure or the enzymatic activity of the kinase. Consistent with our hypothesis (i.e., that phosphorylation at the PQR sites exacerbates the end-blocking effect of the ABCDE>ala mutant), cells expressing the combined mutant are significantly more radioresistant than cells expressing the ABCDE>ala mutant (Fig. 3D). However, in cells expressing the combined mutant, both coding and signal end joining are just as severely impaired as in cells expressing the ABCDE>ala mutant (Table 1). Sequencing of the rare coding joints mediated by the combined mutant demonstrates an intermediate level of nucleotide loss, i.e., a level between those of the ABCDE>ala and PQR>ala mutants (Table 2 and Fig. S1 in the supplemental material). The severe V(D)J recombination defects in cells expressing the combined mutant suggest that the combined mutant is just as defective in NHEJ as the ABCDE>ala mutant. We considered that blocking PQR phosphorylation in the ABCDE>ala mutant reverses primarily the inhibition of HR, explaining the increased radioresistance. Consistent with this interpretation, cells expressing the combined mutant are substantially more resistant to etoposide (etoposide damage is repaired by either HR or NHEJ) than are cells expressing the ABCDE>ala mutant (Fig. 3D). In summary, these data suggest that blocking phosphorylation of PQR allows HR factors access to etoposide-induced damage.
FIG. 3.
Blocking PQR phosphorylation promotes HR. (A) Top panel, immunoblot analyses of WCE (50 μg) from V3 transfectants as indicated. Bottom panel, Western blots of Ni+ agarose fractions of control Sf9 extracts (“c,” lanes 1, 3, 5, and 7) or Sf9 extracts expressing Ku, Artemis, or XRCC4 as indicated (lanes 2, 4, 6, and 8) coincubated with V3 extracts from cells expressing wild-type DNA-PKcs (lanes 1 and 2), mutant ABCDE>ala (lanes 3 and 4), mutant PQR>ala (lanes 5 and 6), or mutant PQR+ABCDE>ala (lanes 7 and 8). (B) WCE prepared from V3 transfectants as indicated were assayed for DNA-PK activity as described in the legend to Fig. 2. (C) Radioresistance of V3 transfectants expressing wild-type DNA-PKcs, vector alone, mutants ABCDE>ala, and ABCDE+PQR>ala was assessed. (D) Etoposide resistance of V3 transfectants expressing wild-type DNA-PKcs, vector alone, mutants ABCDE>ala, and ABCDE+PQR>ala was assessed. (E) Top panel, HR substrate stably integrated (one copy) into the V3 cell line (1). Bottom panel, immunoblot analyses of WCE (50 μg) from V3 transfectants harboring the HR substrate as indicated. (F) DSB-induced HR (calculated as G418-resistant colonies/puromycin-resistant colonies) is depicted for one of four independent experiments. Error bars represent standard deviations for three replicas. Only one clone for each mutant is depicted, but two independent clones for each mutant were tested with similar results. wt, wild type; vect., vector.
Blocking PQR phosphorylation promotes HR.
To directly test the effect of PQR phosphorylation on HR, another series of transfectants was generated in a V3 cell strain that harbors one copy of an integrated HR substrate (Fig. 3E). This substrate contains two nonfunctional neomycin resistance genes (1). The first is nonfunctional because of a frameshift mutation that is coincident with the restriction site for the homing endonuclease, I-SceI. The second is nonfunctional because it lacks a promoter. A DSB can be introduced in the first neo gene by transient expression of I-SceI. It has been shown previously that HR repairs I-SceI-induced DSBs of this substrate. HR was induced in each of the transfectants by transient expression of I-SceI, and the relative HR for each clonal transfectant was determined. Consistent with our previous report, repair via HR is more robust in cells completely deficient in DNA-PKcs (vector controls) than in cells expressing DNA-PKcs, since HR is the only major DSBR pathway intact in DNA-PKcs-deficient cells (1, 8). The ABCDE>ala mutant substantially inhibits HR compared to wild-type DNA-PKcs (Fig. 3F and reference 8). Our model predicts that the PQR>ala mutant will not block HR. To our surprise, blocking PQR phosphorylation significantly potentiates HR (Fig. 3F). These data suggest that the phosphorylation status of DNA-PKcs not only regulates DNA end processing during NHEJ but may also dictate repair pathway choice.
