Abstract
F and R27 are conjugative plasmids of enteric bacteria belonging to the IncF and IncHI1 plasmid incompatibility groups, respectively. Based on sequence analysis, two genes of the F transfer region, traF and trbB, and three genes of the R27 transfer region, trhF, dsbC, and htdT, are predicted to encode periplasmic proteins containing a C-terminal thioredoxin fold. The C-X-X-C active-site motif of thioredoxins is present in all of these proteins except TraFF. Escherichia coli carrying a dsbA mutation, which is deficient in disulfide bond formation, cannot synthesize pili and exhibits hypersensitivity to dithiothreitol (DTT) as monitored by mating ability. Overproduction of the E. coli disulfide bond isomerase DsbC, TrbBF, DsbCR27, or HtdTR27, but not TraFF or TrhFR27, reverses this hypersensitivity to DTT. Site-directed mutagenesis established that the C-X-X-C motif was necessary for this activity. Secretion into the periplasm of the C-terminal regions of TrbBF and DsbCR27, containing putative thioredoxin folds, but not TrhFR27, partially complemented the host dsbA mutation. A trbBF deletion mutant showed a 10-fold-lower mating efficiency in an E. coli dsbC null strain but had no phenotype in wild-type E. coli, suggesting redundancy in function between TrbBF and E. coli DsbC. Our results indicate that TrbBF, DsbCR27, and HtdTR27 are putative disulfide bond isomerases for their respective transfer systems. TraFF is essential for conjugation but appears to have a function other than disulfide bond chemistry.
Bacterial conjugation is a fundamental mechanism for horizontal gene transfer that facilitates the transmission of genetic material, such as antibiotic resistance and other virulence factors, within and between bacterial species (67). It involves the transfer of single-stranded DNA from a donor cell to a recipient cell that has established close contact via mating pair formation. Although the F plasmid of Escherichia coli remains a paradigm for this process, conjugative plasmids have been reported in a variety of species, including many bacterial pathogens (42). More recently, conjugation has been considered to belong to the type IV secretion system (T4SS) family based on the similarities between the proteins involved in both processes (16).
Mating pair formation involves a complex apparatus spanning the donor cell envelope that assembles the conjugative pilus. This filamentous appendage interacts with the recipient cell and is thought to retract by depolymerization into the donor cell, thereby allowing intimate wall-to-wall contact during mating pair stabilization (1). The conjugative plasmid is transferred across both bacterial envelopes by a mechanism that remains poorly understood (17). All genes required for mating pair formation and DNA transfer are typically located in one or two clusters, the transfer regions, on conjugative plasmids.
Many components of the F plasmid transfer apparatus have homologues in other T4SS. For example, eight F transfer proteins are homologous to VirB proteins encoded by the T4SS of the Ti plasmid (42). However, the F plasmid encodes a unique group of proteins that are essential for pilus assembly and DNA transfer in F-like systems: TraF, -H, -N, -U, and -W and TrbC. All of these proteins reside in the periplasm, except the outer membrane protein TraN (24). Mutations in traF, traG, traH, traW, and trbC impair pilus assembly (5), whereas TraG, TraN, and TraU appear to be involved in mating pair stabilization and DNA transfer (38, 54).
Periplasmic and outer membrane proteins often contain disulfide bonds. Although disulfide bonds can form spontaneously in the oxidizing periplasmic environment (3), disulfide bond formation is facilitated in vivo by a number of specialized thiol-disulfide exchange enzymes. In E. coli, DsbA is a soluble, monomeric 21.1-kDa periplasmic protein (10) that randomly and rapidly oxidizes pairs of cysteine residues in secreted proteins through reduction of its own disulfide bond (68). The disulfide bond in DsbA is restored by the inner membrane protein DsbB, using the oxidizing power of the electron transport chain (8). dsbA mutants exhibit a pleiotropic phenotype including the rapid degradation of cell envelope proteins, a lack of motility due to incomplete assembly of the flagellar motor, hypersensitivity to metals and dithiothreitol (DTT), and resistance to M13 bacteriophage infection in F+ cells because of the absence of F pili (35).
DsbC and DsbG are homodimeric disulfide isomerases that resolve incorrectly formed disulfide bonds (12, 50). They are maintained in a reduced state by DsbD, which is regenerated by the cytoplasmic thioredoxin reductase system (12, 61). DsbC stimulates proper folding and correct disulfide bond formation in a number of heterologous proteins (49). Although DsbC null mutants do not have an obvious phenotype, overexpressed dsbC can partially compensate for dsbA null mutants, possibly due to a high level of redundancy among the disulfide isomerase proteins (53).
