Abstract
The cellular responses to double-stranded breaks (DSBs) typically involve the extensive accumulation of checkpoint proteins in chromatin surrounding the damaged DNA. One well-characterized example involves the checkpoint protein Crb2 in the fission yeast Schizosaccharomyces pombe. The accumulation of Crb2 at DSBs requires the C-terminal phosphorylation of histone H2A (known as γ-H2A) by ATM family kinases in chromatin surrounding the break. It also requires the constitutive methylation of histone H4 on lysine-20 (K20). Interestingly, neither type of histone modification is essential for the Crb2-dependent checkpoint response. However, H4-K20 methylation is essential in a crb2-T215A strain that lacks a cyclin-dependent kinase phosphorylation site in Crb2. Here we explain this genetic interaction by describing a previously overlooked effect of the crb2-T215A mutation. We show that crb2-T215A cells are able to initiate but not sustain a checkpoint response. We also report that γ-H2A is essential for the DNA damage checkpoint in crb2-T215A cells. Importantly, we show that inactivation of Cdc2 in γ-H2A-defective cells impairs Crb2-dependent signaling to the checkpoint kinase Chk1. These findings demonstrate that full Crb2 activity requires phosphorylation of threonine-215 by Cdc2. This regulation of Crb2 is independent of the histone modifications that are required for the hyperaccumulation of Crb2 at DSBs.
Phosphorylation of histone H2A(X) is one of the earliest, most striking, and highly conserved cellular responses triggered by DNA double-stranded breaks (DSBs) (17, 31, 41). This phosphorylation, which is localized to chromatin adjacent to the break site and occurs at an SQE motif near the C terminus of H2A(X), is catalyzed by protein kinases of the ATM family. These proteins have central roles in DNA damage response and telomere maintenance. Members of this family include ATM and ATR in mammals (17, 31), Tel1 (ATM) and Mec1 (ATR) in Saccharomyces cerevisiae (10, 32), and Tel1 and Rad3 (ATR) in Schizosaccharomyces pombe (29).
Histone H2A(X) has been one of the most widely studied substrates of the ATM family kinases, and yet the functional significance of the phosphorylated form of histone H2A(X), known as γ-H2A(X), is not yet fully understood (41). Many of the initial studies of γ-H2A(X) focused on its role in checkpoint signaling, culminating in evidence that γ-H2A(X) associates directly with members of a family of DNA damage checkpoint proteins that have tandem C-terminal BRCT domains (16, 29, 44). Included in this family are metazoan 53BP1, S. pombe Crb2/Rhp9, and S. cerevisiae Rad9. Indeed, γ-H2A(X) is required for the large-scale accumulation of Crb2 at ionizing radiation (IR)-induced DSBs in fission yeast (29). In mouse embryo fibroblasts (MEFs), H2A(X) is required for the persistence (but not the initial formation) of 53BP1 nuclear foci in response to IR (6). However, elimination of γ-H2A(X) does not abolish checkpoint responses in fission yeast, budding yeast, and MEFs (10, 29, 32, 41), even though Crb2, Rad9, and 53BP1 (at low doses of IR) are essential for the checkpoint arrest of the cell cycle in response to DNA damage (8, 16, 34, 43, 45, 46). In fact, the checkpoint defect of S. pombe hta1,2-AQE cells, in which both histone H2A genes encode proteins that lack the C-terminal SQE motifs, is quite weak (29). The equivalent mutant strains in S. cerevisiae have no reported checkpoint defects (10, 32).
Recent studies have revealed that additional forms of histone modification are required for formation of 53BP1 and Crb2 foci in response to DNA damage. Specifically, histone H3-K79 methylation by Dot1 methyltransferase is required for formation of 53BP1 foci in U2OS osteosarcoma cells (21), while histone H4 lysine-20 (K20) methylation by Set9 methyltransferase is required for formation of Crb2 foci in S. pombe (35). However, neither Set9 in S. pombe (35) nor Dot1 in S. cerevisiae (19) is required to arrest division in response to DNA damage.
In addition to evidence linking γ-H2A(X) to recruitment of checkpoint signaling proteins, recent studies carried out with S. cerevisiae have uncovered unexpected roles for γ-H2A(X) in the recruitment of protein complexes involved in chromatin modification or higher-order chromatin structure (41). The NuA4 histone acetyltransferase complex was found to associate with DSBs through binding of its subunits to γ-H2A(X), as were the INO80 and SWR1 chromatin remodeling complexes (2, 9, 28, 40). In addition, γ-H2A(X) was found to be important for loading of the cohesin complex onto chromosomes in the vicinity of DSBs (37, 39). These data are interesting in light of evidence that H2AX−/− MEFs are defective in homologous recombination between sister chromatids (47) and with evidence that γ-H2A(X) is essential for the efficient repair of “checkpoint-blind” DNA damage during S phase in S. cerevisiae (32). There is also evidence that γ-H2A(X) is necessary for the recruitment of other BRCT domain-containing proteins involved in DNA damage signaling, such as MDC1/NFBD1 and NBS1 (24, 26, 36).
In view of the evidence for the involvement of γ-H2A(X) in the DSB recruitment of a variety of proteins involved in checkpoint signaling, chromatin modification, and chromatid cohesion, it is surprising that γ-H2A(X)-defective mutants in budding yeast, fission yeast, and MEFs are far less sensitive to DSBs than are mutants that lack proteins required for DSB repair or checkpoints (7, 29, 32). For example, more than half of the cells in a hta1,2-AQE culture survive 300 Gy of IR, whereas only 1 in ∼10,000 of the cells in rad3Δ or crb2Δ cultures survive this dose of IR (29). S. cerevisiae hta1,2-AQE cells are also only very marginally sensitive to IR (32). One possible explanation of these findings is that γ-H2A(X) acts redundantly with another mechanism to transmit checkpoint signals. A recent discovery consistent with this possibility was reported by Sanders et al. (35), who found that combining the set9Δ mutation with the crb2-T215A mutation resulted in a complete elimination of the IR-induced DNA damage checkpoint. The crb2-T215A mutation was first reported by Esahi and Yanagida (15), who found that the cyclin-dependent kinase Cdc2 phosphorylated Crb2 on threonine-215. The crb2-T215A mutation did not impair the checkpoint arrest in response to UV, but it did prevent cells from resuming cell division after the DNA damage was repaired. These findings indicated that phosphorylation of Crb2 by Cdc2 is required to restart the cell cycle from a checkpoint arrest (15). A later study indicated that T215 phosphorylation of Crb2 also controls a putative DNA repair function of Crb2 that is linked to Rqh1 DNA helicase (5).
