Abstract
Groucho (Gro)/TLE transcriptional corepressors are involved in a variety of developmental mechanisms, including neuronal differentiation. They contain a conserved C-terminal WD40 repeat domain that mediates interactions with several DNA-binding proteins. In particular, Gro/TLE1 interacts with forkhead transcription factor brain factor 1 (BF-1; also termed FoxG1). BF-1 is an essential regulator of neuronal differentiation during cerebral cortex development and represses transcription together with Gro/TLE1. Gro/TLE-related gene product 6 (Grg6) shares with Gro/TLEs a conserved WD40 repeat domain but is more distantly related at its N-terminal half. We demonstrate that Grg6 is expressed in cortical neural progenitor cells and interacts with BF-1. In contrast to Gro/TLE1, however, Grg6 does not promote, but rather suppresses, BF-1-mediated transcriptional repression. Consistent with these observations, Grg6 interferes with the binding of Gro/TLE1 to BF-1 and does not repress transcription when targeted to DNA. Moreover, coexpression of Grg6 and BF-1 in cortical progenitor cells leads to a decrease in the number of proliferating cells and increased neuronal differentiation. Conversely, Grg6 knockdown by RNA interference causes decreased neurogenesis. These results identify a new role for Grg6 in cortical neuron development and establish a functional link between Grg6 and BF-1.
Members of the Groucho (Gro)/transducin-like Enhancer of split (TLE) family of transcription factors are involved in a number of developmental pathways in invertebrates and vertebrates (2, 3, 5, 7, 16, 24, 42). In particular, Drosophila Gro plays an important role in regulating the generation of the correct number of central and peripheral neurons in the insect nervous system (5, 16, 29). Loss of gro function results in the differentiation of supernumerary neurons as a consequence of the perturbation of lateral specification mechanisms that normally restrict the number of neural progenitors that differentiate into neurons (16). Vertebrate Gro/TLE proteins are also involved in the regulation of neuronal development (20, 24, 42). In particular, Gro/TLE1 is involved in mechanisms that negatively regulate the generation of postmitotic neurons from undifferentiated neural progenitors in the telencephalon (27, 42).
Gro/TLE proteins are transcriptional corepressors that lack DNA-binding activity of their own. They become recruited to specific gene regulatory sequences in context-dependent manners by forming complexes with a number of DNA-binding transcription factors. Specifically, Drosophila Gro regulates neuronal differentiation together with a family of related basic helix-loop-helix proteins designated Hairy/Enhancer of split (Hes) (5, 10, 16, 29). Mammalian Gro/TLE proteins also interact with Hes family members and are coexpressed with the latter in a number of neural cell populations, including progenitors in the developing cerebral cortex (12, 23, 26, 33, 41). Hes proteins play critical roles in regulating neurogenesis in the cortex and other regions of the nervous system (18, 19). Gro/TLEs are also coexpressed, and interact, with another important regulator of cortical neuron development, the forkhead domain protein brain factor 1 (BF-1; also referred to as FoxG1) (13, 14, 39, 43). Mouse embryos lacking BF-1 function display severe hypoplasia of the cerebral hemispheres resulting from perturbation of both dorsal and ventral telencephalon development (40). Specifically, BF-1 inactivation causes telencephalic neural progenitor cells to differentiate into neurons prematurely. Loss of BF-1 activity also leads to an anticipated lengthening of the cell cycle in cortical progenitors (13), mimicking the slowing of the cell cycle that normally occurs during cortical neurogenesis at later stages of embryonic development (38). The premature lengthening of the progenitor cell cycle and the increase in the number of progenitors that undergo neuronal differentiation are believed to be the combined causes of the decreased size of the telencephalon in BF-1-deficient embryos (13, 40).
BF-1 is endowed with the intrinsic ability to bind to DNA through its forkhead domain. In addition, BF-1 interacts with other DNA-binding proteins (34). Thus, BF-1 can be potentially recruited to DNA in both direct and indirect manners. When recruited to DNA, BF-1 acts as a transcriptional repressor (21, 43). This repressor activity is promoted by Gro/TLE proteins (35, 43), suggesting that Gro/TLEs are involved in the transcription functions of BF-1 during cortical neuron differentiation. In this study, we describe investigations aimed at determining whether a recently identified Gro/TLE-related gene product termed Grg6 (6) might also be involved in BF-1 functions. Grg6 displays approximately 60% conservation with Gro/TLEs at the level of the WD40 repeat (WDR) domain that mediates Gro/TLE binding to BF-1 but otherwise shows only a limited similarity at the level of its N-terminal half (6; see Fig. 1). We provide evidence that Grg6 is expressed in neural progenitors in the developing cerebral cortex and physically interacts with BF-1. However, Grg6 does not act as a transcriptional corepressor and inhibits the interaction of BF-1 with Gro/TLE1. Consistent with these findings, Grg6 suppresses BF-1-mediated transcriptional repression. Moreover, coexpression of BF-1 and Grg6 results in the differentiation of supernumerary cortical neurons, whereas Grg6 inactivation inhibits neuronal differentiation. These results demonstrate for the first time that Grg6 is involved in the regulation of neurogenesis and suggest that Grg6 acts as a regulator of the functions of BF-1 during cortical neuron differentiation.
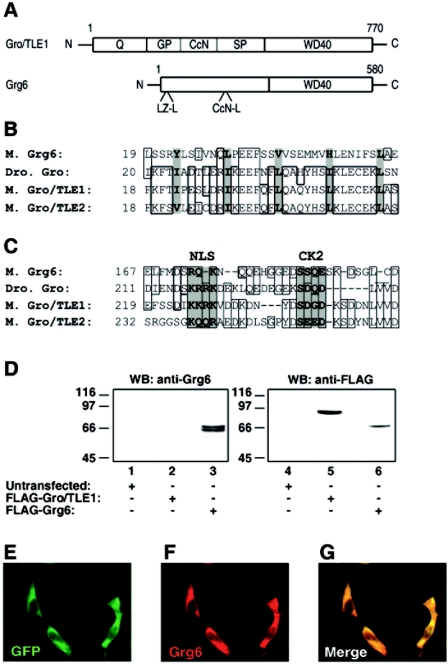
FIG. 1.
Characterization of anti-Grg6 antibody. (A) Top, schematic representation of the domain structure of Gro/TLE1 (37); bottom, Grg6 displays the highest relatedness to Gro/TLEs at the level of the C-terminal WDR domain, with similarity within the N-terminal half limited only to a potential leucine zipper-like motif (LZ-L) and a CcN domain-like region (CcN-L). (B) Comparison of the putative LZ-L motif of mouse (M.) Grg6 and the first LZ-L motif within the Q domain of Drosophila (Dro.) Gro and mouse Gro/TLE1 and Gro/TLE2. Hydrophobic residues considered to form the core of this motif (36) are shaded and in bold. Identical amino acids and conservative substitutions are boxed. (C) Comparison of the CcN motif of Gro/TLE proteins and the CcN-L motif of Grg6. The nuclear localization sequence (NLS) and protein kinase CK2 phosphorylation site (CK2) (27) are shaded and in bold. Identical amino acids and conservative substitutions are boxed. (D) Western blotting analysis. HEK293 cells were either not transfected (lanes 1 and 4) or transfected with FLAG-Gro/TLE1 (lanes 2 and 5) or FLAG-Grg6 (lanes 3 and 6), followed by Western blotting (WB) with anti-Grg6 (lanes 1 to 3) or anti-FLAG (lanes 4 to 6) antibodies (Ab). Here and in succeeding figures, the positions and sizes of standards are indicated in kilodaltons. (E to G) COS7 cells were transfected with GFP-Grg6, fixed, and subjected to double-labeling analysis of GFP expression (E) and anti-Grg6 immunoreactivity (F). (G) Combined GFP and Grg6 staining.
MATERIALS AND METHODS
Plasmids.
PCR was used to amplify the sequence encoding mouse Grg6 (oligonucleotide primers Grg6-1 [5′-GATGACTTCCCACAGACAGAGC-3′] and Grg6-2 [5′-GTGTACCACATCAAGTACTGA-3′]) by using a pMT-CB6-Grg6 plasmid as the template (6). The PCR product was subcloned into pCMV2-FLAG digested with EcoRV. Plasmid pCMV2-HA-Grg6 was obtained by digesting pCMV2-FLAG-Grg6 with HindIII and KpnI, followed by subcloning into pCMV2-HA digested with HindIII and KpnI. pcDNA3-GAL4bd-Grg6 (encoding a fusion protein of the DNA-binding domain of GAL4 [GAL4bd] and Grg6) was generated by subcloning the Grg6 PCR product described above into the filled-in BamHI site of pcDNA3-GAL4bd. Plasmid pGEX3-Grg6(183-287) was obtained by PCR amplification of the region encoding amino acids 183 to 287 of Grg6 and subcloning into the SmaI site of pGEX3. Full-length Grg6 was cloned into pGEX1 by digesting pCMV2-FLAG-Grg6 with BglII and KpnI and subcloning the ensuing fragment into pGEX1. Construct pEGFP-Grg6 was generated by first digesting pCMV2-FLAG-Grg6 with EcoRI and KpnI and then subcloning the resulting fragment into pEGFP-C1 digested with EcoRI and KpnI. Plasmid pEGFP-Gro/TLE1 was obtained by digesting pcDNA3-Gro/TLE1 with BamHI and ApaI and subcloning the ensuing fragment into pEGFP-C1 digested with BglII and ApaI. Plasmids pCMV2-FLAG-Gro/TLE1, pcDNA3-Gro/TLE1, pcDNA3-GAL4bd-Gro/TLE1, pEBG-Gro/TLE1, pEBG-Gro/TLE1(1-135), pMyc-Gro/TLE4, pCMV2-FLAG-BF-1, pCMV2-FLAG-BF-1(NH-AA), pcDNA3-GAL4bd-BF-1(241-336), pCMV2-FLAG-Hes1, pCMV2-FLAG-Hes1ΔWRPW, p6B-CMV-Luc (luciferase gene under the control of the cytomegalovirus [CMV] promoter linked to six BF-1-binding sites), p6N-βactin-Luc (luciferase gene under the control of the β-actin promoter linked to six Hes1-binding sites), and p5XUAS-SV40-Luc (luciferase gene under the control of the simian virus 40 promoter linked to five GAL4 upstream activation sequence sites) were described previously (8, 9, 12, 22, 23, 27, 33, 43).