DsbA, DsbC, and other soluble Dsb proteins all contain a domain with homology to the thioredoxin fold. Thioredoxins form a large family of proteins that catalyze the formation and isomerization of disulfide bonds as well as other redox reactions (59). The thioredoxin fold consists of at least three α-helices and a four-stranded β-sheet (48). The active site consists of two cysteine residues in a C-X-X-C motif and is located in a loop connecting the first β-strand and the first α-helix within the thioredoxin fold. The cysteines are reversibly reduced and oxidized during the catalytic cycle. A conserved proline residue plays an important role in both structure and function of thioredoxins by forming van der Waals interactions between the loop in which it resides and the C-X-X-C disulfide bond (18).
Here we report that the F- and H-like (R27) conjugative plasmids encode proteins with a predicted C-terminal thioredoxin fold. Using assays developed for DsbC, we have determined that TrbBF, DsbCR27, and HtdTR27 contain a functional C-X-X-C active site in their thioredoxin-like domains and that they are possibly disulfide bond isomerases. TrhFR27, which contains a C-X-X-C motif, and TraFF, which does not, apparently have a function not directly related to disulfide bond formation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All strains and plasmids used in this study are listed in Table 1. Bacterial cultures were grown in Luria-Bertani medium with appropriate antibiotics at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 25 μg/ml; nalidixic acid, 40 μg/ml; rifampin, 20 μg/ml; streptomycin, 200 μg/ml; tetracycline, 10 μg/ml.
TABLE 1.
Plasmids and Escherichia coli strains used in this study
| Strain or plasmid | Relevant characteristics | Selective marker(s)a | Reference or source |
|---|---|---|---|
| Bacterial strains | |||
| DH5α | supE44 ΔlacU169 (φ80dlacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Nal | Laboratory collection (29a) |
| DY330 | W3110 ΔlacU169 gal490 λc1857 Δ(cro-bioA) | Nal or Rif | 69 |
| MC4100 | araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB3501 deoC1 ptsF25 rbsR | Sm | Manoil; laboratory collection |
| RI89 | MC1000 phoR Δara714 leu+ | Sm | 60a |
| RI90 | RI89 dsbA::kan1 | Km, Sm | 60a |
| RI179 | RI89 ΔdsbC::cam | Cm, Sm | 60a |
| XK1200 | F−lacΔU124 Δ(nadA aroG gal attλ bio gyrA) | Nal | K. Ippen-Ihler (54) |
| Plasmids | |||
| PCR4Blunt-TOPO | 4-Kb cloning vector with covalently linked topoisomerase | Amp, Km | Invitrogen |
| pBAD30 | Cloning vector for controlled expression from Para-BAD | Amp | 28 |
| pOX38-Tc | pOX38::mini-Tn10 | Tc | 4 |
| pOX38-Tc ΔtraF::kan | pOX38-Tc with kan replacing codons 34 to 229 in traF | Tc, Km | This study |
| pOX38-Tc ΔtrbB::cat | pOX38-Tc with cat replacing codons 14 to 160 in trbB | Tc, Cm | This study |
| pFTraF | F traF in pBAD30 (DPID)b | Amp | This study |
| pFTraF(CPYC) | F traF (D135C, I137Y, D138C)c in pBAD30 (CPYC)b | Amp | This study |
| pFTrbB | F trbB in pBAD30 (CPYC)b | Amp | This study |
| pFTrbB(Y63H) | F trbB (Y63H)c in pBAD30 (CPHC)b | Amp | This study |
| pFTrbB(SPYS) | F trbB (C61S, C64S)c in pBAD30 (SPYS)b | Amp | This study |
| pHTrhF | R27 trhF in pBAD30 (CQFC)b | Amp | This study |
| pHHtdT | R27 htdT in pBAD30 (CDGC)b | Amp | This study |
| pHDsbC | R27 dsbC in pBAD30 (CGFC)b | Amp | This study |
| pHDsbC(SGFS) | R27 dsbC (C107S, C110S)c in pBAD30 (SDGS)b | Amp | This study |
| pKDsbA | E. coli K-12 dsbA in pBAD30 | Amp | This study |
| pKDsbA(NheI) | E. coli K-12 dsbA in pBAD30 with NheI site at codons 17 and 18 | Amp | This study |
| pFBASS | dsbA signal sequence with A34 to K159 of F trbB in pBAD30 | Amp | This study |
| pFBASS(Y63H) | pFBASS using F trbB (Y63H)c | Amp | This study |
| pHFASS | dsbA signal sequence with Q137 to R324 of R27 trhF in pBAD30 | Amp | This study |
| pHCASS | dsbA signal sequence with A82 to K227 of R27 dsbC in pBAD30 | Amp | This study |
Antibiotic resistances: Amp, ampicillin; Cm, chloramphenicol; Km, kanamycin; Nal, nalidixic acid; Rif, rifampin; Sm, streptomycin; Tc, tetracycline.