It is unclear why Set9 is required for the DNA damage checkpoint in crb2-T215A cells, but the fact that set9Δ and hta1,2-AQE mutations have similar effects on Crb2 accumulation at DSBs (29, 35) suggested to us that T215 phosphorylation of Crb2 might act redundantly with γ-H2A(X) to transmit checkpoint signals. In support of this hypothesis, here we report that crb2-T215A hta1,2-AQE cells are unable to arrest division in response to DNA damage. We further show that this genetic interaction, and hence also the checkpoint abolishment in crb2-T215A set9Δ cells, can be explained by a previously unrecognized checkpoint defect caused by the crb2-T215A mutation. We also report that Cdc2 activity is required for efficient checkpoint signaling through Crb2 in hta1,2-AQE cells. These findings show that phosphorylation of Crb2 at T215 is important for checkpoint signaling, and it acts cooperatively with the γ-H2A(X) and Set9-dependent accumulation of Crb2 at DSBs to activate the checkpoint response.
MATERIALS AND METHODS
Fission yeast strains and plasmids.
Strains used in this study were constructed by standard techniques (1), and their genotypes and figure numbers are listed in Table 1. Most of the strains in Table 1 carry previously described mutations or epitope tags (12, 29). The crb2-T215A, crb2-T215E, and crb2-T215D mutations were generated by multistep PCR and were each cloned as a BamHI-SacI fragment to generate pJK148+crb2-T215A (BamHI), pJK148+crb2-T215D (BamHI), and pJK148+crb2-T215E (BamHI). Plasmids pJK148+YFP-crb2 and pJK148+YFP-crb2-T215A were generated by cloning a BamHI-BglII fragment of 2× yellow fluorescent protein (YFP) into pJK148+crb2 (BamHI) and pJK148+crb2-T215A (BamHI), respectively. Mutational primers used are the following: crb2-T215A (BAM150, gtagagactGCTcctactcgactggc; BAM151, gccagtcgagtaggAGCagtctctac [uppercase indicates mutated sequences]), crb2-T215E (BAM152, gtagagactGAAcctactcgactggc; BAM153, gccagtcgagtaggTTCagtctctac), and crb2-T215D (BAM154, gtagagactGACcctactcgactggc; BAM155, gccagtcgagtaggGTCagtctctac).
TABLE 1.
Strains used in this study
| Figure no. | Short genotype | Strain | Full genotype |
|---|---|---|---|
| 1A | wta | TMN3289 | h−leu1-32 ura4-D18 his3-D1 |
| 1A | crb2Δ | TMN2941 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 crb2::ura4+ |
| 1A | crb2-T215A | TMN3543 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 crb2-T215A |
| 1B, C | wt | TMN3311 | h−leu1-32 ura4-D18 ade6-M210 his3-D1 cdc25-22 |
| 1B | crb2-T215A | TMN3557 | h+leu1-32 ura4-D18 ade6-M216 his3-D1 crb2-T215A cdc25-22 |
| 1C | hta1,2-AQE | TMN3312 | h−leu1-32 ura4-D18 ade6-M210 his3-D1 cdc25-22 hta1-S129A::ura4+hta2-S128A::his3+ |
| 1D | hta1,2-AQE crb2-T215A | TMN3556 | h+leu1-32 ura4-D18 ade6-M216 his3-D1 crb2-T215A hta1-S129A::ura4+hta2-S128A::his3+cdc25-22 |
| 1D | crb2Δ | TMN3555 | h+leu1-32 ura4-D18 ade6-M216 his3-D1 crb2::ura4+cdc25-22 |
| 1E | chk1+ | TMN3288 | h+leu1-32 ura4-D18 his3-D1 |
| 1E | chk1-myc | TMN3309 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 chk1+::9myc2HA6His::ura4+ |
| 1E | chk1-myc hta1,2-AQE | TMN3310 | h−leu1-32 ura4-D18 ade6-M210 his3-D1 chk1+::9myc2HA6His::ura4+hta1-S129A:ura4+hta2-S128A:his3+ |
| 1E | chk1-myc crb2-T215A | TMN3558 | h+leu1-32 ura4-D18 his3-D1 crb2-T215A chk1+::9myc2HA6His::ura4+ |
| 1E | chk1-myc hta1,2-AQE crb2-T215A | TMN3559 | h+leu1-32 ura4-D18 his3-D1 crb2-T215A chk1+::9myc2HA6His::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 1E | chk1-myc crb2Δ | TMN3560 | h+leu1-32 ura4-D18 his3-D1 crb2::ura4+chk1+::9myc2HA6His::ura4+ |
| 2A | crb2Δ | LLD3423 | h−leu1-32 ura4-D18 his3-D1 crb2-D2::ura4+ |
| 2A | crb2+ | TMN3288 | See 1Eb |
| 2A | hta1,2-AQE crb2+ | TMN3291 | h−leu1-32 ura4-D18 his3-D1 hta1-S129A::ura4+hta2-S128A::his3+ |
| 2A | crb2-T215A | TMN3561 | h+leu1-32 ura4-D18 ade6-M216 his3-D1 crb2-T215A |
| 2A | hta1,2-AQE crb2-T215A | TMN3562 | h+leu1-32 ura4-D18 ade6-M216 his3-D1 crb2-T215A hta1-S129A::ura4+hta2-S128A::his3+ |
| 2A | rad3Δcrb2+ | TMN3563 | h+leu1-32 ura4-D18 ade6-M216 his3-D1 rad3-D2::LEU2 |
| 2B, C | crb2+ | LLD3260 | h−leu1-32::2xYFP-crb2-leu1+ura4-D18 crb2-D2::ura4+ |
| 2B, C | crb2-T215A | LLD3564 | h−leu1-32::2xYFP-crb2-T215A-leu1+ura4-D18 crb2-D2::ura4+ |
| 2B, C | crb2+hta1,2-AQE | LLD3340 | h−leu1-32::2xYFP-crb2-leu1+ura4-D18 his3-D1 crb2-D2::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 2B, C | crb2-T215A hta1,2-AQE | LLD3565 | h+leu1-32::2xYFP-crb2-T215A-leu1+ura4-D18 his3-D1 crb2-D2::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 3A, C | wt | TMN3289 | See 1Ac |
| 3A, C | hta1,2-AQE | TMN3293 | h−leu1-32 ura4-D18 ade6-M210 his3-D1 hta1-S129A::ura4+hta2-S128A::his3+ |
| 3A-C | crb2-T215A | TMN3543 | See 1A |
| 3A-C | chk1Δ | TMN2943 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 chk1::ura4+ |
| 3A, C | crb2Δhta1,2-AQE | TMN3297 | h−leu1-32 ura4-D18 ade6-M210 his3-D1 crb2::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 3A, C | crb2-T215A hta1,2-AQE | TMN3546 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 crb2-T215A hta1-S129A::ura4+hta2-S128A::his3+ |
| 3A, C | crb2-T215A chk1Δhta1,2-AQE | TMN3554 | h+leu1-32 ura4-D18 ade6-M216 his3-D1 crb2-T215A chk1::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 3A, C | crb2Δ | TMN2941 | See 1A |
| 3A, C | crb2Δchk1Δ | TMN3305 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 crb2::ura4+chk1::ura4+ |
| 3A-C | crb2-T215A chk1Δ | TMN3549 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 crb2-T215A chk1::ura4+ |