Affinity purification of Grg6 antibodies.
The previously described anti-Grg6 serum BIO-81 (6) was first preadsorbed on a protein powder prepared from adult mouse kidneys as previously described (15). The treated serum was then affinity purified on nitrocellulose-immobilized FLAG-Grg6 protein expressed in transfected HEK293 cells. For each purification, lysate from transfected cells was fractionated by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by transfer to a nitrocellulose membrane. A vertical strip was cut from the middle portion of the nitrocellulose, followed by Western blotting with anti-FLAG antibody to visualize the position of migration of FLAG-Grg6. A thin horizontal strip containing the region where FLAG-Grg6 migrated was then excised, cut into small pieces, and incubated with preadsorbed Grg6 serum in buffer 1 (25 mM Tris-HCl, pH 7.8, 150 mM NaCl, 0.05% Triton X-100, 5% milk powder), followed by two washes in buffer 1 and three or four washes in phosphate-buffered saline. Bound antibodies were recovered with buffer 2 (100 mM glycine, 1 M acetic acid, pH 3.0) and immediately mixed with an equal volume of 1 M KH2PO4.
In situ hybridization and RT-PCR.
Sense and antisense Grg6 riboprobes were generated by first amplifying by PCR an ∼0.8-kb product corresponding to the 3′ end of the Grg6 cDNA (oligonucleotide primers Grg6 forward [5′-TGAAGCACCAGGAACTGCTA-3′] and Grg6 reverse [5′-CCGTTCTCAGTCATGTCGAA-3′]). The PCR product was cloned into pCRII-TOPO (Invitrogen). Riboprobes were then generated in the presence of digoxigen-11-UTP with T7 or SP6 RNA polymerase (Roche) and 1 μg of linearized plasmid. In situ hybridization was performed as previously described (31), with the hybridization temperature set at 70°C. High-stringency washes were used to remove unbound probe. Sections were subsequently blocked with 10% fetal bovine serum-1% blocking reagent (Roche) and incubated with anti-digoxigenin-alkaline phosphatase antibody (1:1,000). Slides were washed and color developed with BM purple as the substrate (Roche). Reverse transcription (RT)-PCR was performed as previously described (6), with sense primer 1 (5′-CCTAGCACAGCACTCTG-3′), antisense primer 2 (5′-TGGATACAACTTACCTG-3′), or antisense primer 3 (5′-GTGTACCACATCAAGTACTGA-3′). Primers 1 and 3 amplify a product of ∼1.0 kb, while primers 1 and 2 amplify a product of ∼0.3 kb.
Transient transfection, protein-protein interaction assays, and Western blotting analysis.
HeLa and HEK293 cells were grown and, when appropriate, transfected with the Superfect reagent (QIAGEN) as previously described (23, 25). Coprecipitation assays with plasmids pEBG-Gro/TLE1 and pEBG-Gro/TLE1(1-135) (or pEBG as a control) and immunoprecipitation experiments with anti-FLAG or anti-Gro/TLE1 antibodies were performed as previously described (22, 23, 25, 43). Forebrain and midbrain tissues were dissected from embryonic day 15.5 (E15.5) mouse embryos and lysed in buffer 3 (150 mM NaCl, 25 mM Tris/HCl, pH 7.8, 1% Triton X-100, Complete proteinase inhibitor cocktail [Roche]). Western blotting studies were performed with the following antibodies: anti-FLAG (1:10,000; Sigma), anti-green fluorescent protein (anti-GFP; 1:7,000; Molecular Probes), antihemagglutinin (anti-HA; 1:1,000; Roche), anti-glutathione S-transferase (anti-GST; 1:500), anti-GAL4bd (1:1,000; Santa Cruz Biotechnology), anti-Myc (1:500; BD Pharmingen), anti-glyceraldehyde-3-phosphate dehydrogenase; 1:2,000 (anti-GAPDH; Abcam), anti-Gro/TLE (panTLE; 1:10 [27, 28, 37]), anti-Gro/TLE1 (1:1,000 [17, 27, 41]), and affinity-purified anti-Grg6 prepared as described above (1:200).
In vitro GST fusion protein interaction assays.
Fusion proteins of GST and either full-length Grg6 or Grg6(183-287) were purified from bacteria as previously described (12, 22). GAL4bd-BF-1(241-336) was in vitro translated and incubated in the presence of the purified fusion proteins, followed by pulldown, gel electrophoresis, and autoradiography as previously described (12, 22).
Immunofluorescence.
COS7 cells were cultured on four-well chamber slides (Nalgene Nunc Int.), fixed in HEPES-buffered saline containing 4% paraformaldehyde, and permeabilized in 0.1% IGEPAL as previously described (11, 27). Primary antibodies for immunofluorescence included monoclonal anti-FLAG (mouse; 1:5,000), affinity-purified anti-Grg6 (rabbit; 1:200), monoclonal anti-Ki67 (mouse; 1:25; BD Pharmingen), and monoclonal anti-NeuN (mouse; 1:50; Chemicon) antibodies. Detection was done as previously described (27). Double-label immunohistochemistry was performed on transverse sections through the forebrains of E14.5 mouse embryos as previously described (27). Cryostat sections were incubated with anti-Grg6 and anti-Ki67 antibodies. All images were captured with a black-and-white Digital Video Company camera mounted on an Axioskop fluorescence microscope (Zeiss, Toronto, Ontario, Canada). Grayscale images were digitally assigned to the appropriate red (Cy3) or green (fluorescein isothiocyanate) channel with Northern Eclipse software (Empix, Mississauga, Ontario, Canada).
Transcription assays.
HeLa or HEK293A cells were transiently transfected with the Superfect reagent. The total amount of transfected DNA was adjusted in each case to 3 μg per well with the pEF-BOS vector. Assays were performed with reporter plasmids p6B-CMV-Luc (500 ng/transfection), p6N-βactin-Luc (1 μg/transfection), and p5XUAS-SV40-Luc (2 μg/transfection). Effector plasmids included pCMV2-FLAG-BF-1 or pCMV2-FLAG-BF-1(NH-AA) (15 ng/transfection), pCMV2-FLAG-Grg6 (25 or 50 ng/transfection), pcDNA3-Gro/TLE1 (50 ng/transfection), pCMV2-FLAG-Hes1 (50 ng/transfection), pcDNA3-GAL4bd (500 ng/transfection), pcDNA3-GAL4bd-Grg6 (50, 200, or 500 ng/transfection), and pcDNA3-GAL4bd-Gro/TLE1 (50, 200, or 500 ng/transfection). In each case, a pCMV-β-galactosidase plasmid was cotransfected to provide a means of normalizing for transfection efficiency (11). Luciferase activity is expressed as the mean ± the standard deviation.
Cortical neural progenitor cell culture, transfection, and analysis of neuronal differentiation.
Primary neural progenitor cell cultures were established from dorsal telencephalic cortices dissected from E11.5 to E12.5 mouse embryos as previously described (11, 27). Cells were cultured, transfected with Lipofectamine 2000, and subjected to double-labeling studies as previously described (11, 27). The amounts of plasmids used for cotransfections were as follows (in nanograms per transfection): pEGFP, 200; pCMV2-FLAG-Gro/TLE1, 500; pCMV2-FLAG-Grg6, 500; pCMV2-FLAG-BF-1, 300. Following transfection, cells were allowed to differentiate until day 4 to 5 in vitro, when they were fixed and subjected to double-label immunocytochemical analysis of the expression of GFP, Ki67, and NeuN. Results from separate experiments were quantified as the percentage of GFP+ cells that were positive for either Ki67 or NeuN. Results were expressed as the mean ± the standard deviation (n = ≥4).
siRNA.
Validated small interfering RNA (siRNA) duplexes targeting Grg6 (identification no. 172273 to 172275) and negative control siRNA (identification no. 4615) were obtained from Ambion. For transfection of HEK293 cells, 8 μl of Superfect reagent (QIAGEN) was added to OPTI-MEM medium (Invitrogen) (total volume, 100 μl) and allowed to sit at room temperature for 10 min. pEGFP, pEGF-Grg6, or pEGFP-Gro/TLE1 DNA (each at 25 ng/transfection) was added to OPTI-MEM medium (total volume, 100 μl) in the absence or presence of either 15 nM or 30 nM Grg6 or control siRNA duplexes for each transfection. Plasmid pcDNA3 (2.0 μg/transfection) was used as the carrier DNA. The transfection agent mixture and the nucleic acid mixture were then combined and allowed to sit at room temperature for an additional 10 min. Each transfection agent-nucleic acid complex was dispensed into six-well tissue culture plates, followed by addition of 2.0 × 105 cells/well. Forty-eight hours after transfection, cells were lysed and processed for Western blotting. Primary cortical progenitor cells were cultured and transfected as previously described (11, 27), with pEGFP (300 ng/transfection) and 30 nM either Grg6 or control siRNA duplexes per transfection. Seventy-two hours after transfection, cells were subjected to immunocytochemistry and quantitation of the percentage of GFP+ Ki67+ or GFP+ NeuN+ cells as described above.
RESULTS
Characterization of anti-Grg6 antibodies.