Blocking ABCDE phosphorylation alters the kinetics of γH2AX focus resolution.
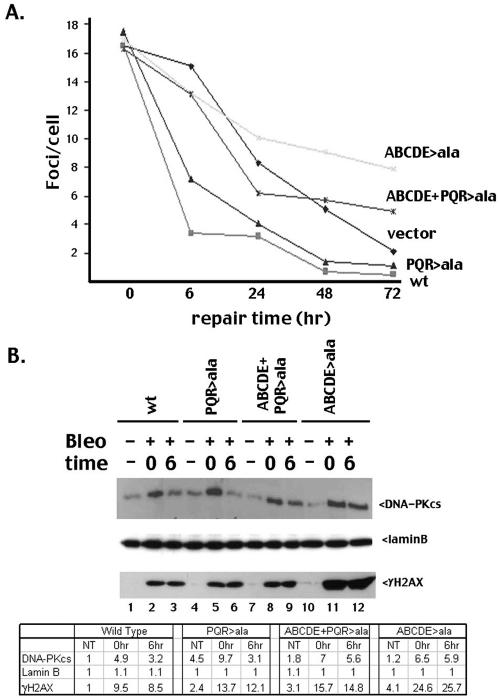
A cell's initial response to a DSB (whether repaired by NHEJ or HR) is to alter the local chromatin structure around the break; this alteration results in well-characterized foci that contain high concentrations of repair factors (for example, RAD51, RAD52, ATM, the MRN complex, the FANC complex, autophosphorylated DNA-PKcs (4, 7; reviewed in reference 15). Modification of a specialized histone (H2AX) by phosphorylation (γH2AX) appears to be the best marker of DSB-induced foci (15). Furthermore, recent reports indicate that a single γH2AX focus forms at each individual DSB (31). We considered that the autophosphorylation mutants might alter the induction or resolution of γH2AX foci; thus, V3 transfectants expressing wild-type, mutant, or no DNA-PKcs were treated with bleomycin to induce DSBs, stained with antibodies specific for γH2AX, and visualized by microscopy, and the average number of foci per cell for each cell strain was determined at various times after treatment. γH2AX foci appear within minutes after irradiation; the foci remain for several hours but are largely resolved in cells expressing wild-type DNA-PKcs by 6 h (Fig. 4A). Consistent with previous reports (29), resolution of γH2AX foci is significantly delayed in cells lacking DNA-PKcs. However, resolution of γH2AX foci in cells expressing the ABCDE mutant is even slower than in cells expressing no DNA-PKcs at all, consistent with our hypothesis that blocking ABCDE phosphorylation limits DNA end access.
FIG. 4.
Mutant ABCDE fails to exit the DSB-associated nuclear compartment with normal kinetics. (A) V3 transfectants were treated (or not) with 7 μM bleomycin for 1 h on coverslips and were stained at the indicated time points as described in Materials and Methods. Average numbers of foci/cell are presented for each time point for the indicated cell strains. Only one clone for each mutant is depicted, but two independent clones for each mutant were tested with similar results. (B) V3 transfectants were treated (or not) with 140 μM bleomycin for 1 h. Cells were harvested either immediately or after 6 h of culture in fresh medium. A triton-insoluble nuclear fraction was isolated as described in reference 13 and analyzed by immunoblotting as indicated. Densitometric quantification of the immunoblot is presented in the bottom panel. This experiment is representative of three independent assays. wt, wild type.