| 3B, C | rqh1Δ | TMN3301 | h+leu1-32 ura4-D18 his3-D1 rqh1::ura4+ |
| 3B, C | crb2-T215A rqh1Δ | TMN3547 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 crb2-T215A rqh1::ura4+ |
| 3B, C | chk1Δrqh1Δ | TMN3304 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 chk1::ura4+rqh1::ura4+ |
| 3B, C | crb2Δchk1Δrqh1Δ | TMN3306 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 crb2::ura4+chk1::ura4+rqh1::ura4+ |
| 3B, C | crb2-T215A chk1Δrqh1Δ | TMN3552 | h+leu1-32 ura4-D18 ade6-M216 his3-D1 crb2-T215A chk1::ura4+rqh1::ura4+ |
| 3B, C | crb2-T215A rqh1Δhta1,2-AQE | TMN3553 | h+leu1-32 ura4-D18 ade6-M210 his3-D1 crb2-T215A rqh1::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 3C | rhq1Δhta1,2-AQE | TMN3302 | h−leu1-32 ura4-D18 ade6-M210 his3-D1 rqh1::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 3C | chk1Δhta1,2-AQE | TMN3298 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 chk1::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 3C | crb2Δchk1Δhta1,2-AQE | TMN3308 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 crb2::ura4+chk1::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 3C | crb2Δrqh1Δ | TMN3303 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 crb2::ura4+rqh1::ura4+ |
| 3C | crb2Δrqh1Δhta1,2-AQE | TMN3307 | h−leu1-32 ura4-D18 ade6-M210 his3-D1 crb2::ura4+rqh1::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 3C | chk1Δrqh1Δhta1,2-AQE | TMN3422 | h−leu1-32 ura4-D18 ade6-M216 his3-D1 chk1::ura4+rqh1::ura4+hta1-S129A::ura4+hta2-S128A::his3+ |
| 4 | wt | TMN3289 | See 1A |
| 4 | crb2-T215A | TMN3543 | See 1A |
| 4 | crb2-T215D | BAM4090 | h−leu1-32 ura4-D18 crb2-T215D |
| 4 | crb2-T215E | BAM4089 | h−leu1-32 ura4-D18 crb2-T215E |
| 5 | chk1+ | OM810 | h−leu1-32 |
| 5 | chk1-myc hta1,2-AQE | TMN3310 | See 1E |
| 5 | chk1-myc | TMN3309 | See 1E |
| 5 | chk1-myc cdc2-L7 | TN4057 | h−leu1-32 ura4-D18 cdc2-L7 chk1+::9myc2HA6His::ura4+ |
| 5 | chk1-myc cdc2-L7 hta1,2-AQE | TN4073 | h−leu1-32 ura4-D18 ade6-M210 cdc2-L7 chk1+::9myc2HA6His::ura4+hta1-S129A:ura4+hta2-S128A:his3+ |
| 5 | chk1-myc cdc2-M26 | TN4063 | h−leu1-32 ura4-D18 cdc2-M26 chk1+::9myc2HA6His::ura4+ |
| 5 | chk1-myc cdc2-M26 hta1,2-AQE | TN4077 | h−leu1-32 ura4-D18 ade6-M210 cdc2-M26 chk1+::9myc2HA6His::ura4+hta1-S129A:ura4+hta2-S128A:his3+ |
The crb2-T215A allele was generated by integrating a BamHI-SacI fragment derived from the pJK-148+crb2-T215A plasmid at the crb2 locus in crb2::ura4+ cells. The crb2-T215D and crb2-T215E alleles were generated in a similar manner by utilizing pJK148+crb2-T215D(BamHI) and pJK148+crb2-T215E(BamHI), respectively. YFP-crb2-T215A (leu1-32::2xYFP-crb2-T215A-leu1+) was generated by integrating pJK-148+YFP-crb2-T215A at the leu1 locus in leu1-32 cells. Integrated crb2-T215A, crb2-T215D, crb2-T215E, and YFP-crb2-T215A mutations were also confirmed by sequencing genomic DNA.
Microscopy.
For YFP-Crb2 fluorescence microscopy, images were photographed at eight 0.5-μm-interval Z-sections, projected into one image as described previously (12), and then quantified for focus number.
IR sensitivity analysis.
Exponentially growing cells were irradiated with IR in YES (yeast extract-supplemented) medium (1) with a 3.2-Gy/min 137Cs source at room temperature (∼26°C) and then plated onto YES plates at 32°C. Colony numbers were counted 4 days after plating to determine the cell survival rate.
Cell cycle analysis.
For cdc25-22 synchronization studies, cells were incubated at 35.5°C for 2.5 h to arrest in G2, irradiated with IR, allowed to recover at 25°C for various time points, fixed with 2.5% glutaraldehyde, stained with calcofluor, and then examined for cell cycle progression under a microscope.
Western blots.
Whole-cell extracts were prepared from exponentially growing cells and processed for Western blot analysis as described previously (29). For cdc2ts experiments, cells were shifted from 25°C to 36°C for 1 h, treated with 5 mU/ml bleomycin for 40 min, and then harvested for preparation of whole-cell extract. For Chk1-myc, a mouse anti-Myc antibody (9E10; Santa Cruz Biotechnology) was used. For Crb2, a rabbit polyclonal anti-Crb2 antibody (12) was used. Western blots were quantified with NIH ImageJ software to analyze TIFF files generated by a Bio-Rad gel documentation system or TIFF files of scanned X-ray films.
RESULTS
The DNA damage checkpoint is partially defective in crb2-T215A cells.
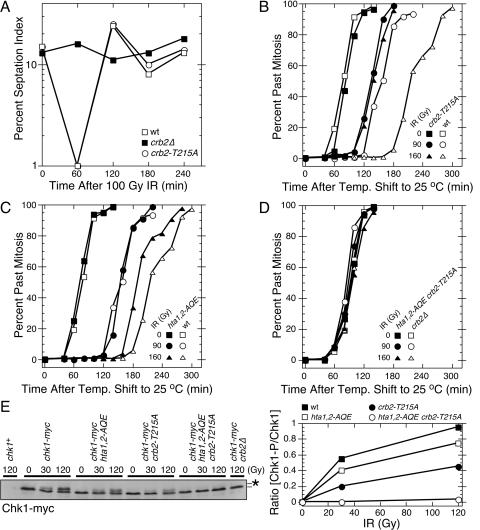
The absence of a DNA damage checkpoint in set9Δ crb2-T215A cells (35) is not readily explained by the phenotypes attributed to crb2-T215A cells, which are an inability to recover from a checkpoint arrest (15) and an increase in Rqh1-dependent recombination following DNA damage (5). We hypothesized that the crb2-T215A mutation might cause a hitherto-unrecognized defect in the checkpoint function of Crb2 that was synergistic with the effect of the set9Δ mutation. To explore this possibility, checkpoint studies were performed with a crb2-T215A strain that was created by replacing the genomic copy of crb2+ with crb2-T215A. The septation index, which is a marker of cell division, was measured in crb2+, crb2Δ, and crb2-T215A cultures treated with 100 Gy of IR. This amount of IR caused a transient checkpoint arrest in wild-type (crb2+) cells, while crb2Δ cells failed to arrest division (Fig. 1A). In this assay the behavior of the crb2-T215A cells was identical to that of the wild type (Fig. 1A).