Gro/TLE proteins have a conserved structure (Fig. 1A) that includes an N-terminal region (the Q domain) containing two coiled-coil motifs involved in protein oligomerization, an internal segment containing adjacent nuclear localization and phosphorylation sequences (the CcN domain), and a C-terminal WDR domain involved in protein-protein interactions (5, 12, 25-27, 30, 36, 37, 43). Compared to Gro/TLEs, Grg6 is a shorter protein with a similar WDR domain (6) but little conservation outside of this region, except for the presence of one putative N-terminal leucine zipper-like motif resembling the coiled-coil motifs of Gro/TLEs (Fig. 1B) and a short region of similarity to the Gro/TLE CcN domain (Fig. 1C). To elucidate the functions of Grg6, we first characterized further a previously described anti-Grg6 antibody (6). This antibody was affinity purified and tested in Western blot assays of lysates from HEK293 cells left untransfected or transfected with FLAG epitope-tagged forms of Grg6 or Gro/TLE1. Both FLAG-Grg6 and FLAG-Gro/TLE1 were recognized by an anti-FLAG antibody (Fig. 1D, lanes 5 and 6). In contrast, the anti-Grg6 antibody decorated only Grg6 and not Gro/TLE1 (Fig. 1D, lanes 2 and 3). No immunoreactive bands were observed in untransfected cells (Fig. 1D, lane 1). The anti-Grg6 antibody reacted with a doublet of roughly 66 to 68 kDa, while the anti-FLAG antibody decorated only a single band that comigrated with the slower species of ∼68 kDa (Fig. 1D, cf. lanes 3 and 6). This finding suggests that this slower species corresponds to full-length Grg6 and that the faster form may represent a breakdown product of Grg6 that lacks the N-terminal FLAG epitope. To characterize further the anti-Grg6 antibody, COS7 cells were transfected with a fusion protein of GFP and Grg6, followed by double-labeling analysis of GFP and Grg6 expression. We found that the GFP fluorescence and Grg6 immunoreactivity overlapped, suggesting further that the anti-Grg6 antibody specifically recognizes Grg6 (Fig. 1E to G). Grg6 was mostly localized to the cytosol of the transfected cells. This intracellular distribution is in contrast to the predominantly nuclear localization of canonical Gro/TLE proteins (37, 41, 42). Taken together, these results show that the anti-Grg6 antibody is specific to Grg6 and that this protein is not localized to the nucleus in transfected COS7 cells.
Expression of Grg6 in cortical progenitor cells of the developing mouse forebrain.
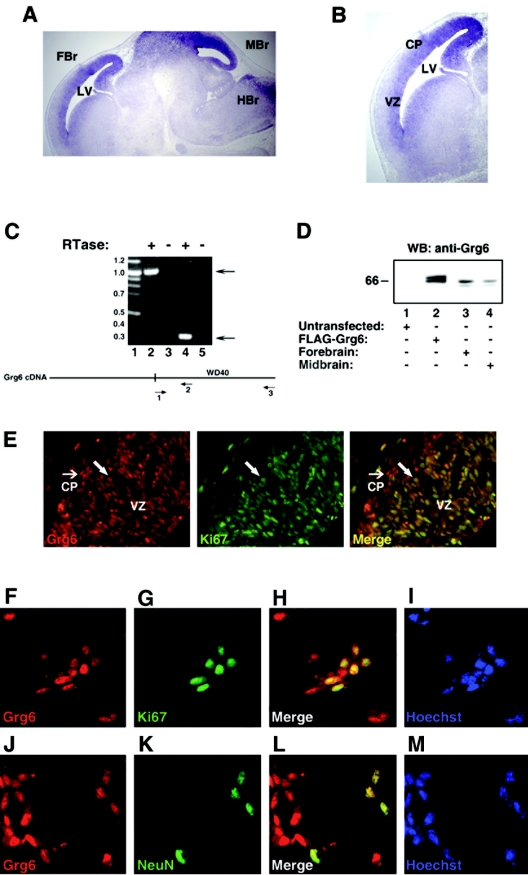
Previous studies have shown the presence of Grg6 transcripts in adult mouse brain (6), suggesting that Grg6 may also be expressed in the developing nervous system. In situ hybridization studies showed that Grg6 was expressed in several regions of the brain at E14.5, including the forebrain, midbrain, and caudal hindbrain (Fig. 2A and B and data not shown). In the dorsal telencephalon, Grg6 gene expression was particularly robust in the ventricular zone, where cortical neural progenitor cells are located (Fig. 2B). In addition, Grg6 transcripts were also detectable in the more superficial cortical plate, where postmitotic neurons are found (Fig. 2B). In agreement with these observations, RT-PCR experiments revealed the presence of Grg6 transcripts in RNA isolated from dorsal telencephalon dissected from the forebrain of E15.5 mouse embryos (Fig. 2C).
FIG.2.
Grg6 expression in the telencephalon. (A and B) In situ hybridization studies. Sagittal sections through the brains of E14.5 mouse embryos were analyzed with an antisense Grg6 riboprobe. Hybridization signals are observed in the forebrain (FBr), dorsal midbrain (Mbr), and caudal hindbrain (HBr); LV, lateral ventricle. No hybridization was detected with control sense riboprobes (not shown). (B) Higher-magnification view of the telencephalon showing robust Grg6 expression in both the ventricular zone (VZ) and the cortical plate (CP). (C) RT-PCR analysis. Each RNA was incubated with (lanes 2 and 4) or without (lanes 3 and 5) reverse transcriptase (RTase). The ensuing cDNA mixture was subjected to PCR with either primers 1 and 3 (lanes 2 and 3) or primers 1 and 2 (lanes 4 and 5). Lane 1 was loaded with the indicated DNA size standards (sizes are in kilobases). A schematic of the Grg6 cDNA is shown, indicating the location of the region encoding the WDR domain and the positions of the oligonucleotide primers. (D) Western blotting analysis. Lysates from HEK293 cells left untransfected (lane 1) or transfected with FLAG-Grg6 (lane 2) were subjected to SDS-PAGE together with lysates from forebrain (lane 3) or midbrain (lane 4) tissue dissected from E15.5 mouse embryos, followed by Western blotting (WB) with affinity-purified anti-Grg6 antibody. (E) Double-labeling immunohistochemical analysis of the dorsal telencephalon from E14.5 mouse embryos with antibodies against Grg6 (left) and Ki67 (middle). Combined Grg6-Ki67 staining is shown on the right; the large arrow points to an example of a double-labeled cell, while the small arrow points to cells positive for Grg6 but not Ki67 expression. Mitotic cells of the surface ectoderm are also visible in the top left corner. (F to M) Primary cultures of E13.5 mouse embryonic cortical progenitor cells grown for 4 days in vitro were fixed and subjected to double-labeling analysis of the expression of Grg6 (F and J), Ki67 (G), and NeuN (K). Combined Grg6-Ki67 (H) or Grg6-NeuN (L) staining is shown. Hoechst staining was used to visualize nuclei (I and M).
To examine the expression of Grg6 proteins, forebrain and midbrain tissues were dissected from E15.5 embryos, followed by preparation of protein extracts and Western blotting analysis with the affinity-purified anti-Grg6 antibody. An immunoreactive band of about 64 kDa was detected in both forebrain and midbrain extracts (Fig. 2D, lanes 3 and 4). This band exhibited an electrophoretic mobility similar to that of the 66- to 68-kDa doublet decorated by the anti-Grg6 antibody in lysates from HEK293 cells transfected with FLAG-Grg6 (Fig. 2D, lane 2). We next tested if Grg6 proteins were expressed in cortical neural progenitor cells. Immunohistochemical analysis of E14.5 dorsal telencephalon showed that Grg6 exhibited a nuclear immunoreactivity that overlapped with the expression of the mitotic cell marker protein Ki67 in the ventricular zone (Fig. 2E). These results show that Grg6 is expressed in cortical progenitors. They show further that Grg6 is localized to the nuclei of those cells. A number of Grg6+ cells located outside of the ventricular zone did not express Ki67, suggesting that they correspond to postmitotic neurons. In agreement with this possibility, Grg6+ cells in the cortical plate expressed the neuronal tissue-specific protein NeuN (data not shown). To confirm these results, the dorsal telencephalon was dissected from E13.5 mouse embryos and primary cultures of cortical neural progenitor cells were established and allowed to proliferate and differentiate into neurons in vitro as previously described (27). We found that Grg6 was expressed in most of the cultured cells and that the Grg6 immunoreactivity was nuclear and overlapped with both Ki67 (Fig. 2F to I) and NeuN (Fig. 2J to M) expression. Together, these studies show that Grg6 is expressed in both mitotic progenitor cells and postmitotic neurons in the developing cerebral cortex and that it is localized to the nuclei of those cells.
Interaction of Grg6 with BF-1.