Sequence of the four amino acids at the C-X-X-C motif indicated in Fig. 1 (in parentheses).
The first letter indicates the wild-type amino acid; the second letter indicates the mutant amino acid; the number refers to the position of the amino acid within the mature protein.
Recombinant DNA techniques, DNA sequencing, and PCR.
Techniques for cloning restriction fragments or PCR fragments were as described previously (64). Blunt-end PCR products for cloning were purified from an agarose gel using the Qiaquick gel extraction kit (QIAGEN) to remove plasmid templates and were directly inserted into pCR4Blunt-TOPO (Invitrogen) according to the manufacturer's instructions. The sequences of PCR insertions into pCR4Blunt-TOPO (Invitrogen) were verified using the M13 Forward (5′ GTAAAACGACGGCCAG) and M13 Reverse (5′ CAGGAAACAGCTATGAC) primers from Invitrogen. Restriction endonuclease digestion and ligation of DNA molecules were performed as described by Ausubel et al. (7). All enzymes were supplied by Roche Applied Science except Vent polymerase (New England Biolabs) and Pfu Turbo DNA polymerase (Stratagene). The method for automated sequencing has been described previously (38). Based on the method of Jonda et al. (34), the plasmid pKDsbA (NheI), which contains a single NheI restriction site at the end of the DsbA signal sequence without change to the amino acid sequence, was used in the construction of plasmids for periplasmic localization of thioredoxin fold variants. Thus the portion of trbBF, trhFR27, or dsbCR27 encoding the thioredoxin fold could be amplified by PCR and cloned into pKDsbA (NheI).
Gene deletion techniques.
PCR amplification of the kanamycin resistance gene (including the native promoter) from the pUC4K plasmid (Amersham Pharmacia Biotech) was obtained using primers TEL-traF-kan-For (5′ GATGCAGGCTGGCAGTGGTATAACGAGAAAATAAATCCGAAAAGCCACGTTGTG TCTCAA) and TEL-traF-kan-Rev (5′ TCTTCAGAAACGTTCAGGAACTGTTTTGCCAGGTCGTCCTCGCTGAGGTCTGCCTCGTGA). The resulting kanlinear DNA cassette had overhanging arms homologous to either nucleotides 15142 to 15182 or nucleotides 15767 to 15806 of traF. The PCR product was purified from an agarose gel using the Qiaquick gel extraction kit (QIAGEN) to remove the plasmid template and electroporated into E. coli strain DY330/F according to the procedure outlined by Yu et al. (69). The E. coli strain MC4100 was added as recipient cells and allowed to mate with DY330/F for 1 h. Recombinants were selected on kanamycin and streptomycin. To verify insertion of the kan cassette into F, clones were sequenced with TEL1 (5′ GGATCCAA AGATGCAGGCTGGCAG) and TEL2 (5′ CAGAATTCCTCAGAAAAGAAATAACCGG). PCR amplification of the Tn9 chloramphenicol acetyltransferase gene (including the native promoter) from the pBAD33 plasmid (28) was obtained using primers TEL-trbB-cat-For (5′ CATGTCTCTCACTAAATCACTGCTGTTCACCCTGTTGCTGCTGTGACGGAAGATCACTTC) and TEL-trbB-cat-Rev (5′ CGTACATCTGCAAAACGGTATCCACCCGCGCCATAAAACCTTATTCAGGCGTAGCACCAG). The resulting cat linear DNA cassette had overhanging arms homologous to nucleotides 16855 to 16894 and nucleotides 17333 to 17372 of trbB. The remainder of the protocol was completed as above. To verify insertion of the cat cassette into F, clones were sequenced with TEL26 (5′ GGTACCGAAGGGCAGCAGGAGGGC) and TEL27 (5′ GAAGCTTCCGGCAAT GAGTAACACCAC).