FIG. 1.
The DNA damage checkpoint is abolished in hta1,2-AQE crb2-T215A cells. (A) Septation index analysis of asynchronous cultures of wild-type (wt), crb2-T215A, and crb2Δ cells exposed to 100 Gy IR. (B to D) Mitotic progression analysis of synchronous cultures of wild-type (wt), crb2-T215A, hta1,2-AQE, or hta1,2-AQE crb2-T215A cells. Cells were synchronized by cdc25-22 mutation at 35.5°C, mock treated or irradiated with the indicated dose of IR at room temperature, and analyzed for cell cycle progression. (E) Immunoblot analysis of Chk1-myc in asynchronous cultures of cells harvested immediately after irradiation with the indicated dose of IR. The position of phosphorylated Chk1 is indicated by the asterisk. The immunoblot was quantified using ImageJ software. All the data in all panels are representative of repeated experiments.
We were concerned that checkpoint assays performed with unsynchronized cultures were insufficiently sensitive to detect a partial checkpoint defect in crb2-T215A cells. To improve the sensitivity of the assay, we used the temperature-sensitive cdc25-22 mutation to synchronize cells in late G2 phase. In response to 90 Gy of IR, we found that crb2+ cells experienced an ∼85-min checkpoint delay, whereas the isogenic crb2-T215A strain delayed mitosis for only ∼50 min (Fig. 1B). The checkpoint defect of the crb2-T215A strain was further accentuated in response to 160 Gy of IR, in which crb2+ cells delayed mitosis for ∼140 min while the crb2-T215A strain delayed mitosis for ∼50-min (Fig. 1B).
We carried out a similar set of experiments with hta1,2-AQE cells. In response to 90 Gy of IR, there was no obvious difference between the hta1,2-AQE cells and wild-type cells (Fig. 1C). However, in response to 160 Gy of IR, the hta1,2-AQE cells resumed division ∼30 min earlier than the hta1,2+ control cells (Fig. 1C). These data are consistent with our earlier studies of hta1,2-AQE cells (29).
From these results we conclude that both crb2-T215A and hta1,2-AQE cells have a partial checkpoint defect. These mutants are able to establish a checkpoint arrest but are unable to maintain it for the proper time.
Abolition of the DNA damage checkpoint in hta1,2-AQE crb2-T215A cells.
To determine whether the hta1,2-AQE and crb2-T215A mutations have additive or synergistic effects on the DNA damage checkpoint, we created a cdc25-22 hta1,2-AQE crb2-T215A strain and performed the IR-induced checkpoint assay with synchronized cells. The checkpoint was completely abolished in this strain (Fig. 1D). This result was confirmed in a checkpoint assay carried out with an asynchronous culture of cells in a cdc25+ background, in which we found that the IR-induced checkpoint was abolished in hta1,2-AQE crb2-T215A cells (data not shown).
To confirm these findings by an independent biochemical assay of checkpoint signaling, we measured the phosphorylation of Chk1 that is induced by DNA damage. Chk1 phosphorylation correlates with checkpoint activation and requires Crb2 (34, 42). As shown in the Chk1 immunoblot (Fig. 1E, left panel) and confirmed by quantification of the immunoblot signals (Fig. 1E, right panel), the hta1,2-AQE and crb2-T215A mutations impaired phosphorylation of Chk1 at both 30 and 120 Gy of IR. The crb2-T215A mutation had the greater effect (Fig. 1E, right panel), reducing the amount of phosphorylated Chk1 by ∼50%. These data correlate with checkpoint arrest assays in which the crb2-T215A mutation had the greater effect (Fig. 1B and C). Strikingly, the IR-induced phosphorylation of Chk1 was completely abolished in hta1,2-AQE crb2-T215A cells (Fig. 1E). These findings confirm those of the checkpoint arrest assays.
From these results we conclude that the hta1,2-AQE and crb2-T215A mutations independently impair the checkpoint function of Crb2. When combined together they abolish the checkpoint function of Crb2. This situation is analogous to genetic interaction described for the set9Δ and crb2-T215A mutations (35).
Analysis of Crb2-T215A phosphorylation and localization.
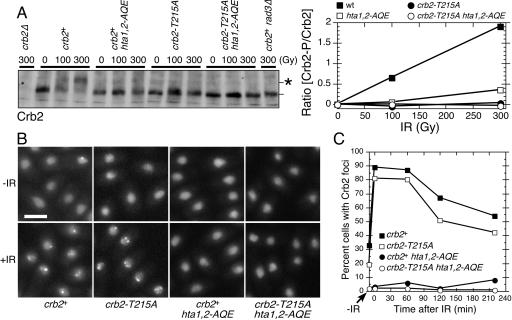
The hta1,2-AQE and crb2-T215A mutations do not noticeably effect the abundance of Crb2 (15, 29), but we were concerned that when combined together these mutations might destabilize Crb2. To address this possibility, we monitored the abundance of Crb2 by immunoblotting with mutant strains exposed to IR. This analysis showed that abundance of Crb2 was unaffected by hta1,2-AQE and crb2-T215A mutations, either separately or when combined in the same strain (Fig. 2A). The immunoblots confirmed earlier studies in which both the hta1,2-AQE and crb2-T215A mutations were shown to impair the Rad3-dependent hyperphosphorylation of Crb2 that is induced by DNA damage (15, 29, 34). This hyperphosphorylation causes Crb2 to have reduced electrophoretic mobility (Fig. 2A).
FIG. 2.
The crb2-T215A mutation decreases IR-induced Crb2 phosphorylation but not focus formation. (A) Immunoblot analysis of Crb2 in asynchronous cultures of cells harvested immediately after irradiation with the indicated dose of IR. The position of hyperphosphorylated Crb2 is indicated by the asterisk. The immunoblot was quantified using ImageJ software. (B) YFP-Crb2 localization was observed for indicated cells harvested immediately after irradiation with IR (36 Gy). Size bar, 5 μm. (C) Time-dependent changes in YFP-Crb2 foci after exposure to IR (36 Gy). Numbers of cells with Crb2 foci at various times after IR treatment were determined and plotted as percentages of cells with Crb2 foci.