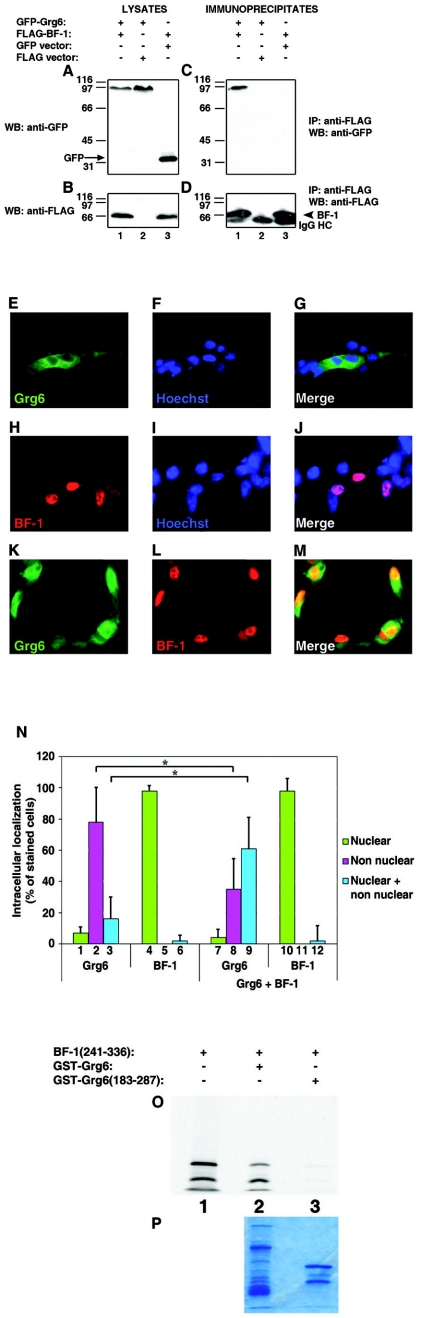
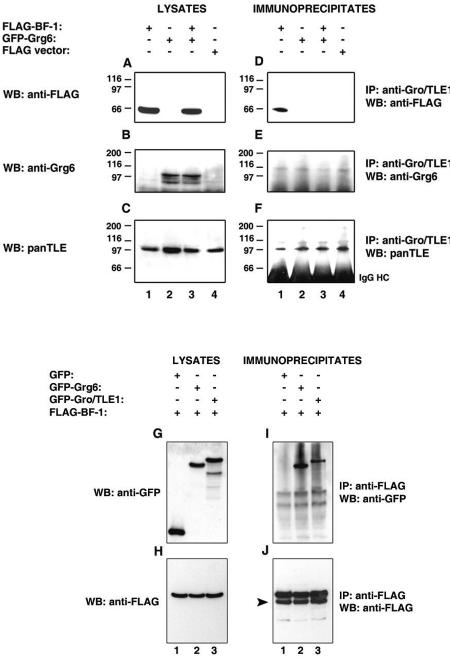
Previous studies have shown that Gro/TLEs are coexpressed, and form transcription repression complexes, with BF-1 and Hes1, two DNA-binding proteins that play important roles in cortical neurogenesis (25, 27, 43). These interactions involve the C-terminal WDR domain of Gro/TLE (43). Based on the similarity of the WDR domains of Gro/TLE and Grg6, and the expression of Grg6 in cortical progenitor cells, we next tested if Grg6 might physically interact with BF-1 and/or Hes1. HEK293 cells were transfected with FLAG-BF-1 (Fig. 3B) in the presence of GFP-Grg6 or GFP alone (Fig. 3A). The choice of GFP-Grg6 was suggested by the fact that the increase in size caused by fusion with GFP eliminates the problem of Grg6 and BF-1 having almost overlapping electrophoretic mobilities. After immunoprecipitation with an anti-FLAG antibody (Fig. 3C and D), GFP-Ggr6 coimmunoprecipitated with FLAG-BF-1 (Fig. 3C, lane 1) but not when cells were transfected with the FLAG vector alone (Fig. 3C, lane 2). In contrast, GFP alone did not coimmunoprecipitate with FLAG-BF-1 (Fig. 3C, lane 3). Determination of whether endogenous Grg6 and BF-1 present in cortical neural progenitor cells would coimmunoprecipitate with each other was not possible because the available anti-Grg6 and anti-BF-1 (43) antibodies proved to be unsuited for immunoprecipitation studies (data not shown). We therefore examined further the possibility that Grg6 might interact with BF-1 by comparing the intracellular localization of Grg6 in the absence or presence of BF-1. COS7 cells were transfected with GFP-Grg6 (Fig. 3E to G), FLAG-BF-1 (Fig. 3H to J), or a combination of these two proteins (Fig. 3K to M), followed by double-labeling analysis of GFP and BF-1 expression. As shown above, Grg6 displayed a predominantly nonnuclear localization when expressed in COS7 cells in the absence of BF-1 (Fig. 3E to G and N, bars 1 to 3), whereas transfected BF-1 was localized to nuclei (Fig. 3H to J and N, bars 4 to 6). We found that a considerable number of cells exhibited overlapping nuclear localization of BF-1 and Grg6 when these proteins were coexpressed (Fig. 3K to M and N, bars 7 to 9). These findings suggest that Grg6 can associate with BF-1 in COS7 cells and that this interaction results in recruitment of Grg6 to nuclei. We next examined if Grg6 bound to BF-1 directly and, more specifically, if amino acids 276 to 336 of BF-1 would mediate this interaction, as was shown to be the case for the binding of BF-1 to Gro/TLE (43). An in vitro-translated protein containing amino acids 241 to 336 of BF-1 interacted with a bacterially purified GST-Grg6 fusion protein (Fig. 3O, lane 2) but not with a fusion protein of GST and amino acids 183 to 287 of Grg6 (this region of Grg6 does not include the WDR domain) (Fig. 3O, lane 3). Taken together, these studies show that Grg6 binds to BF-1 directly and this interaction involves the same domain of BF-1 that interacts with Gro/TLEs.
FIG.3.
Interaction of Grg6 and BF-1. (A to D) Coimmunoprecipitation studies. HEK293 cells were transfected with FLAG-BF-1 in the absence or presence of GFP-Grg6 or GFP alone, as indicated. Each cell lysate was subjected to immunoprecipitation (IP) with anti-FLAG antibody (C and D), followed by analysis of the immunoprecipitated material, together with 1/10 of each input lysate (A and B), by Western blotting (WB) with anti-GFP (A and C) or anti-FLAG (B and D) antibodies. In panel A, the arrow points to the position of migration of GFP alone. In panel D, the arrowhead points to the position of migration of BF-1. Here and in succeeding figures, IgG HC indicates the immunoglobulin G heavy chain. (E to M) Immunocytochemical analysis. COS7 cells were transfected with GFP-Grg6 (E to G), FLAG-BF-1 (H to J), or the two proteins together (K to M), followed by double-labeling analysis of the expression of GFP-Grg6 (E and K) or BF-1 (H and L). (M) Combined GFP-Grg6/BF-1 staining. Hoechst staining is shown alone (F and I) or in combination with GFP-Grg6 (G) or BF-1 (J). (N) Separate immunocytochemical experiments (n = 7) were used to quantitate the intracellular distribution of Grg6 in the absence (bars 1 to 3) or presence (bars 7 to 9) of BF-1. The localization of BF-1 in the absence (bars 4 to 6) or presence (bars 10 to 12) of Grg6 is also shown. Results are depicted as the mean ± the standard deviation. *, P < 0.01 by analysis of variance. (O and P) In vitro interaction of Grg6 and BF-1. Amino acids 241 to 336 of BF-1 were in vitro translated as a fusion protein with GAL4bd and incubated with ∼2.0 μg of GST-Grg6 (lane 2) or GST-Grg6(183-287) (lane 3) purified from bacteria. (O) Precipitates recovered with glutathione-Sepharose beads were subjected to SDS-PAGE and autoradiography together with 10% of the amount of the in vitro-translated protein used in the incubation mixture (lane 1). GAL4-BF-1(241-336) consistently migrated as a doublet. (P) Coomassie blue staining of the GST fusion proteins used in these assays.
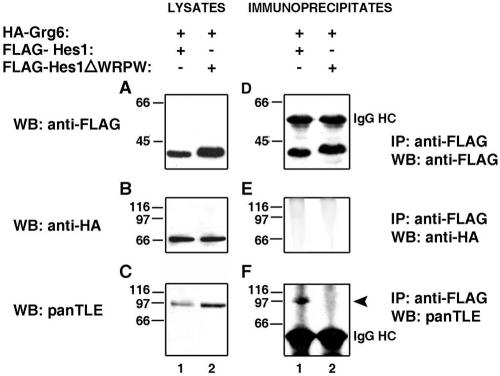
To determine the specificity of its interaction with BF-1, we tested if Grg6 would also interact with Hes1. HEK293 cells, which express endogenous Gro/TLEs (Fig. 4C), were transfected with HA epitope-tagged Grg6 (Fig. 4B, lanes 1 to 2) together with FLAG-Hes1 (Fig. 4A, lane 1) or a truncated form of Hes1 lacking the WRPW motif necessary for Gro/TLE binding (FLAG-Hes1ΔWRPW) (23) (Fig. 4A, lane 2). Immunoprecipitation with an anti-FLAG antibody resulted in the coimmunoprecipitation of endogenous Gro/TLEs with Hes1 (Fig. 4F, lane 1) but not Hes1ΔWRPW (Fig. 4F, lane 2). In contrast, HA-Grg6 did not coimmunoprecipitate with Hes1 (Fig. 4E). These findings strongly suggest that Grg6 interacts specifically with BF-1 and does not associate with either Hes1 alone or complexes of Hes1 and Gro/TLE.
FIG. 4.
Interaction of Hes1 with Gro/TLE but not Grg6. HEK293 cells were cotransfected with HA-Grg6 and either FLAG-Hes1 (lane 1) or FLAG-Hes1ΔWRPW (lane 2). Each cell lysate was subjected to immunoprecipitation (IP) with anti-FLAG antibody (D to F), followed by analysis of the immunoprecipitates, together with 1/10 of each input lysate collected prior to immunoprecipitation (A to C), by Western blotting (WB) with anti-FLAG (A and D), anti-HA (B and E), or panTLE (C and F) antibodies. In panel F, the arrowhead points to the position of migration of endogenous Gro/TLE proteins.
Inhibition of BF-1-mediated transcriptional repression by Grg6.
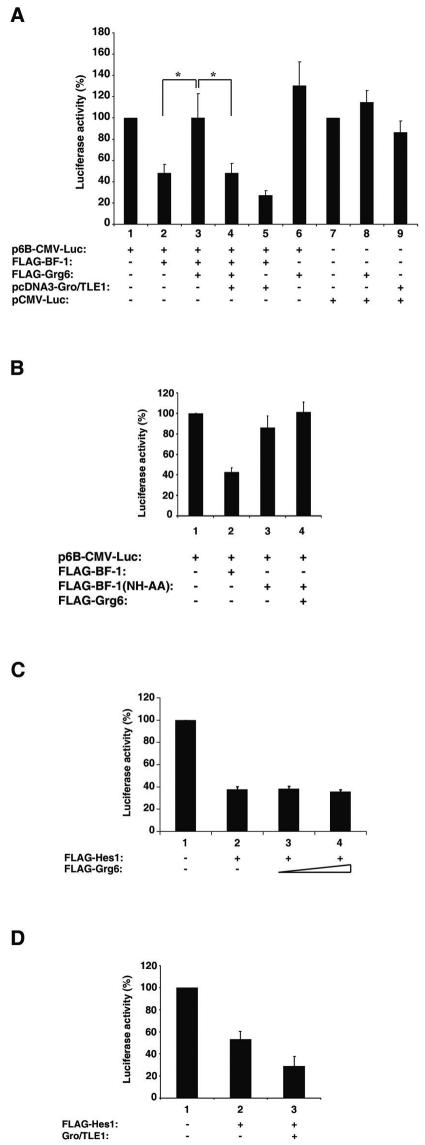
BF-1 mediates transcriptional repression, and Gro/TLE1 can act as a corepressor for BF-1 (35, 43). We therefore tested whether Grg6 might also be involved in the regulation of BF-1 transcription repression activity. HEK293 cells were transfected with a reporter construct containing the luciferase gene under the control of the CMV promoter linked to six tandem copies of a BF-1-binding site (43). The basal level of promoter activity in the absence of BF-1 was designated 100% (Fig. 5A, bar 1). As shown previously (43), cotransfection of BF-1 led to repression of basal transcription (Fig. 5A, bar 2) and this effect was potentiated by the coexpression of Gro/TLE1 (Fig. 5A, bar 5). In contrast, coexpression of Grg6 resulted in suppression of BF-1-mediated transcriptional repression and restored reporter gene expression to basal levels (Fig. 5A, cf. bars 1 to 3). This derepressive effect of Grg6 was antagonized by the coexpression of Gro/TLE1 (Fig. 5A, bar 4). Grg6 had no significant effect on reporter gene expression when it was cotransfected with a mutated form of BF-1, BF-1(NH-AA) (8), that does not bind to DNA and does not mediate transcriptional repression (Fig. 5B). Neither Grg6 nor Gro/TLE1 had a significant effect on the activity of the CMV promoter alone (Fig. 5A, bars 7 to 9). Moreover, Grg6 expression did not cause changes in FLAG-BF-1 expression levels (data not shown).