Site-directed mutagenesis.
All mutagenesis was completed using QuikChange (Stratagene) site-directed mutagenesis according to the manufacturer's instructions. Briefly, 25 ng of pBAD30-trbB was used as a template for 125 ng of mutated nucleotide primers described in Table 2. PCR conditions were as follows: initial denaturing at 95°C for 2 min, followed by 18 cycles of denaturing at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 7 min. After digestion with DpnI, 5 μl of PCR product was used to transform XL-1 Blue competent cells. Every mutant was sequenced to ensure that mutants were correct and in frame.
TABLE 2.
Primers used in this studya
| Construct | Primer |
|---|---|
| pFTraF For | 5′ GCTAAGGTACCCTCCGGTAATTATCTGG-3′ |
| pFTraF Rev | 5′ GGCATAAGCTTCCTGTCATTACGCTCAG-3′ |
| pFTraF(CPYC) For | 5′ CATGTTTTTTTACCGGGGGCAGTGCCCCTACTGCGGGCAACTGGCGCAGGTC-3′ |
| pFTraF(CPYC)Rev | 5′ GACCTGCGCCAGTTGCCCGCAGTAGGGGCACTGCCCCCGGTAAAAAAACATG-3′ |
| pFTrbB For | 5′ GGTACCGAAGGGCAGCAGGAGGGC-3′ |
| pFTrbB Rev | 5′ GAAGCTTCCGGCAATGAGTAACAGCAC-3′ |
| pFTrbB(Y63H) For | 5′ CAGGGGCATTGCCCTCACTGTCACCAGTTTGAC-3′ |
| pFTrbB(Y63H) Rev | 5′ GTCAAACTGGTGACAGTGAGGGCAATGCCCCTG-3′ |
| pFTrbB(SPYS) For | 5′ GTTTATGCAGGGGCATTCCCCTTACTCTCACCAGTTTGACCCGG-3′ |
| pFTrbB(SPYS) Rev | 5′ CCGGGTCAAACTGGTGAGAGTAAGGGGAATGCCCCTGCATAAAC-3′ |
| pHTrhF For | 5′ GGTACCATCATACCTCTATTCCATG-3′ |
| pHTrhF Rev | 5′ CAAGCTTCAAACGGCTAACGATAAAAG-3′ |
| pHHtdT For | 5′ GGTACCAGGCGAACCTAAAGAAAGTGAG-3′ |
| pHHtdT Rev | 5′ GAAGCTTGATGAACCAACAACCAGAGCGC-3′ |
| pHDsbC For | 5′ GGTACCTTCGCATGATAGTATTAACACTCC-3′ |
| pHDsbC Rev | 5′ GAAGCTTCATAAACACCTCTTGCAGTCG-3′ |
| pHDsbC(SGFS) For | 5′ GTTTACGGATATTACATCCGGCTTTTCCCAGAAATTGCATCAAG-3′ |
| pHDsbC(SGFS) Rev | 5′ CTTGATGCAATTTCTGGGAAAAGCCGGATGTAATATCCGTAAAC-3′ |
| pKDsbA For | 5′ GGTACCAGTTCTACAAGAACCCCCTTTGC-3′ |
| pKDsbA Rev | 5′ GAAGCTTCAGCGGCAGGATGCATTATCAG-3′ |
| pKDsbA(NheI) For | 5′ GTTTAGTTTTAGCGTTTAGCGCTAGCGCGGCGCAGTATGAAGATGG-3′ |
| pKDsbA(NheI) Rev | 5′ CCATCTTCATACTGCGCCGCGCTAGCGCTAAACGCTAAAACTAAAC-3′ |
| pFBASS For | 5′ GCTAGCGCGGCTCCCCGCTGGTTCCG-3′ |
| pHFASS For | 5′ GCTAGCGCGGCTCAGCAGTCGGTTATGAAAGATATTTTC-3′ |
| pHCASS For | 5′ GCTAGCGCCGCCACTAAAACAAAATCCATCG-3′ |
Numbers present in the primer names indicate amino acid mutations in the active site. Restriction sites are underlined; KpnI or NheI were used for forward primers, whereas HindIII was used for reverse primers. Nucleotide changes for mutagenic primers are in boldface.
Mating assays.