We also examined the localization of Crb2 in the mutant strains. These experiments were performed by microscopic analysis of live cells that expressed an integrated copy of YFP-tagged Crb2 or Crb2-T215A. Crb2 with an N-terminal YFP tag is a fully functional protein that localizes in the nucleus and forms bright foci at sites of DNA damage (12). These foci appear within a few minutes after exposure to IR. In crb2-T215A cells, mutant YFP-Crb2 localized in the nucleus and formed foci in response to IR (Fig. 2B). This pattern of Crb2 localization was not significantly different from that for the wild type (Fig. 2C). As seen previously in hta1,2-AQE cells (29), YFP-Crb2 was properly localized in the nucleus, but it failed to form foci in response to IR (Fig. 2B and C). The pattern of Crb2 localization in crb2-T215A hta1,2-AQE cells was identical to that in hta1,2-AQE cells (Fig. 2B and C).
From these results we conclude that both the crb2-T215A and hta1,2-AQE mutations cause a reduction in the DNA damage-induced hyperphosphorylation of Crb2, but they are likely to have these effects for different reasons because only the hta1,2-AQE mutations cause a defect in the accumulation of Crb2 at sites of DNA damage.
Equivalent effects of the crb2-T215A and crb2Δ mutations in hta1,2-AQE cells.
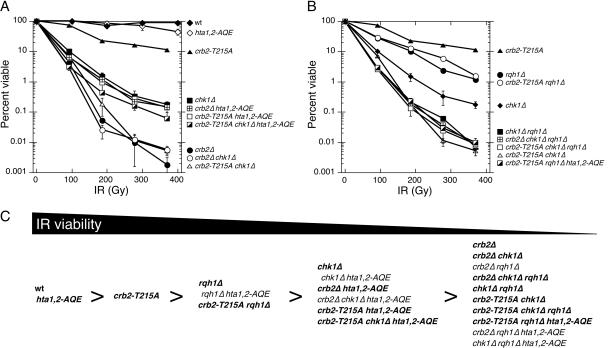
Having found that crb2-T215A hta1,2-AQE cells are unable to arrest division in response to DNA damage (Fig. 1D), we proceeded to carry out IR survival assays to determine if Crb2 function is fully defective in these cells. We found that crb2-T215A hta1,2-AQE and crb2Δ hta1,2-AQE strains were equally sensitive to IR (Fig. 3A and C). These findings demonstrate that all functions of Crb2 that contribute to IR survival are fully eliminated in the crb2-T215A hta1,2-AQE background.
FIG. 3.
IR survival experiments involving crb2-T215A. (A and B) Genetic interactions of crb2-T215A with chk1Δ, rqh1Δ, and hta1,2-AQE in response to IR treatment. Experiments are repeated at least three times, and error bars correspond to standard deviations. (C) Summary of IR sensitivity for various mutant combinations. Bold type indicates results shown in this study, while normal type indicates results from a previous study (29).
It should be noted that crb2Δ cells are substantially more sensitive to IR than crb2Δ hta1,2-AQE cells (Fig. 3A). This suppressive effect of the hta1,2-AQE mutations was noted in previous studies (29). These genetic interactions indicate that phosphorylation of the C terminus of histone H2A is counterproductive to survival in the absence of Crb2. The damage-induced cell cycle arrest is fully eliminated in crb2Δ cells; therefore, we conclude that in the absence of Crb2, γ-H2A(X) must hinder DNA repair or promote a nonproductive mechanism of DNA repair.
In the hta1,2-AQE background, the crb2-T215A and crb2Δ mutations caused a degree of IR sensitivity that was approximately equivalent to that of chk1Δ cells (Fig. 3A). The hta1,2-AQE mutations do not suppress chk1Δ (29); hence, the IR sensitivity of crb2-T215A hta1,2-AQE and crb2Δ hta1,2-AQE strains can be explained by an inability to activate Chk1. It should be noted that the suppressive effect of hta1,2-AQE was also observed in crb2-T215A chk1Δ and crb2Δ chk1Δ strains, which appeared to be equal to crb2Δ strains in IR sensitivity (Fig. 3A).
We previously observed that the suppressive effect of the hta1,2-AQE mutations in a crb2Δ background depended on the DNA helicase Rqh1 (29). We observed the same effect with the crb2-T215A hta1,2-AQE cells, in which the presence of the rqh1Δ mutation reduced IR resistance to a level comparable to that of crb2Δ cells (Fig. 3B and C). Therefore, in the absence of Crb2 function in a hta1,2-AQE background, Rqh1 plays an important role in IR survival.
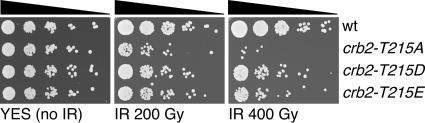
Rescue of cdc2-T215A by amino acid substitutions that mimic phosphorylation.
Our studies indicated that phosphorylation of Crb2 on threonine-215 by Cdc2 plays a significant role in the DNA damage checkpoint. We carried out additional experiments to test this hypothesis. We first addressed whether the effect of the crb2-T215A mutation is caused by an absence of phosphorylation at residue 215. Aspartic acid (D) and glutamic acid (E) substitutions have been used with occasional success to mimic phosphorylated threonine (11, 25). If the phosphorylation of T215 is required for the proper function of Crb2, we would expect that the crb2-T215D and crb2-T215E alleles might have effects that are less severe than those of crb2-T215A. Accordingly, we replaced crb2+ with crb2-T215D and crb2-T215E alleles. As observed for crb2-T215A cells, the plating efficiencies and growth rates of crb2-T215D and crb2-T215E strains did not noticeably differ from those of the wild type in the absence of genotoxic stress (Fig. 4). When exposed to 200 or 400 Gy IR, the survival of the crb2-T215D and crb2-T215E strains was reduced relative to that of the wild type but increased relative to that of the isogenic crb2-T215A cells (Fig. 4). The intermediate effects of the crb2-T215D and crb2-T215E alleles indicate that these charged amino acids cannot fully mimic phosphorylated threonine at position 215. Nevertheless, the improved survival of crb2-T215D and crb2-T215E strains relative to that of the crb2-T215A strain supports the conclusion that phosphorylation of T215 is required for the full activity of Crb2.
FIG. 4.
Analysis of crb2-T215D and crb2-T215E mutants. Fivefold serial dilutions of the indicated strains were exposed to 0, 200, or 400 Gy IR.
Inactivation of Cdc2 in hta1,2-AQE cells impairs checkpoint signaling.
T215 phosphorylation of Crb2 requires Cdc2 (15). If this phosphorylation acts cooperatively with γ-H2A(X) in checkpoint signaling, we would expect that inactivation of Cdc2 kinase in combination with hta1,2-AQE should synergistically impair checkpoint signaling. This proposition was tested by combining hta1,2-AQE mutations with two well-characterized temperature-sensitive cdc2 mutant alleles, cdc2-L7 and cdc2-M26 (3). Direct observation of the cell cycle arrest phenotype was not possible because the cdc2ts strains arrest in G2 phase at the restrictive temperature. Instead, the damage checkpoint was assayed by monitoring the DNA damage-induced phosphorylation of Chk1. Bleomycin was used to damage DNA because the restrictive temperature could not be maintained in the gamma irradiator. Control experiments confirmed that the bleomycin-induced checkpoint arrest was abolished in crb2-T215A hta1,2-AQE cells (data not shown).