FIG. 5.
Effect of Grg6 on BF-1-mediated transcriptional repression. (A and B) HEK293 cells were transfected with either the reporter construct p6B-CMV-Luc (A, bars1 to 6; B, bars 1 to 4) or the control plasmid pCMV-Luc (A, bars 7 to 9) in the absence or presence of the indicated combinations of expression vectors. (C) HEK293 cells were transfected with the reporter construct p6N-βactin-Luc in the absence (bar 1) or presence (bar 2) of FLAG-Hes1 and either 25 ng/transfection (bar 3) or 50 ng/transfection (bar 4) of FLAG-Grg6. (D) Cells were transfected as in panel C, except that a Gro/TLE1 expression plasmid (50 ng/transfection) was used instead of Grg6. In all panels, basal luciferase activity in the absence of effector plasmids was considered 100% and values in the presence of effector plasmids are expressed as the mean ± the standard deviation of at least three independent experiments performed in duplicate. (A) *, P < 0.05 by analysis of variance; the difference between bars 1 and 6 was not statistically significant.
To test the specificity of the inhibitory effect of Grg6 on BF-1, we next examined Hes1-mediated transcriptional repression in the absence or presence of Grg6. A reporter construct containing the luciferase gene under the control of the β-actin promoter linked to six tandem copies of a Hes1-binding site (33) was transfected into HEK293 cells in the absence or presence of Hes1 and Grg6. Hes1 expression resulted in repression of reporter gene expression (Fig. 5C, bars 1 and 2), and this effect was not influenced by coexpression of increasing amounts of Grg6 (Fig. 5C, bars 2 to 4). As previously reported (23), Gro/TLE1 enhanced Hes1-mediated repression in the same assays (Fig. 5D). These results are in agreement with the fact that Grg6 does not physically bind to Hes1 and suggest further that Grg6 interacts specifically with BF-1. Moreover, Grg6 does not appear to antagonize the interaction of Hes1 with corepressor factors required for Hes1-mediated repression. Taken together, these results strongly suggest that, in contrast to Gro/TLE proteins, Grg6 does not act as a transcriptional corepressor for BF-1 and instead negatively regulates the transcription repression activity of the latter. They also suggest that Grg6 and Gro/TLE1 may compete with each other for BF-1 binding.
Reduced interaction of BF-1 with Gro/TLE1 in the presence of Grg6.
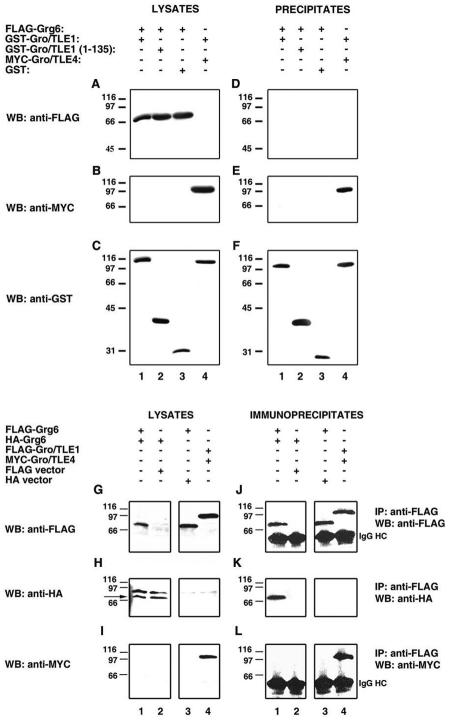
To test directly if Grg6 might interfere with the interaction of BF-1 with Gro/TLE1, HEK293 cells were transfected with FLAG-BF-1 alone or in combination with GFP-Grg6 (Fig. 6A and B), followed by immunoprecipitation with anti-Gro/TLE1 antibodies to precipitate endogenous Gro/TLE1. BF-1 was coimmunoprecipitated with Gro/TLE1 in the absence of Grg6 (Fig. 6D, lane 1). This coimmunoprecipitation was inhibited when Grg6 was cotransfected (Fig. 6D, lane 3), even though the expression of BF-1 was not altered (Fig. 6A, lane 3). Grg6 was not coimmunoprecipitated with Gro/TLE1 in either the absence or the presence of BF-1, suggesting that Grg6 and Gro/TLE1 do not interact (Fig. 6E, lanes 2 and 3). In agreement with this possibility, separate coprecipitation studies showed that Grg6 failed to interact with both Gro/TLE1 and the N-terminal Q domain of Gro/TLE1 that mediates homo- or heterodimerization (Fig. 7A and D, lanes 1 and 2). In contrast, Gro/TLE4 coprecipitated with Gro/TLE1 (Fig. 7B and E, lane 4), in agreement with previous studies (5, 12, 28, 36). We found, however, that Grg6 formed homodimers (Fig. 7G to K, lane 1). Taken together with the demonstration that Grg6 and BF-1 associate with each other, these results suggest that Grg6 does not interact with Gro/TLE but can compete with the latter for binding to BF-1.
FIG. 6.
Decreased interaction of BF-1 with Gro/TLE in the presence of Grg6. (A to F) HEK293 cells were transfected with the indicated combinations of expression vectors, followed by immunoprecipitation (IP) of endogenous Gro/TLE1 proteins with anti-Gro/TLE1 antibodies. The immunoprecipitates (D to F), together with 1/10 of each input lysate collected prior to immunoprecipitation (A to C), were subjected to Western blotting (WB) with either anti-FLAG (A and D), anti-Grg6 (B and E), or panTLE (C and F) antibodies. (G to J) HEK293 cells were transfected with the indicated combinations of expression vectors, followed by immunoprecipitation of FLAG-BF-1. The immunoprecipitates (I and J), together with 1/10 of each input lysate collected prior to immunoprecipitation (G and H), were subjected to Western blotting with either anti-GFP (G and I) or anti-FLAG (H and J) antibodies. In panel J, the arrowhead points to the immunoglobulin G heavy chain.
FIG. 7.
Failure of Grg6 to interact with Gro/TLE1. (A to F) HEK293 cells were transfected as indicated, followed by isolation of either GST-Gro/TLE1 (lanes 1 and 4), GST-Gro/TLE1(1-135) (lane 2), or GST alone (lane 3) on glutathione-Sepharose beads. The precipitated material (D to F), together with 1/10 of each input lysate collected prior to precipitation (A to C), was subjected to Western blotting (WB) with either anti-FLAG (A and D), anti-Myc (B and E), or anti-GST (C and F) antibodies. (G to L) HEK293 cells were transfected with the indicated combinations of expression vectors, followed by immunoprecipitation (IP) of FLAG-Grg6 (lanes 1 and 3) or FLAG-Gro/TLE1 (lane 4). The immunoprecipitates (J to L), together with 1/10 of each input lysate collected prior to immunoprecipitation (G to I), were subjected to Western blotting with either anti-FLAG (G and J), anti-HA (H and K), or anti-Myc (I and L) antibodies. In panel H, the arrow points to the position of migration of HA-Grg6.
To determine if Grg6 might have a higher affinity for BF-1 than Gro/TLE1, HEK293 cells were cotransfected with FLAG-BF-1 and either GFP, GFP-Grg6, or GFP-Gro/TLE1, followed by immunoprecipitation of BF-1 (Fig. 6G to J). Roughly equivalent amounts of GFP-Grg6 and GFP-Gro/TLE1 were specifically coimmunoprecipitated with BF-1 (Fig. 6I, lanes 2 and 3). This result shows that, at least under those experimental conditions, Grg6 and Gro/TLE1 have similar affinities for BF-1. This finding suggests that other mechanisms including, but not limited to, regulated differences in expression levels or nonoverlapping expression may determine whether BF-1 associates preferentially with Grg6 or Gro/TLE proteins.
Lack of transcription repression activity by Grg6.