Recently, it has been shown that γH2AX and associated factors can be biochemically fractionated into a detergent-insoluble fraction, providing further support for the idea that γH2AX foci mark a specialized nuclear subcompartment that is the site of DSB repair (13). Thus, we treated cells expressing wild-type and mutant DNA-PKcs with bleomycin and isolated the detergent-insoluble nuclear fraction (termed P3). This fraction also contains laminB, a protein associated with the nuclear matrix. As can be seen, whereas the three mutants (PQR>ala, ABCDE>ala, and the combined mutant) are present at similar levels in this fraction immediately after DSB induction, only the ABCDE>ala mutant (that inhibits HR) remains in this fraction at substantial levels 6 h after DNA damage (Fig. 4B). Similarly, the γH2AX response is dramatically accentuated in the P3 fraction isolated from this mutant but not the others. Thus, we conclude that the ABCDE mutant does not exit the specialized DSB repair compartment with normal kinetics. These data are consistent with the hypothesis that the ABCDE>ala mutant blocks access of DNA ends to various DNA repair factors.
DISCUSSION
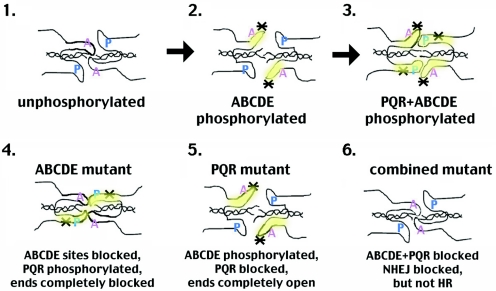
DNA-PK has many targets; to date, the sites studied here and the previously identified ABCDE sites are the only targets shown unequivocally to be functionally relevant for NHEJ. As with the ABCDE mutant, mutations in the PQR sites do not affect the in vitro protein kinase activity of DNA-PK; thus, the observed NHEJ deficits are likely the consequence of DNA-PK's inability to autophosphorylate and alter the function of this region of the large catalytic subunit. Experiments presented here and previously (2, 9, 28) provide strong evidence that the PQR and ABCDE clusters function reciprocally to regulate end access. In fact, data from three independent biologic assays [HR, V(D)J joining, and I-SceI joining] support this conclusion. We propose the following model (Fig. 5). Autophosphorylation of DNA-PKcs results in a series of conformational changes. After initial binding, unphosphorylated (at ABCDE) DNA-PKcs protects DNA ends (panel 1). Although the ABCDE region is depicted as contacting the DNA ends, we are suggesting only that the conformation of unphosphorylated DNA-PKcs (not necessarily the ABCDE region) physically protects DNA ends. In this conformation, the DNA termini are not able to dissociate to facilitate alignment of short sequence homologies, and the XRCC4/ligase IV complex cannot access the DNA termini, consistent with our previous biochemical analyses of the ABCDE mutant (2, 9, 28). Phosphorylation within ABCDE results in a more open conformation, leaving DNA ends accessible to DNA modification and open for alignment of DNA ends (panel 2). Subsequent phosphorylation within PQR induces a conformational change that blocks access of DNA ends to end processing but still allows (or promotes) interaction with XRCC4/ligase IV (panel 3). With alanine substitutions at the ABCDE sites, the initial “opening” conformational change would not occur, leaving the DNA ends protected from nibbling. Moreover, since this mutant maintains kinase activity, the PQR sites would be phosphorylated and the PQR closed conformation would occur, exacerbating the lack of DNA end processing (panel 4). With the PQR>ala mutant, the ABCDE sites would still be phosphorylated (panel 5) and thus “opened,” but the second “closing” conformational change would not occur, accentuating nucleotide loss as evidenced by the sequence analysis of coding joints mediated by the PQR mutant. This model would predict (and is supported by the fact) that blocking phosphorylation at both clusters should partially relieve the end-blocking effect of the ABCDE>ala mutant. In fact, blocking phosphorylation at PQR in the ABCDE mutant substantially enhances HR, suggesting that phosphorylation of PQR is critical to DNA-PK's ability to inhibit HR.
FIG. 5.
Proposed model of reciprocal regulation of end access by DNA-PK's autophosphorylation status. Panels 1 to 6 represent distinct conformations induced by autophosphorylation. Certainly, we have no information that these particular sites are physically associated with DNA ends; we are proposing that a conformational change induces the described structures that regulate end processing. Asterisks denote phosphorylated sites (either ABCDE or PQR). The hypothesized model of a continuous conformational change from nonphosphorylation to complete autophosphorylation is presented in panels 1 to 3. The bottom panels represent the proposed configuration of the three alanine mutants: panel 4, ABCDE>A; panel 5, PQR>A; and panel 6, ABCDE+PQR>A.