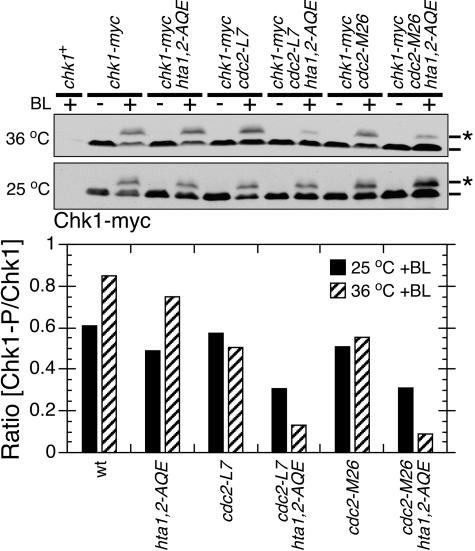
Cells incubated at 36°C or at the permissive temperature (25°C) were treated with bleomycin (5 mU/ml) for 40 min prior to harvest for immunoblotting. In wild-type and hta1,2-AQE cells, incubation at 36°C caused an ∼35% increase in the amount of phosphorylated Chk1 (Fig. 5). The reason for this temperature effect is unknown. It was not observed with the cdc2ts strains (Fig. 5), indicating that there was less-efficient checkpoint signaling in these cells. Relative to the wild type, there was a moderate reduction of Chk1 phosphorylation in hta1,2-AQE cells at both temperatures (Fig. 5). Most noticeably, there was a synergistic effect of combining the hta1,2-AQE and cdc2ts alleles, particularly at 36°C (Fig. 5). Relative to the wild type at 36°C, Chk1 phosphorylation was reduced ∼5.7-fold in cdc2-L7 hta1,2-AQE cells and ∼8.5-fold in cdc2-M26 hta1,2-AQE cells.
FIG. 5.
Cdc2 activity is required for efficient phosphorylation of Chk1 in hta1,2-AQE cells. Inactivation of Cdc2 in the hta1,2-AQE background specifically reduces DNA damage-induced Chk1 mobility shift in response to 5 mU/ml bleomycin treatment for 40 min. Cells were treated at 25°C or 36°C as indicated. The Chk1 mobility change caused by phosphorylation was quantified as described in the legend to Fig. 1.
The synergistic effects of hta1,2-AQE and cdc2ts alleles on Chk1 phosphorylation were consistent with results of DNA checkpoint and cell survival assays of crb2-T215A hta1,2-AQE cells. Taken together, they support the conclusion that Cdc2-dependent phosphorylation of Crb2 acts cooperatively with the Rad3/Tel1-dependent phosphorylation of histone H2A in checkpoint signaling.
DISCUSSION
We have discovered that crb2-T215A cells are unable to maintain a checkpoint arrest in response to DNA damage (Fig. 1). This phenotype of crb2-T215A cells correlates with a sensitivity to DNA damage that is intermediate between those of the wild type and checkpoint null mutants (Fig. 3). Strikingly, crb2-T215A has a strong synergistic interaction with the set9Δ (35) and hta1,2-AQE mutations (Fig. 1), which themselves cause a modest checkpoint defect. The DNA damage checkpoint is fully abrogated in hta1,2-AQE crb2-T215A cells (Fig. 1). Consistent with these findings and the evidence that Cdc2 catalyzes the phosphorylation of Crb2 at threonine-215 (15), signaling to Chk1 is substantially impaired when Cdc2 is inactivated in a hta1,2-AQE background (Fig. 5). Moreover, the effect of crb2-T215A on IR survival can be largely rescued by replacement of alanine with positively charged amino acids that partially mimic phosphorylated threonine-215 (Fig. 4). From these results we conclude that T215 phosphorylation of Crb2 is important for the DNA damage checkpoint and it acts cooperatively with the mechanisms that are required for accumulation of Crb2 at sites of DNA damage, namely, the C-terminal phosphorylation of histone H2A by Rad3/Tel1 and the methylation of histone H4-K20 by Set9 (29, 35).
A revised view of the role of γ-H2A(X) in the DNA damage response.
γ-H2A(X) has been implicated in many of the cellular responses to DNA damage, ranging from cell cycle arrest (the classic checkpoint response) to the recruitment of chromatin remodeling factors to sites of DNA damage. The perceived importance of γ-H2A(X) in the checkpoint response has faded with reports that γ-H2A(X)-defective mutants in S. cerevisiae, S. pombe, and mouse cells have only subtle checkpoint defects. However, as our studies now show, γ-H2A(X) is essential for the DNA damage checkpoint in crb2-T215A cells. These findings point towards γ-H2A(X)-dependent accumulation of Crb2 at sites of DNA damage as being one of the mechanisms that ensure the sufficient duration of a checkpoint arrest.
These findings may have implications for the role of γ-H2A(X) in the recruitment of chromatin remodeling factors. We are struck by the fact that elimination of γ-H2A(X) partially rescues crb2Δ mutants in S. pombe, implying that in the absence of Crb2, C-terminal phosphorylation of histone H2A interferes with DNA repair or promotes pathways of repair that are counterproductive for cell survival. From these results it appears that the noncheckpoint functions of γ-H2A(X), be they recruitment of chromatin remodeling factors or something else, are able to promote cell survival only when done in the context of accumulation of Crb2 at sites of DNA damage. These findings suggest that there may be intimate association of Crb2 and chromatin remodeling factors at sites of DNA damage.
Threonine-215 phosphorylation and checkpoint recovery.
Esashi and Yanagida (15) found that the crb2-T215A mutation prevented or delayed recovery from a UV-induced checkpoint arrest, whereas we observed the opposite effect in the response to IR (Fig. 1A). The different findings are not explained by the use of UV versus IR (our unpublished data) but may be explained by differences in the amount of crb2-T215A expression. Esashi and Yanagida used crb2Δ cells carrying crb2-T215A on a multicopy plasmid (15), whereas genomic crb2+ was replaced with crb2-T215A in our strain.
Regulation of Crb2 by Cdc2.
The DNA damage checkpoint delays mitosis by preventing Cdc25 from activating Cdc2 (33); thus, it was unexpected that Cdc2 and Rad3 would act cooperatively to enforce the G2-M checkpoint. Cdc2 has low but biologically significant activity in G2 phase that is required to prevent licensing of replication origins (20). It now appears this activity also plays an important role in the DNA damage checkpoint. This mechanism may be used for cell cycle regulation of checkpoints and DNA repair. To our knowledge, this is the first Cdc2 phosphorylation site found to be critical for the DNA damage checkpoint. This function of Cdc2 is distinct from the role of budding yeast Cdc28 (Cdk1) in modulating activation of the checkpoint kinase Rad53 by controlling the resection rate at a DSB (22).