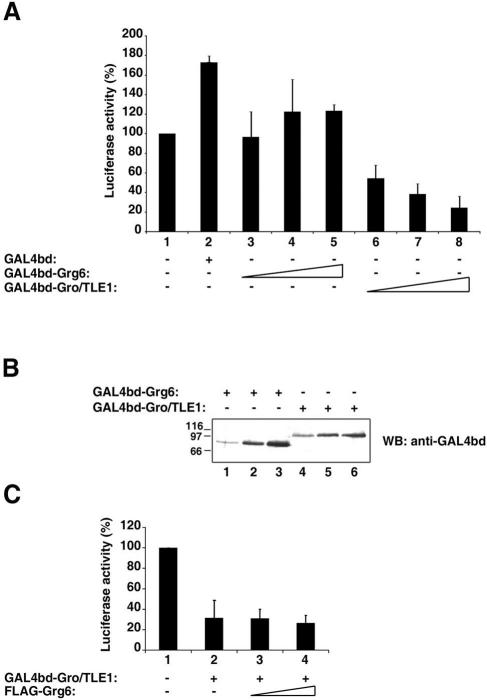
The formation of Grg6-BF-1 complexes at the expense of Gro/TLE-BF-1 complexes may negatively regulate BF-1-mediated transcriptional repression if Grg6 is unable to provide a corepressor function to BF-1. To examine this possibility, we tested if Grg6 is able to act as a transcriptional repressor. Previous studies have shown that Gro/TLE1 represses transcription from a basally active promoter when targeted to DNA as a fusion protein with GAL4bd (12, 26). In agreement with those results, expression of increasing amounts of GAL4bd-Gro/TLE1 resulted in dose-dependent repression of transcription from a simian virus 40 promoter linked to tandem GAL4bd-binding sites (10) (Fig. 8A, cf. bars 1, 2, and 6 to 8). In contrast, expression of equivalent levels of GAL4bd-Grg6 did not cause significant suppression of basal transcription (Fig. 8A, cf. bars 1, 2, and 3 to 5, and B). These results strongly suggest that Grg6 does not mediate transcriptional repression. We next tested if Grg6 would affect Gro/TLE1-mediated transcriptional repression by transfecting GAL4bd-Gro/TLE1 in the absence or presence of increasing amounts of FLAG-Grg6. The repressive effect of Gro/TLE1 was neither decreased nor increased by Grg6 (Fig. 8C). Together, these results suggest that Grg6 does not inhibit the transcription repression ability of Gro/TLE1 but competes with the latter for binding to BF-1, resulting in the formation of complexes in which Grg6 does not provide BF-1 with a corepressor activity.
FIG. 8.
Transcriptional repression by Gro/TLE1 but not Grg6. (A and C) Transient transfection-transcription assays. HeLa cells were transfected with the reporter construct p5XUAS-SV40-Luc in the absence or presence of the indicated proteins. Basal luciferase activity in the absence of effector plasmids was considered 100%, and values in the presence of effector plasmids are expressed as the mean ± the standard deviation of at least three independent experiments performed in duplicate. (B) Western blotting (WB) analysis. Lysates from cells transfected with either 50 (lanes 1 and 4), 200 (lanes 2 and 5), or 500 (lanes 3 and 6) ng/transfection of either GAL4bd-Grg6 (lanes 1 to 3) or GAL4bd-Gro/TLE1 (lanes 4 to 6) were probed with anti-GAL4bd antibodies.
Promotion of cortical neuron differentiation by Grg6 and BF-1.
Based on the demonstrated role of BF-1 in the regulation of cortical neurogenesis (13, 40) and the ability of Grg6 to regulate BF-1-mediated transcriptional repression, we next tested if Grg6 might also be involved in cortical neuron differentiation. To examine this possibility, an siRNA-based RNA interference approach was used to knock down the expression of endogenous Grg6 in primary cultures of cortical progenitor cells undergoing proliferation and neuronal differentiation. siRNA oligonucleotides specific for Grg6, but not negative control siRNA, were first shown to cause a significant decrease in the GFP-Grg6 protein level in transfected HEK293 cells (Fig. 9A, cf. lanes 1 to 3). The Grg6 siRNA duplexes had no effect on the expression of either transfected GFP (Fig. 9B) or endogenous GAPDH (Fig. 9C). When transfected into cultures of dividing cortical progenitors (together with enhanced GFP to mark the transfected cells), the Grg6 siRNA caused an increase in the number of mitotic cells expressing the Ki67 protein and a parallel decrease in NeuN+ differentiated neurons, compared to GFP alone (Fig. 9D and E, cf. bars 1 and 2). No effects were observed when control siRNA was transfected (Fig. 9D and E, bar 3). These findings show that endogenous Grg6 is involved in mechanisms that promote the differentiation of cortical progenitors into neurons.
FIG.9.
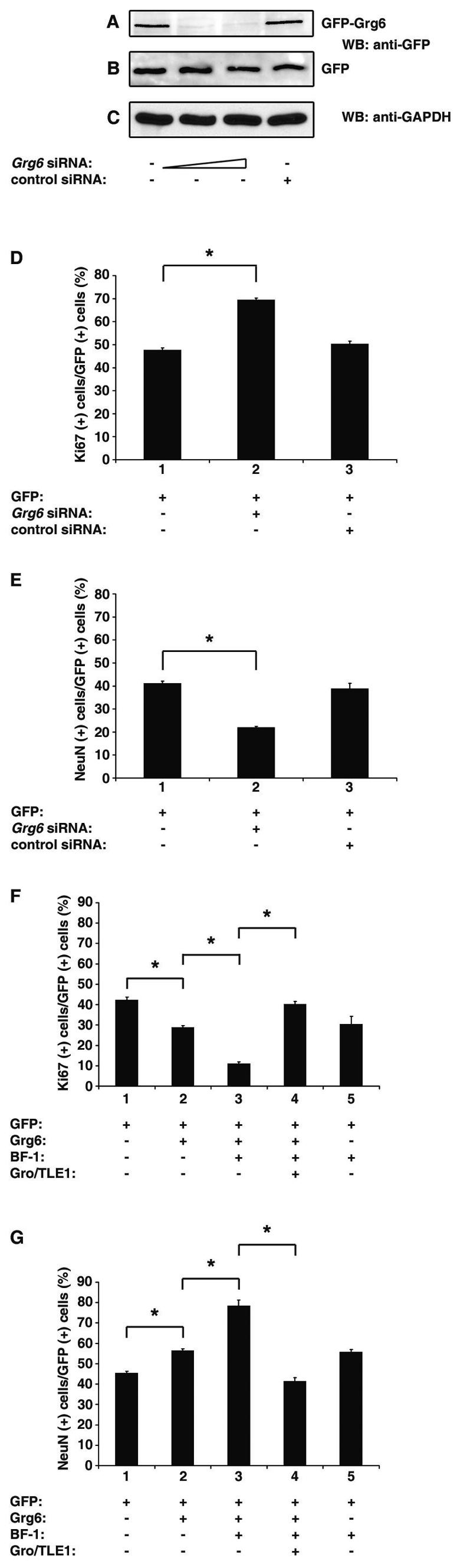
Involvement of Grg6 in cortical neurogenesis. (A to C) HEK293 cells were transfected with either GFP-Grg6 (A) or GFP alone (B) in the absence (lane 1) or presence of Grg6 siRNA (lane 2, 15 nM/transfection; lane 3, 30 nM/transfection) or control siRNA (lane 4, 30 nM/transfection). Forty-eight hours later, cell lysates were subjected to Western blotting (WB) analysis with anti-GFP (A and B) or anti-GAPDH (C) antibodies. (D and E) Primary cultures of mouse E11.5 to E12.5 cortical progenitor cells were transfected with GFP either alone (bar 1) or together with Grg6 (bar 2) or control (bar 3) siRNA (30 nM/transfection). Seventy-two hours later, cells were subjected to double-labeling analysis of the expression of GFP, Ki67, or NeuN. Results were quantitated as the percentage of GFP+ cells that were also positive for either Ki67 (D) or NeuN (E). The results are shown as the mean ± the standard deviation (*, P < 0.0001). (F and G) Primary cultures of mouse E11.5 to E12.5 cortical progenitor cells were transfected with either GFP alone (bar 1) or a combination of GFP and the indicated proteins (bars 2 to 5). Forty-eight hours later, cells were subjected to double-labeling analysis of the expression of GFP, Ki67, or NeuN and quantitation as described above (*, P < 0.0001).
To investigate if Grg6 acts together with BF-1 during cortical neurogenesis, gain-of-function studies were performed by transfecting exogenous Grg6 into cortical progenitor cells. Immunocytochemical analysis of the GFP+ cells showed that exogenous Grg6 caused a moderate increase in the number of differentiated neurons and a concomitant decrease in mitotic cells, compared to GFP alone (Fig. 9F and G, cf. bars 1 and 2). Exogenous expression of BF-1 had a similar effect (Fig. 9F and G, bar 5). Importantly, the coexpression of Grg6 and BF-1 resulted in a considerable increase in the number of differentiated neurons and a parallel decrease in progenitor cells compared to the expression of either protein alone (Fig. 9F and G, cf. bars 2, 3, and 5). These findings strongly suggest that Grg6 and BF-1 associate with each other in cortical progenitor cells and that their interaction results in promotion of the transition of cortical progenitors into neurons.
To test if the promotion of neurogenesis caused by BF-1 and Grg6 (which does not have transcriptional corepressor activity) could be antagonized by coexpression of Gro/TLE1 (which can act as a transcriptional corepressor for BF-1), these three proteins were cotransfected into progenitor cells. Coexpression of Gro/TLE1 blocked the proneuronal effect of Grg6 and BF-1 and restored proliferation and differentiation to control levels (Fig. 9F and G, bar 4). These findings show that Grg6 and Gro/TLE1 have opposite roles during cortical neurogenesis. They suggest further that Grg6 and Gro/TLE1 contribute antagonistic activities to the neural functions of BF-1.
DISCUSSION
Grg6 and Gro/TLE have both similar and different functional properties.
Gro/TLE proteins lack DNA-binding activity of their own but become recruited to specific gene-regulatory sequences in context-dependent manners by forming complexes with a number of different DNA-binding transcription factors. Most, if not all, of these interactions require the C-terminal WDR domain conserved in all Gro/TLE family members. Grg6 shares with Gro/TLEs a conserved WDR domain but otherwise shows little similarity. Although this situation suggested that Grg6 might share at least certain protein-protein interaction properties with Gro/TLEs, the molecular and cellular functions of Grg6 had not been investigated prior to the present work.
We have found that, similar to Gro/TLEs, Grg6 forms homodimers. However, it does not heterodimerize with either Gro/TLE1 (this study) or Gro/TLE4 (N.M and S.S., unpublished data), in contrast to the ability of Gro/TLEs to homo- and heterodimerize with each other (5, 12, 28, 36). The inability of Grg6 to bind to Gro/TLEs may be due to the lack of the two conserved N-terminal leucine zipper-like motifs that were shown to mediate Gro/TLE oligomerization (36). It is possible that the single putative leucine zipper-like motif at its N terminus may be sufficient to mediate Grg6 homodimerization but not interaction with Gro/TLEs. However, the structural elements that underlie Grg6 homodimerization remain to be fully elucidated.