The question remains: why have two reciprocally acting autophosphorylation site clusters evolved? Reciprocally regulated access of DNA ends would be a beneficial mechanism if downstream repair factors were limiting compared to DNA-PK. In fact, this is probably the case. The three component polypeptides of DNA-PK are abundant proteins, whereas XRCC4/ligase IV are expressed at more limiting amounts. If autophosphorylation of DNA-PKcs allowed progressive end modification, excessive nucleolytic processing might occur until XRCC4/ligase IV were correctly targeted to the repair complex. However, autophosphorylation of PQR (presumably after ABCDE phosphorylation) would prevent excessive nucleotide loss prior to XRCC4/ligase IV-mediated ligation. This would provide a beneficial mechanism to prevent excessive DNA end degradation; it would also provide a mechanism to exclude HR.
These data also provide evidence that the autophosphorylation state of DNA-PKcs may affect whether a cell utilizes HR versus NHEJ to repair DSBs. Whereas blocking phosphorylation at ABCDE inhibits HR, blocking phosphorylation at PQR significantly enhances HR. It follows that phosphorylation at ABCDE will promote HR, but phosphorylation at PQR will block HR. This model would predict that DNA-PKcs-deficient cells would have an increase in HR; in fact, two different studies have documented increased HR in cells deficient in DNA-PK (1, 27). Consistent with this model, it has recently been demonstrated that autophosphorylation of DNA-PKcs is cell cycle regulated, occurring primarily in G0/G1 (7) when NHEJ predominates and HR is limited (30). PQR phosphorylation in G0/G1 would provide a mechanism to limit HR in the absence of a sister chromatid. Our data suggest that phosphorylation of these sites blocks HR (HR would, of course, be detrimental in G0/G1). Together, these data suggest that DNA-PK's autophosphorylation status blocks HR in G0/G1 but allows either HR or NHEJ in later stages of the cell cycle when both pathways have been shown to be active (30). In contrast, the nonphosphorylated status of PQR in S and G2 should promote HR. Thus, in the presence of a sister chromatid, DNA-PKcs may equally potentiate either HR or NHEJ. Though our data suggest DNA-PK as a potential mediator of repair pathway choice, it is not necessarily the only mediator. During preparation of the manuscript, Esashi et al. described cell cycle-regulated phosphorylation of BRCA2 that disrupts its interaction with RAD51; the consequence of this posttranslational modification is to limit RAD51's function (and thus HR) to S and G2 (14).
It has been shown previously that autophosphorylation of DNA-PK is affected by the type of DNA to which it is bound (34). It will be of interest to determine the extent of ABCDE and PQR autophosphorylation when DNA-PK is activated by lesions that are normally repaired via HR (for example, interstrand cross-links) or by binding to Holliday junctions (10) versus the extent of ABCDE and PQR autophosphorylation when DNA-PK is activated by simple double-stranded ends.
Finally, several lines of experimentation strongly suggest that kinase inactivation and dissociation are not mediated by autophosphorylation within either of the two major clusters. Studies to define the autophosphorylation events responsible for kinase inactivation are ongoing.
Supplementary Material
Acknowledgments
We thank Martin Gellert for many years of scientific encouragement, for perceptive suggestions, and for his meticulous review of the manuscript. Similarly, we thank Dale Ramsden for insightful discussions, sharing of ideas, and critical readings of the manuscript. We thank Nick Morrice (University of Dundee) for valiant attempts to identify additional phosphorylation sites in DNA-PKcs.