Potential mechanism of Cdc2's contribution for DNA damage checkpoint.
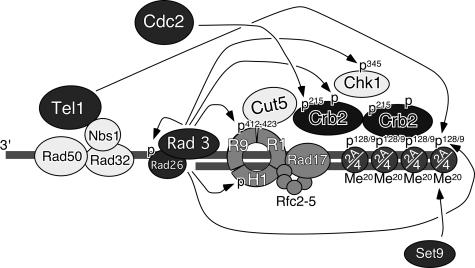
Based on our current data and those of previous papers, we propose a model for the checkpoint response as illustrated in Fig. 6. In this model, Rad3 and Tel1 kinases act in association with their cofactors (Rad26 and Rad32 [Mre11]-Rad50-Nbs1 complex, respectively) to recognize DSBs and generate γ-H2A. γ-H2A, in conjunction with histone K4-K20 methylation by Set9, recruits and retains Crb2 at DSBs. We have previously reported that Rad3-Rad26, Rad9-Rad1-Hus1, and Rad17-Rfc2-5 complexes are dispensable for initial recruitment of Crb2 to DNA damage sites but are required for retention of Crb2 at sites of DNA damage (12). Previous studies have established that Rad3 kinase is also involved in phosphorylation of Rad26, Rad9, Hus1, Crb2, and Chk1 (4, 14, 18, 23, 34, 42). Threonine-215 phosphorylation of Crb2 by Cdc2, which appears to occur independently of Crb2 focus formation, is neither necessary nor sufficient for Crb2 focus formation and retention, but it is then required for efficient hyperphosphorylation of Crb2 by Rad3. This hyperphosphorylation appears to allow Crb2 to act as a more-efficient adaptor protein to facilitate the phosphorylation and resultant activation of Chk1 by Rad3.
FIG. 6.
A model for redundant regulation of Crb2 and DNA damage checkpoints by histone H2A phosphorylation-dependent focus formation and Cdc2- and Rad3-dependent hyperphosphorylation of Crb2. Arrows represent phosphorylation of various substrates by Rad3, Tel1, or Cdc2 kinases or methylation of histone H4 K20 by Set9 histone methyltransferase. Histone H2A (abbreviated as 2A), histone H4 (abbreviated as 4), and Set9 are required for Crb2 focus formation and retention (29, 35). Phosphorylation is abbreviated as P, while methylation is abbreviated as Me. Rad9, Rad1, and Hus1 are abbreviated as R9, R1, and H1, respectively.
Phosphorylation of Rad9 at T412 and S423 by Rad3 is required for DNA damage-induced association of Rad9 to BRCT domains of Cut5 (18). Additionally, the association of Crb2 with both Cut5 and Chk1 is induced by DNA damage (27). Cut5 is considered to be a homolog of mammalian TopBP1 protein, and Crb2 was originally identified in a genetic screen for proteins that interact with Cut5 (15, 18). Using a yeast two-hybrid assay, we mapped the domain of Crb2 responsible for Cut5 interaction to amino acids 1 to 275, which includes threonine-215 (13). Perhaps the Rad9-Rad1-Hus1 complex recruits Cut5 to sites of DNA damage, and Cut5 then associates with Crb2 and Chk1 to facilitate Rad3-dependent phosphorylation and activation of Chk1 kinase (18) (Fig. 6). The phosphorylation of Crb2 at threonine-215 may promote an interaction with Cut5 or other checkpoint proteins.
Our findings may also explain why H2A phosphorylation is not essential for DNA damage checkpoints in budding yeast and mice. S. cerevisiae Rad9, the functional homolog of Crb2, may be cooperatively regulated by its H2A-dependent retention of Rad9 at DSBs and its phosphorylation by the cyclin-dependent kinase Cdc28. The similarity of regulation for 53BP1 and Crb2 is very compelling (16, 29, 30, 44); thus, we also suggest that a similar redundancy in the DNA damage response is likely to be important in higher eukaryotes. MDC1/NFBD1, a mammalian checkpoint adaptor protein that accumulates at sites of DNA damage and has C-terminal BRCT domains in common with Crb2, Rad9, and 53BP1 (38), might be similarly regulated. Analysis of cyclin-dependent kinase regulation of 53BP1 and MDC1 might uncover a more direct and essential nature of histone H2AX phosphorylation in regulation of DNA damage responses in mammals.
Conclusions.
We have presented evidence that phosphorylation of Crb2 at threonine-215 by Cdc2 is required for maintenance of the DNA damage checkpoint. There is a similar requirement for the C-terminal phosphorylation of histone H2A by ATM family kinases, which is needed for the accumulation of Crb2 at DSBs. The checkpoint is abolished when both pathways regulating Crb2 are inactivated. These findings may explain why elimination of γ-H2A(X) has only modest effects on the DNA damage checkpoint in other organisms. A fascinating implication of these findings is that Cdc2 promotes the activity of a checkpoint protein, Crb2, which is required to prevent Cdc2 activation when DNA is damaged.
Acknowledgments
We thank members of the Russell laboratory and the Scripps Cell Cycle Groups for discussions. We thank the Yeast Resource Center at the University of Washington for the YFP vector.
T.M.N. was supported in part by the Damon Runyon Cancer Research Foundation, DRG-1565. T.M.N. and B.A.M. were also supported by a start-up fund from University of Illinois at Chicago. T.M.N. is a Sidney Kimmel Scholar. L.L.D. is a fellow of the Leukemia and Lymphoma Society. This work was funded by NIH grant CA77325, awarded to P.R.