Mating assays have been described by Anthony et al. (4) and Klimke and Frost (38). To test transfer of pOX38-Tc, an F derivative, from RI89 (wild-type), RI90 (dsbA), or RI179 (dsbC) donor cells to XK1200 recipient cells, donor cells were grown to mid-log phase and mixed with various concentrations of DTT (0 mM to 7 mM) for 1 h prior to the mating assay. Two hundred microliters of mid-log-phase donor cells and recipient cells was incubated at 37°C for 45 min. The cultures were serially diluted in saline (between 10−2 and 10−7), and 10-μl droplets were plated out on either tetracycline and streptomycin or tetracycline and nalidixic acid. If protein overproduction for vector constructs was required, then donor cells were induced with 0.1% arabinose 2 h prior to adding the recipient cells. When testing the DTT hypersensitivity of a dsbA null mutant, donor cells were grown to mid-log phase and mixed with 1 mM dithiothreitol for 1 h prior to the mating assay.
RESULTS
F- and H-like transfer regions encode putative thiol-disulfideoxidoreductases.
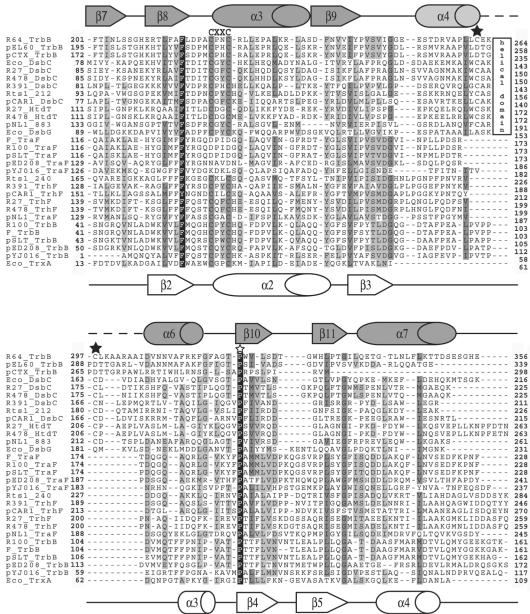
The nonredundant protein sequence database was searched with the program PSIBLAST (2) to find homologues of TraFF encoded by the F plasmid (24, 27). PSIBLAST detected several TraF homologues encoded by the transfer regions of other F-like (E values = 4e−21 to e−136) and H-like (E-values = 6e−4 to 3e−18) plasmids (Table 3). Using the position-sensitive scoring matrix (PSSM) based on this set of TraF homologues, a second cycle of PSIBLAST found a group of proteins that displayed homology to approximately 100 amino acids of the C-terminal region of TraFF (E values = 2e−6 to 4e−8) (Table 3). This group of proteins included TrbBF and its homologues encoded by R100 (5), pSLT (51), pED208 (45), and pYJ016 (19), as well as a few proteins annotated as thiol-disulfide isomerases. Subsequent cycles of PSIBLAST using a PSSM inclusion threshold of 0.001 revealed extensive homology to the thioredoxin/thiol-disulfide isomerase family (E values = <8e−10) (Table 3). A search for conserved domains within homologues of TraFF using the program CD-Search (47) revealed thioredoxin-like regions (COG0526, TrxA) within TraF encoded by R391 (14) and SXT (11). Although TraFF lacks the C-X-X-C motif that forms the catalytic site of thioredoxins, closely related TraF orthologues encoded by the H-like T4SS as well as all TrbB proteins contain this active-site motif (Fig. 1). Interestingly, TraF orthologues which lack the C-X-X-C motif are encoded only by plasmids that encode TrbB, whereas TrbB is not encoded by plasmids encoding a TraF homologue that contains a C-X-X-C motif. This suggests that TrbB may compensate for the lost redox function of TraF.
TABLE 3.