This work was supported by Public Health Service grant AI048758 (K.M.). Work in S.P.L.-M.'s laboratory is supported by grant 13639 from the Canadian Institutes of Health Research. S.P.L.-M. is supported by the Alberta Heritage Foundation for Medical Research and the Canadian Institutes of Health Research.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allen, C., J. Halbrook, and J. A. Nickoloff. 2003. Interactive competition between homologous recombination and non-homologous end joining. Mol. Cancer Res. 1:913-920. [PubMed] [Google Scholar]
- 2.Block, W., Y. Yu, D. Merkle, J. Gifford, Q. Ding, K. Meek, and S. P. Lees-Miller. 2004. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) regulates ligation of DNA ends. Nucleic Acids Res. 32:4351-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calsou, P., C. Delteil, P. Frit, J. Drouet, and B. Salles. 2003. Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment. J. Mol. Biol. 326:93-103. [DOI] [PubMed] [Google Scholar]
- 4.Chan., D. W., B. P. Chen, S. Prithivirajsingh, A. Kurimasa, M. D. Story, J. Qin, and D. J. Chen. 2002. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 16:2333-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, D. W., and S. P. Lees-Miller. 1996. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem. 271:8936-8941. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. W., R. Ye, C. J. Veillette, and S. P. Lees-Miller. 1999. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry 38:1819-1828. [DOI] [PubMed] [Google Scholar]
- 7.Chen, B. P., D. W. Chan, J. Kobayashi, S. Burma, A. Asaithamby, K. Morotomi-Yano, E. Botvinick, J. Qin, and D. J. Chen. 2005. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 280:14709-14715. [DOI] [PubMed] [Google Scholar]
- 8.Convery, E., E. K. Shin, Q. Ding, W. Wang, P. Douglas, J. A. Nickoloff, S. P. Lees-Miller, and K. Meek. 2005. Inhibition of homologous recombination by variants of DNA-PKcs. Proc. Natl. Acad. Sci. USA 102:1345-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding, Q., Y. V. Reddy, W. Wang, T. Woods, P. Douglas, D. A. Ramsden, S. P. Lees-Miller, and K. Meek. 2003. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 23:5836-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dip, R., and H. Naegeli. 2004. Binding of the DNA-dependent protein kinase catalytic subunit to Holliday junctions. Biochem. J. 381:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglas, P., G. B. Moorhead, R. Ye, and S. P. Lees-Miller, S. P. 2001. Protein phosphatases regulate DNA-dependent protein kinase activity. J. Biol. Chem. 276:18992-18998. [DOI] [PubMed] [Google Scholar]
- 12.Douglas, P., G. P. Sapkota, N. Morrice, Y. Yu, A. A. Goodarzi, D. Merkle, K. Meek, D. R. Alessi, and S. P. Lees-Miller. 2002. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J. 368:243-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drouet, J., C. Delteil, J. Lefrancois, P. Concannon, B. Salles, and P. Calsou. 2005. DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. J. Biol. Chem. 280:7060-7069. [DOI] [PubMed] [Google Scholar]
- 14.Esashi, F., N. Christ, J. Gannon, Y. Liu, T. Hunt, M. Jasin, and S. C. West. 2005. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 434:598-604. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Capetillo, O., A. Lee, M. Nussenzweig, and A. Nussenzweig. 2004. H2AX: the histone guardian of the genome. DNA Repair 3:959-967. [DOI] [PubMed] [Google Scholar]
- 16.Ferrin, L. J., and R. D. Camerini-Otero. 1991. Selective cleavage of human RecA-assisted restriction endonuclease (RARE) cleavage. Science 54:1494-1497. [DOI] [PubMed] [Google Scholar]
- 17.Goodarzi, A. A., and S. P. Lees-Miller. 2004. Biochemical characterization of the ataxia-telangiectasia mutated (ATM) protein from human cells. DNA Repair 3:753-767. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, H. L., S. M. Yannone, and D. J. Chen. 2002. Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair 1:225-235. [DOI] [PubMed] [Google Scholar]