Another similarity between Grg6 and Gro/TLEs is the fact that both are able to interact with BF-1, but only the latter bind to Hes1 with high affinity. These observations suggest that Grg6 and Gro/TLEs share the ability to bind to certain common transcription factors through their conserved WDR domain. The sequence dissimilarities within the WDR domain of Grg6 and Gro/TLEs may be responsible for the different protein-protein interaction properties of these molecules. This situation may facilitate our understanding of how specific regions of the Gro/TLE WDR domain contribute to the binding of different cofactors.
In contrast to Gro/TLEs, Grg6 does not mediate transcriptional repression when recruited to DNA. This conclusion is suggested by the finding that Grg6 does not repress transcription when fused to GAL4bd and does not promote transcriptional repression mediated by BF-1. It is likely that Grg6 cannot repress transcription due to the lack of significant relatedness to the N-terminal Q and GP domains of Gro/TLEs. Both of these regions are involved in functions that are believed to be important for Gro/TLE-mediated transcriptional repression, namely, protein oligomerization and interactions with histone deacetylases and components of the basal transcriptional machinery (4, 28, 36, 44). It is likely that Grg6 is unable to form complexes with histone deacetylases and/or other general transcriptional regulators that associate with Gro/TLEs. This possibility is suggested by our observation that Grg6 suppresses neither Gro/TLE- nor Hes1-mediated transcriptional repression and thus does not seem able to interact with, and titrate away, cofactors required by Gro/TLE and/or Hes1. It is also entirely possible that Grg6 does not participate in other, yet to be characterized, mechanisms underlying transcriptional repression by canonical Gro/TLEs.
Also in contrast to Gro/TLEs, Grg6 does not appear to be generally localized to nuclei and its intracellular localization is cell type dependent. Grg6 was localized to the cytosol in COS7 and 293 cells when transfected alone but was detected in the nucleus when cotransfected with BF-1. In contrast, endogenous Grg6 immunoreactivity was detected in the nuclei of cortical progenitor cells, where BF-1 is endogenously expressed. These observations suggest that Grg6 may depend on interactions with other factors, not necessarily limited to BF-1, to become localized to the nucleus. The nuclear association of Gro/TLEs is mediated by their CcN motif, which includes a nuclear localization sequence and phosphorylation sites for the protein kinases CK2 and cdc2 (26, 27). Grg6 harbors a domain that exhibits limited similarity to the CcN domain of Gro/TLEs but does not contain a defined nuclear localization sequence. Thus, Grg6 intracellular localization may be dependent on the cellular environment and may be influenced by its association with other proteins.
Grg6 acts as a negative regulator of the transcription repression activity of BF-1.
Our previous (43) and present studies have identified a specific case where Grg6 and Gro/TLE1 exhibit similar biochemical properties but mediate different functional effects. Although they both interact with BF-1, transcriptional repression mediated by BF-1 is promoted by Gro/TLE1 (35, 43) and reduced by Grg6. We have demonstrated further that Grg6 has no detectable transcription repression activity of its own. Grg6 does not associate with either Gro/TLE1 or complexes of Gro/TLE1 and BF-1 but instead antagonizes the interaction of BF-1 with Gro/TLE1. Together, these findings suggest that Grg6 can negatively regulate the ability of BF-1 to mediate transcriptional repression by competing for BF-1 binding with Gro/TLE, thereby depriving BF-1 of the corepressor function provided by the latter. This possibility is consistent with our finding that the inhibitory effect of Grg6 on BF-1-mediated transcriptional repression can be alleviated by overexpressing Gro/TLE1. Moreover, overexpression of Gro/TLE1 suppresses the promotion of cortical neurogenesis induced by the coexpression of Grg6 and BF-1, consistent with a functional competition between these proteins (see below for further discussion). It should also be emphasized that we cannot rule out the possibility that Grg6 binding prevents the association of BF-1 with transcriptional corepressors other than Gro/TLE. However, the identity of these possible cofactors remains unknown.
Grg6 promotes cortical neurogenesis.
BF-1 is essential for the development of the cerebral hemispheres. Its function is required to prevent premature slowing of the rate of telencephalic neural progenitor cell growth and to ensure the correct timing of precursor cell cycle exit and neuronal differentiation (13, 40). The role of BF-1 in controlling cortical progenitor proliferation is believed to be mediated by mechanisms in which protein-protein interactions, rather than BF-1's own DNA-binding ability, are of critical importance (8, 13, 32, 34). In contrast, BF-1 requires an intact DNA-binding domain to inhibit or delay the neuronal differentiation of telencephalic precursor cells (13). Whether through its own DNA-binding ability or through interactions with other DNA-binding proteins (34), the recruitment of BF-1 to DNA is believed to result in transcriptional repression of the targeted genes (21, 39). Consistent with this notion, BF-1 is coexpressed with Gro/TLEs in the developing telencephalon (43) and the latter can act as transcriptional corepressors for BF-1 (35, 43). Taken together with the demonstration that exogenous Gro/TLE1 expression in cortical progenitor cells causes accumulation of proliferating cells and a decrease in the number of progenitors that differentiate into neurons (27), these findings suggest that BF-1 works together with Gro/TLEs to prevent premature precursor cell cycle exit and differentiation in the telencephalon.
Based on our finding that Grg6 can antagonize the interaction of BF-1 with Gro/TLE1 and reduce BF-1-mediated transcriptional repression, we tested if Grg6 might be involved in modulating BF-1 functions during the cortical progenitor-to-neuron transition. RNA interference studies have demonstrated that reduction of endogenous Grg6 expression in proliferating cortical progenitor cells is correlated with decreased generation of postmitotic neurons and expansion of the progenitor cell pool. This finding shows that Grg6 is an important positive regulator of the transition from proliferation to differentiation in the telencephalon. Furthermore, converse studies have shown that exogenous Grg6 expression in cortical progenitor cells leads to increased neuronal differentiation and, more importantly, that this effect is significantly enhanced when Grg6 and BF-1 are coexpressed. The latter observation suggests that the formation of Grg6-BF-1 complexes is correlated with an increased rate of precursor cell cycle exit and neuronal differentiation. The weaker proneuronal effect observed with Grg6 alone is possibly the result of the fact that the interaction of Grg6 and BF-1 is likely promoted when both of these proteins are cotransfected, whereas exogenous expression of Grg6 alone may not always result in interaction with BF-1 because the latter is not expressed in all cortical progenitor cells (13, 14). Taken together, these observations suggest that Grg6 works together with BF-1 to increase the rate of neuronal differentiation in the telencephalon.
The possibility that BF-1 can promote the transition of cortical progenitors into neurons when complexed with Grg6 may appear in contrast to the general view that BF-1 acts a negative regulator of neuronal differentiation in the mammalian telencephalon. However, previous studies with Xenopus embryos showed that amphibian BF-1, termed XBF-1, can act both as an activator and a repressor of neuronal differentiation (1). Exogenous expression of high levels of XBF-1 leads to suppression of neuronal differentiation in the injected area, whereas small amounts of XBF-1 result in supernumerary neuronal differentiation (1). The latter results are similar to the moderate increase in neuronal differentiation that we observed when cortical progenitor cells were transfected with BF-1 alone. Studies with Xenopus have suggested further that XBF-1 may act not only as a transcriptional repressor but also as an activator. Bourguignon et al. (1) used chimeric proteins where XBF-1 was fused to either a transcription repression or an activation domain to examine its function as either a dedicated repressor or activator, respectively. They found that neither of these fusion proteins fully reproduced the phenotype associated with wild-type XBF-1 expression, but both exhibited a subset of the full phenotype, suggesting that XBF-1 may utilize a combination of transcriptional mechanisms. Taken together, these observations suggest that BF-1 may be able to both inhibit and promote neuronal differentiation through regulated interactions with different transcriptional cofactors. More specifically, we propose that BF-1 interacts with either Gro/TLE or Grg6 protein to perform different functions during cortical neurogenesis. In this scenario, BF-1 may preferentially interact with Gro/TLEs during phases of active proliferation (i.e., earlier than E11.5) to inhibit or delay neuronal differentiation via transcription repression mechanisms. As neurogenesis becomes progressively more active, Grg6 may become a preferred partner of BF-1 over Gro/TLEs. This situation may prevent BF-1 from repressing the expression of genes that promote cell cycle exit. This effect, coordinated with concomitant neurogenic stimuli, may promote neuronal differentiation. In that regard, it is interesting that BF-1 can suppress the transforming growth factor β-induced activation of the cell cycle inhibitory gene p21Cip1 and cell cycle arrest in epithelial cells (34). It is possible that BF-1 performs a similar function in cortical progenitors and that Gro/TLEs and Grg6 contribute to that activity in opposite manners. Although our transcription studies with transfected cells do not suggest that Grg6 acts as a transcriptional coactivator for BF-1 (and not simply as an inhibitor), it is possible that Grg6 plays such a role in the context of differentiating neural progenitor cells. This possibility would likely depend on the Grg6-mediated recruitment of other factors because Grg6 did not exhibit detectable transactivation ability in our studies.
The mechanisms that may favor the formation of Grg6-BF-1 complexes versus Gro/TLE-BF-1 complexes are unknown. It is conceivable that they involve (i) physiologically regulated changes in the levels of Gro/TLE and/or Grg6 during different phases of neural development, (ii) nonoverlapping expression of these proteins in different subpopulations of telencephalic progenitor or precursor cells, and/or (iii) developmentally regulated influences from other cofactors that may be associated with protein complexes containing these molecules. Future investigations aimed at addressing these possibilities will shed light on the antagonistic functions of Grg6 and Gro/TLEs during cortical neurogenesis.
Acknowledgments
We thank T. Kennedy for invaluable advice, M. Busslinger for the pMyc-Gro/TLE4 plasmid, and L. Liu, R. Lo, Y. Tang, and T. Ruiz de Algara for excellent technical assistance.
This study was supported by grants MOP-13957 and MGC-14971 from the Canadian Institutes for Health Research (CIHR) to S.S., who is a Senior Scholar of the Fonds de la Recherche en Sante du Quebec, and by a Genome Canada grant to C.D.H., who is a CIHR New Investigator and a Michael Smith Foundation for Health Research Scholar.