Homologies among TraF, TrhF, TrbB, DsbC, and HtdT proteins encoded by F-like, H-like, and I-like plasmids
| Plasmid | Inc group | Organism | Plasmid reference(s) or accession no. | BLAST query
|
|||
|---|---|---|---|---|---|---|---|
| F TraF
|
R27 DsbC
|
||||||
| Protein | E value | Protein | E value | ||||
| F-like | |||||||
| F | FI | Escherichia coli | 24 and 27 | TraF | e−136 (1)a | ||
| TrbB | 4e−07 (2) | ||||||
| R100 | FII | Salmonella enterica serovar Typhimurium | 5 | TraF | e−133 (1) | ||
| TrbB | 8e−08 (2) | ||||||
| pSLT | Salmonella enterica serovar Typhimurium | 51 | TraF | e−107 (1) | |||
| TrbB | 2e−06 (2) | ||||||
| pED208 | FV | Salmonella enterica serovar Typhi | 45 | TraF | 1e−51 (1) | ||
| TrbB | 4e−08 (2) | ||||||
| pYJ016 | Vibrio vulnificus | 19 | TraF | 4e−21 (1) | |||
| TrbB | 1e−07 (2) | ||||||
| H-like | |||||||
| pNL1 | Novosphingomonas aromaticivorans | 63 | TraF | 3e−18 (1) | Orf883 | 2e−21 (2) | |
| Rts1 | T | Proteus vulgaris | 55 | Orf240 | 8e−09 (1) | Orf212 | 2e−16 (2) |
| R391 | J | Providencia rettgeri | 14 | TrhF | 5e−13 (1) | DsbC | 3e−23 (2) |
| SXT | Vibrio cholerae | 11 | TrhF | 4e−13 (1) | S054 | 3e−23 (2) | |
| pCAR1 | Pseudomonas resinovorans | 46 | TrhF | 1e−09 (1) | DsbC | 2e−14 (2) | |
| pHCM1 | HI1 | Salmonella enterica serovar Typhi | 57 | TrhF | 7e−05 (1) | DsbC | e−140 (1) |
| HtdT | 4e−04 (2) | ||||||
| R27 | HI1 | Salmonella enterica serovar Typhi | 40, 41, and 65 | TrhF | 7e−05 (1) | DsbC | e−140 (1) |
| HtdT | 3e−04 (2) | ||||||
| R478 | H2 | Serratia marcescens | 26 | TrhF | 6e−04 (1) | DsbC | e−120 (1) |
| HtdT | 1e−04 (2) | ||||||
| I-like | |||||||
| R64 | I1 | Salmonella enterica serovar Typhimurium | 25 | TrbB | 5e−15 (3) | ||
| ColIb-P9 | I1 | Shigella sonnei | 60 | TrbB | 5e−15 (3) | ||
| pEL60 | Erwinia amylovora | 23 | TrbB | 3e−07 (2) | |||
| pCTX-M3 | Citrobacter freundii | NC_004464 | TrbB | 4e−06 (3) | |||
| None | None | Escherichia coli | TrxA | 8e−10 (3) | |||
| None | None | Escherichia coli | DsbC | 5e−32 (1) | |||
| None | None | Escherichia coli | DsbG | 5e−16 (2) | |||
The numbers in parentheses indicate the number of cycles of PSI-BLAST.
FIG. 1.
Multiple sequence alignment of TraF, TrhF, TrbB, HtdT, and DsbC encoded by the F-like, H-like, and I-like transfer regions with the TrxA, DsbC, and DsbG proteins from E. coli. Each protein in the alignment is preceded by the plasmid name or a three-letter abbreviation for the organism if the gene is carried on the chromosome (Eco = Escherichia coli). The following information regarding these proteins is arranged as plasmid name (if necessary), protein name, GenBank accession number, and host organism: R64, TrbB, BAB91646, S. enterica serovar Typhimurium; pEL60, TrbB, AAQ97939, Erwinia amylovora; pCTX-M3, TrbB, AAN87722, Citrobacter freundii; DsbC, AAC75931, E. coli; R27, DsbC, AAF69969, S. enterica serovar Typhi; R478, DsbC, CAE51723, Serratia marcescens; R391, DsbC, AAM08034, Providencia rettgeri; Rts1, Orf212, BAB93774, Phaseolus vulgaris; pCAR1, DsbC, BAC41659, Pseudomonas resinovorans; R27, HtdT, AAF69865, S. enterica serovar Typhi; R478, HtdT, CAE51541, S. marcescens; pNL1, Orf883, AAD03960, Novosphingobium aromaticivorans; DsbG AAC73705, E. coli; F, TraF, BAA97961, E. coli; R100, TraF, BAA78873, E. coli; pSLT, TraF, AAL23500, S. enterica serovar Typhimurium; pED208, TraF, AAM90720, S. enterica serovar Typhi; pYJ016, TraF, BAC97739, Vibrio vulnificus; Rts1, Orf240, BAB93802, P. vulgaris; R391, TraF, AAM08018, P. rettgeri; pCAR1, TraF, BAC41668, P. resinovorans; R27, TrhF, AAF69964, S. enterica serovar Typhi; R478, TrhF, CAE51730, S. marcescens; pNL1, TraF, AAD03949, N. aromaticivorans; R100, TrbB, BAA78876, E. coli; F, TrbB, BAA97965, E. coli; pSLT, TrbB, AAL23502, S. enterica serovar Typhimurium; pED208, TrbB, AAM90721, S. enterica