- 19.Kabotyanski, E. B., L. Gomelsky, J. O. Hanc, T. D. Stamato, and D. B. Roth. 1998. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 26:5333-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kienker, L. J., E. K. Shin, and K. Meek. 2000. Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res. 28:2752-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurimasa, A., S. Kumano, N. V. Boubnov, M. D. Story, C. S. Tung, S. R. Peterson, and D. J. Chen. 1999. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 19:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leber, R., T. W. Wise, R. Mizuta, and K. Meek. 1998. The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J. Biol. Chem. 273:1794-1801. [DOI] [PubMed] [Google Scholar]
- 23.Lees-Miller, S. P., and K. Meek. 2003. Repair of DNA double strand breaks by non-homologous end joining. Biochemie 85:1161-1173. [DOI] [PubMed] [Google Scholar]
- 24.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 25.Meek, K., S. Gupta, D. A. Ramsden, and S. P. Lees-Miller. 2004. DNA-PK: activity director at the end. Immunol. Rev. 200:132-141. [DOI] [PubMed] [Google Scholar]
- 26.Merkle, D., P. Douglas, G. B. Moorhead, Z. Leonenko, Y. Yu, D. Cramb, D. P. Bazett-Jones, and S. P. Lees-Miller. 2002. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry 41:12706-12714. [DOI] [PubMed] [Google Scholar]
- 27.Pierce, A. J., P. Hu, M. Han, N. Ellis, and M. Jasin. 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15:3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy, V., Q. Ding, S. P. Lees-Miller, K. Meek, and D. A. Ramsden. 2004. Nonhomologous end-joining requires that the DNA-PK complex undergo an autophosphorylation-dependent rearrangement at DNA ends. J. Biol. Chem. 279:39408-39413. [DOI] [PubMed] [Google Scholar]
- 29.Riballo, E., M. Kuhne, N. Rief, A. Doherty, G. C. Smith, M. J. Recio, C. Reis, K. Dahm, A. Fricke, A. Krempler, A. R. Parker, S. P. Jackson, A. Gennery, P. A. Jeggo, and M. Lobrich. 2004. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol. Cell 16:715-724. [DOI] [PubMed] [Google Scholar]
- 30.Rothkamm, K., I. Kruger, L. H. Thompson, and M. Lobrich. 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 23:5706-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sedelnikova, O. A., E. P. Rogakou, I. G. Panyutin, and W. M. Bonner. 2002. Quantitative detection of 125IdU-induced DNA double-strand breaks with γ-H2AX antibody. Radiat. Res. 158:486-492. [DOI] [PubMed] [Google Scholar]
- 32.Shin, E. K., T. Rijkers, A. Pastink, and K. Meek. 2000. Analyses of TCRB rearrangements substantiate a profound deficit in recombination signal sequence joining in SCID foals: implications for the role of DNA-dependent protein kinase in V(D)J recombination. J. Immunol. 164:1416-1424. [DOI] [PubMed] [Google Scholar]
- 33.Soubeyrand, S., L. Pope, B. Pakuts, and R. J. Hache. 2003. Threonines 2638/2647 in DNA-PK are essential for cellular resistance to ionizing radiation. Cancer Res. 63:1198-1201. [PubMed] [Google Scholar]
- 34.Soubeyrand, S., H. Torrance, W. Giffin, W. Gong, C. Schild-Poulter, and R. J. Hache. 2001. Activation and autoregulation of DNA-PK from structured single-stranded DNA and coding end hairpins. Proc. Natl. Acad. Sci. USA 98:9605-9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wechsler, T., B. P. Chen, R. Harper, K. Morotomi-Yano, B. C. Huang, K. Meek, J. E. Cleaver, D. J. Chen, and M. Wabl. 2004. DNA-PKcs function regulated specifically by protein phosphatase 5. Proc. Natl. Acad. Sci. USA 101:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weterings, E., N. S. Verkaik, H. T. Bruggenwirth, J. H. J. Hoeijmakers, and D. C. van Gent. 2003. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res. 31:7238-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu, Y., W. Wang, Q. Ding, R. Ye, D. Chen, D. Merkle, D. Schriemer, K. Meek, and S. P. Lees-Miller. 2003. DNA-PK phosphorylation sites in XRCC4 are not required for survival after radiation or for V(D)J recombination. DNA Repair 2:1239-1252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.