REFERENCES
- 1.Bourguignon, C., J. Li, and N. Papalopulu. 1998. XBF-1, a winged helix transcription factor with dual activity, has a role in positioning neurogenesis in Xenopus competent ectoderm. Development 125:4899-4900. [DOI] [PubMed] [Google Scholar]
- 2.Brinkmeier, M. L., M. A. Potok, K. B. Cha, T. Gridley, S. Stifani, J. Meeldijk, H. Clevers, and S. A. Camper. 2003. TCF and Groucho-related genes influence pituitary growth and development. Mol. Endocrinol. 17:2152-2161. [DOI] [PubMed] [Google Scholar]
- 3.Carvalho, L. R., K. S. Woods, B. B. Mendonca, N. Marcal, A. L. Zamparini, S. Stifani, J. M. Brickman, I. V. J. Arnhold, and M. Dattani. 2003. A homozygous mutation in HESX1 is associated with evolving hypopituitarism due to impaired repressor-corepressor interaction. J. Clin. Investig. 112:1192-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, G., J. Fernandez, S. Mische, and A. J. Courey. 1999. A functional interaction between the histone deacetylase rpd3 and the corepressor Groucho in Drosophila development. Genes Dev. 13:2218-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, G., and A. J. Courey. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249:1-16. [DOI] [PubMed] [Google Scholar]
- 6.Dang, J., T. Inukai, H. Kurosawa, K. Goi, T. Inaba, N. T. Lenny, J. R. Downing, S. Stifani, and A. T. Look. 2001. The E2A-HLF oncoprotein activates Groucho-related genes and suppresses Runx1. Mol. Cell. Biol. 21:5935-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasen, J. S., J. P. Barbera, T. S. Herman, S. O. Connell, L. Olson, B. Ju, J. Tolkuhn, S. H. Back, D. W. Rose, and M. G. Rosenfeld. 2001. Temporal regulation of a paired-like homeodomain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis. Genes Dev. 15:3193-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dou, C., J. Lee, B. Liu, J. Massague, S. Xuan, and E. Lai. 2000. BF-1 interferes with transforming growth factor β signaling by associating with Smad partners. Mol. Cell. Biol. 20:6201-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eberhard, D., G. Jimenez, B. Heavy, and M. Busslinger. 2000. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 19:2292-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher, A. L., S. Ohsako, and M. Caudy. 1996. The WRPW motif of the Hairy-related helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16:2670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gratton, M. O., E. Torban, S. Belanger-Jasmin, F. Theriault, M. S. German, and S. Stifani. 2003. Hes6 promotes cortical neurogenesis and inhibits Hes1 transcription repression activity by multiple mechanisms. Mol. Cell. Biol. 23:6922-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grbavec, D., R. Lo, Y. Liu, and S. Stifani. 1998. Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur. J. Biochem. 258:339-349. [DOI] [PubMed] [Google Scholar]
- 13.Hanashima, C., L. Shen, S. C. Li, and E. Lai. 2002. Brain Factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J. Neurosci. 22:6526-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanashima, C., S. C. Li, L. Shen, E. Lai, and G. Fishell. 2004. Foxg1 suppresses early cortical cell fate. Science 303:56-59. [DOI] [PubMed] [Google Scholar]
- 15.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Heitzler, P., M. Bourouis, L. Ruel, C. Carteret, and P. Simpson. 1996. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signaling in Drosophila. Development 122:161-171. [DOI] [PubMed] [Google Scholar]
- 17.Husain, J., R. Lo, D. Grbavec, and S. Stifani. 1996. Affinity for the nuclear compartment and expression during cell differentiation implicate phosphorylated Groucho/TLE1 forms of higher molecular mass in nuclear functions. Biochem. J. 317:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishibashi, M., K. Moriyoshi, Y. Sasai, K. Shiota, S. Nakanishi, and R. Kageyama. 1994. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 13:1799-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishibashi, M., S. L. Ang, K. Shiota, S. Nakanishi, R. Kageyama, and F. Guillemot. 1995. Targeted disruption of mammalian hairy and Enhancer of split homologue 1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 9:3136-3148. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, M., K. Nishikawa, T. Suzuki, and M. Yamamoto. 2001. The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev. Biol. 232:315-326. [DOI] [PubMed] [Google Scholar]
- 21.Li, J., H. Chang, E. Lai, E. Parker, and P. Vogt. 1995. The oncogene qin codes for a transcriptional repressor. Cancer Res. 55:5540-5544. [PubMed] [Google Scholar]
- 22.McLarren, K. W., R. Lo, D. Grbavec, K. Thirunavukkarasu, G. Karsenty, and S. Stifani. 2000. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the Runt-related factor Cbfa1. J. Biol. Chem. 275:530-538. [DOI] [PubMed] [Google Scholar]
- 23.McLarren, K. W., F. Theriault, and S. Stifani. 2001. Association with the nuclear matrix and interaction with Groucho and RUNX proteins regulate the transcription repression ability of the basic helix loop helix factor Hes1. J. Biol. Chem. 276:1578-1584. [DOI] [PubMed] [Google Scholar]
- 24.Muhr, J., E. Andersson, M. Persson, T. M. Jessell, and J. Ericson. 2001. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 104:861-873. [DOI] [PubMed] [Google Scholar]
- 25.Nuthall, H. N., J. Husain, K. W. McLarren, and S. Stifani. 2002. Role for Hes1-induced phosphorylation in Groucho-mediated transcriptional repression. Mol. Cell. Biol. 22:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nuthall, H. N., K. Joachim, A. Palaparti, and S. Stifani. 2002. A role for cell cycle-regulated phosphorylation in Groucho-mediated transcriptional repression. J. Biol. Chem. 277:51049-51057. [DOI] [PubMed] [Google Scholar]
- 27.Nuthall, H. N., K. Joachim, and S. Stifani. 2004. Phosphorylation of serine 239 of Groucho/TLE1 by protein kinase CK2 is important for inhibition of neuronal differentiation. Mol. Cell. Biol. 24:8395-8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palaparti, A., A. Baratz, and S. Stifani. 1997. The Groucho/Transducin-like Enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem. 272:26604-26610. [DOI] [PubMed] [Google Scholar]
- 29.Paroush, Z., R. L. Finley, T. Kidd, S. M. Wainwright, P. W. Ingham, R. Brent, and D. Ish-Horowicz. 1994. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79:805-815. [DOI] [PubMed] [Google Scholar]
- 30.Pickles, L. M., S. M. Roe, E. J. Hemingway, S. Stifani, and L. H. Pearl. 2002. Crystal structure of the C-terminal WD40 repeat domain of the human Groucho/TLE1 transcriptional corepressor. Structure 10:751-761. [DOI] [PubMed] [Google Scholar]
- 31.Prado, C. L., A. E. Pugh-Bernard, L. Elghazi, B. Sosa-Pineda, and L. Sussel. 2004. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc. Natl. Acad. Sci. USA 101:2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez, C., L. J. Huang, J. K. Son, A. McKee, Z. Xiao, and H. F. Lodish. 2001. Functional cloning of the proto-oncogene brain factor-1 (BF-1) as a Smad-binding antagonist of transforming growth factor-beta signaling. J. Biol. Chem. 276:30224-30230. [DOI] [PubMed] [Google Scholar]
- 33.Sasai, Y., R. Kageyama, Y. Tagawa, R. Shigemoto, and S. Nakanishi. 1992. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 5:2620-2634. [DOI] [PubMed] [Google Scholar]
- 34.Seoane, J., H. V. Le, L. Shen, S. A. Anderson, and J. Massague. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117:211-223. [DOI] [PubMed] [Google Scholar]
- 35.Sonderegger, C. K., and P. K. Vogt. 2003. Binding of the corepressor TLE1 to Qin enhances Qin-mediated transformation of chicken embryo fibroblasts. Oncogene 22:1749-1757. [DOI] [PubMed] [Google Scholar]
- 36.Song, H., P. Hasson, Z. Paroush, and A. J. Courey. 2004. Groucho oligomerization is required for repression in vivo. Mol. Cell. Biol. 24:4341-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stifani, S., C. M. Blaumueller, N. J. Redhead, R. E. Hill, and S. Artavanis-Tsakonas. 1992. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat. Genet. 2:119-127. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi, T., R. Nowakowski., and V. J. Caviness. 1993. Cell cycle parameters and patterns of nuclear movement in the neocortical proliferative zone of the fetal mouse. J. Neurosci. 13:820-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao, W., and E. Lai. 1992. Telencephalon-restricted expression of BF-1, a new member of the HNF-2/fork head gene family, in the developing rat brain. Neuron 8:957-966. [DOI] [PubMed] [Google Scholar]
- 40.Xuan, S., C. Baptista, G. Balas, W. Tao, V. Soares, and E. Lai. 1995. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 14:1141-1152. [DOI] [PubMed] [Google Scholar]
- 41.Yao, J., Y. Liu, J. Husain, R. Lo, A. Palaparti, J. Henderson, and S. Stifani. 1998. Combinatorial expression patterns of individual TLE proteins during cell determination and differentiation suggest non-redundant functions for mammalian homologs of Drosophila Groucho. Dev. Growth Differ. 40:133-146. [DOI] [PubMed] [Google Scholar]
- 42.Yao, J., Y. Liu, R. Lo, I. Tretjakoff, A. Peterson, and S. Stifani. 2000. Disrupted development of the cerebral hemispheres in transgenic mice expressing the mammalian Groucho homologue Transducin-like Enhancer of split 1 in postmitotic neurons. Mech. Dev. 93:105-115. [DOI] [PubMed] [Google Scholar]
- 43.Yao, J., E. Lai, and S. Stifani. 2001. The winged-helix protein brain factor 1 interacts with Groucho and Hes proteins to repress transcription. Mol. Cell. Biol. 21:1962-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, H., and S. W. Emmons. 2002. Caenorhabditis elegans unc-37/groucho interacts genetically with components of the transcriptional mediator complex. Genetics 160:799-803. [DOI] [PMC free article] [PubMed] [Google Scholar]