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. [DOI] [PubMed] [Google Scholar]
- 3.Carr, A. M., S. A. MacNeill, J. Hayles, and P. Nurse. 1989. Molecular cloning and sequence analysis of mutant alleles of the fission yeast cdc2 protein kinase gene: implications for cdc2+ protein structure and function. Mol. Gen. Genet. 218:41-49. [DOI] [PubMed] [Google Scholar]
- 4.Caspari, T., M. Dahlen, G. Kanter-Smoler, H. D. Lindsay, K. Hofmann, K. Papadimitriou, P. Sunnerhagen, and A. M. Carr. 2000. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 20:1254-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caspari, T., J. M. Murray, and A. M. Carr. 2002. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celeste, A., O. Fernandez-Capetillo, M. J. Kruhlak, D. R. Pilch, D. W. Staudt, A. Lee, R. F. Bonner, W. M. Bonner, and A. Nussenzweig. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675-679. [DOI] [PubMed] [Google Scholar]
- 7.Celeste, A., S. Petersen, P. J. Romanienko, O. Fernandez-Capetillo, H. T. Chen, O. A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M. J. Difilippantonio, C. Redon, D. R. Pilch, A. Olaru, M. Eckhaus, R. D. Camerini-Otero, L. Tessarollo, F. Livak, K. Manova, W. M. Bonner, M. C. Nussenzweig, and A. Nussenzweig. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiTullio, R. A., Jr., T. A. Mochan, M. Venere, J. Bartkova, M. Sehested, J. Bartek, and T. D. Halazonetis. 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 4:998-1002. [DOI] [PubMed] [Google Scholar]
- 9.Downs, J. A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger, N. Bouchard, S. J. Kron, S. P. Jackson, and J. Cote. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16:979-990. [DOI] [PubMed] [Google Scholar]
- 10.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001-1004. [DOI] [PubMed] [Google Scholar]
- 11.Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630-639. [DOI] [PubMed] [Google Scholar]
- 12.Du, L.-L., T. M. Nakamura, B. A. Moser, and P. Russell. 2003. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 23:6150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du, L. L., B. A. Moser, and P. Russell. 2004. Homo-oligomerization is the essential function of the tandem BRCT domains in the checkpoint protein Crb2. J. Biol. Chem. 279:38409-38414. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, R. J., N. J. Bentley, and A. M. Carr. 1999. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1:393-398. [DOI] [PubMed] [Google Scholar]
- 15.Esashi, F., and M. Yanagida. 1999. Cdc2 phosphorylation of Crb2 is required for reestablishing cell cycle progression after the damage checkpoint. Mol. Cell 4:167-174. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Capetillo, O., H. T. Chen, A. Celeste, I. Ward, P. J. Romanienko, J. C. Morales, K. Naka, Z. Xia, R. D. Camerini-Otero, N. Motoyama, P. B. Carpenter, W. M. Bonner, J. Chen, and A. Nussenzweig. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4:993-997. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Capetillo, O., A. Lee, M. Nussenzweig, and A. Nussenzweig. 2004. H2AX: the histone guardian of the genome. DNA Repair (Amsterdam) 3:959-967. [DOI] [PubMed] [Google Scholar]
- 18.Furuya, K., M. Poitelea, L. Guo, T. Caspari, and A. M. Carr. 2004. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 18:1154-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannattasio, M., F. Lazzaro, P. Plevani, and M. Muzi-Falconi. 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280:9879-9886. [DOI] [PubMed] [Google Scholar]
- 20.Hayles, J., D. Fisher, A. Woollard, and P. Nurse. 1994. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell 78:813-822. [DOI] [PubMed] [Google Scholar]
- 21.Huyen, Y., O. Zgheib, R. A. Ditullio, Jr., V. G. Gorgoulis, P. Zacharatos, T. J. Petty, E. A. Sheston, H. S. Mellert, E. S. Stavridi, and T. D. Halazonetis. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406-411. [DOI] [PubMed] [Google Scholar]
- 22.Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani, W. Carotenuto, G. Liberi, D. Bressan, L. Wan, N. M. Hollingsworth, J. E. Haber, and M. Foiani. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431:1011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaur, R., C. F. Kostrub, and T. Enoch. 2001. Structure-function analysis of fission yeast hus1-rad1-rad9 checkpoint complex. Mol. Biol. Cell 12:3744-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi, J., H. Tauchi, S. Sakamoto, A. Nakamura, K. Morishima, S. Matsuura, T. Kobayashi, K. Tamai, K. Tanimoto, and K. Komatsu. 2002. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 12:1846-1851. [DOI] [PubMed] [Google Scholar]
- 25.Leger, J., M. Kempf, G. Lee, and R. Brandt. 1997. Conversion of serine to aspartate imitates phosphorylation-induced changes in the structure and function of microtubule-associated protein tau. J. Biol. Chem. 272:8441-8446. [DOI] [PubMed] [Google Scholar]
- 26.Lukas, C., F. Melander, M. Stucki, J. Falck, S. Bekker-Jensen, M. Goldberg, Y. Lerenthal, S. P. Jackson, J. Bartek, and J. Lukas. 2004. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 23:2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mochida, S., F. Esashi, N. Aono, K. Tamai, M. J. O'Connell, and M. Yanagida. 2004. Regulation of checkpoint kinases through dynamic interaction with Crb2. EMBO J. 23:418-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt, J. E. Haber, and X. Shen. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767-775. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura, T. M., L. L. Du, C. Redon, and P. Russell. 2004. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 24:6215-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 31.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 32.Redon, C., D. R. Pilch, E. P. Rogakou, A. H. Orr, N. F. Lowndes, and W. M. Bonner. 2003. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 4:678-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell, P. 1998. Checkpoints on the road to mitosis. Trends Biochem. Sci. 23:399-402. [DOI] [PubMed] [Google Scholar]
- 34.Saka, Y., F. Esashi, T. Matsusaka, S. Mochida, and M. Yanagida. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders, S. L., M. Portoso, J. Mata, J. Bahler, R. C. Allshire, and T. Kouzarides. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603-614. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, G. S., B. Wang, C. R. Bignell, A. M. Taylor, and S. J. Elledge. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961-966. [DOI] [PubMed] [Google Scholar]
- 37.Strom, L., H. B. Lindroos, K. Shirahige, and C. Sjogren. 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16:1003-1015. [DOI] [PubMed] [Google Scholar]
- 38.Stucki, M., and S. P. Jackson. 2004. MDC1/NFBD1: a key regulator of the DNA damage response in higher eukaryotes. DNA Repair (Amsterdam) 3:953-957. [DOI] [PubMed] [Google Scholar]
- 39.Unal, E., A. Arbel-Eden, U. Sattler, R. Shroff, M. Lichten, J. E. Haber, and D. Koshland. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16:991-1002. [DOI] [PubMed] [Google Scholar]
- 40.van Attikum, H., O. Fritsch, B. Hohn, and S. M. Gasser. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777-788. [DOI] [PubMed] [Google Scholar]
- 41.Vidanes, G. M., C. Y. Bonilla, and D. P. Toczyski. 2005. Complicated tails: histone modifications and the DNA damage response. Cell 121:973-976. [DOI] [PubMed] [Google Scholar]
- 42.Walworth, N. C., and R. Bernards. 1996. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271:353-356. [DOI] [PubMed] [Google Scholar]
- 43.Wang, B., S. Matsuoka, P. B. Carpenter, and S. J. Elledge. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science 298:1435-1438. [DOI] [PubMed] [Google Scholar]
- 44.Ward, I. M., K. Minn, K. G. Jorda, and J. Chen. 2003. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278:19579-19582. [DOI] [PubMed] [Google Scholar]
- 45.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 46.Willson, J., S. Wilson, N. Warr, and F. Z. Watts. 1997. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic. Acids Res. 25:2138-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie, A., N. Puget, I. Shim, S. Odate, I. Jarzyna, C. H. Bassing, F. W. Alt, and R. Scully. 2004. Control of sister chromatid recombination by histone H2AX. Mol. Cell 16:1017-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]