serovar Typhi; pJY016, TrbB, BAC97741, V. vulnificus; TrxA, AAA24694, E. coli. Proteins from other plasmid systems with sequences identical to a protein listed above were not included in Fig. 1, but their accession numbers are as follows: ColIb-P9, TrbB, BAA75142, Shigella sonnei; pHCM1, DsbC, CAD09838, S. enterica serovar Typhi; SXT, S054, AAL59715, V. cholerae; pHCM1, HtdT, CAD09683, S. enterica serovar Typhi; SXT, TraF, AAL59678, V. cholerae; pHCM1, TrhF, CAD09843, S. enterica serovar Typhi. The secondary structures of DsbC (top) and thioredoxin (bottom) are as determined by X-ray structure and nuclear magnetic resonance analysis (33, 50). The active sites are labeled as CXXC. The black stars indicate cysteines known or proposed to be important in forming a disulfide bond. The white star indicates a conserved proline residue that is found in close proximity to the active sites of DsbA, DsbC, and thioredoxin from E. coli. The box marked “helical domain” indicates the presence of an extra helical domain in some DsbC-like proteins. Residues in black indicate 100% conservation, residues in dark gray indicate more than 80% conservation, and residues in light gray indicate at least 50% conservation.
The IncHI1 R27-encoded transfer protein DsbCR27 is annotated as a putative disulfide interchange protein (accession number, AAF69969) (40, 41, 65). A PSIBLAST search with this protein sequence revealed DsbCH homologues encoded by the transfer regions of H-like plasmids pHCM1 (57) and R478 (26) (E values = e−140 and e−120, respectively) as well as thewell-characterized E. coli chromosomally encoded DsbC (Evalue = 5e−32) (Table 3). The second cycle of PSIBLAST identified another group of DsbCH homologues from other H-like plasmids (E values = 2e−14 to 3e−23), establishing homology to H-like HtdT proteins (E value = 4e−4) and the chromosomally encoded E. coli DsbG (E value = 5e−16) (Table 3). Subsequent cycles of PSIBLAST using a PSSM inclusion threshold of 0.001 revealed homology to the TrbB proteins encoded by the IncI plasmids R64 (25), ColIb-P9 (60), pEL60 (23), and pCTX-M3 (accession number, NC_004464) (E values = 4e−6 to 5e−15). The naming of R64 TrbB and F TrbB was coincidental; discerning the relationship between these two proteins required many iterations of PSIBLAST, suggesting that they are, at best, distantly related. P-like T4SS (42), such as that of RP4 or the Vir region of the Ti plasmid of Agrobacterium tumefaciens, do not encode these homologues. Searches with CD-Search also revealed that all DsbCH and HtdTH homologues, with the exception of HtdTR478, share homology with COG1651 (DsbG), showing E values between 7e−16 and 0.004. All these proteins contain the C-X-X-C active-site motif of thioredoxins (Fig. 1). The thioredoxin domain of E. coli DsbC contains an extra 43-residue α-helical subdomain (50). Cys141 and Cys163 within this subdomain form an intramolecular disulfide bond that is important for the stability of the molecule (44). Despite variations in length, all DsbCH and HtdTH proteins contain this extra domain, which includes the two conserved cysteine residues (Fig. 1).
Disulfide isomerization activity requires hydrophobic or aromatic amino acids in the third position of the C-X-X-C active site (13). All DsbCH and TrbBF proteins contain aromatic amino acids in this position. Most TraFF or TrhFH proteins with an active site have either aromatic or hydrophobic amino acids in this position, with the exception of TraFpNL1, which has an alanine, and the HtdTH proteins, which have glycine. A proline in close structural proximity to the active site is required for maintaining an active conformation in E. coli thiol-disulfide oxidoreductases, (36) and is conserved in all TraFF, TrhFH, TrbBF, HtdTH, and DsbCH proteins discussed here (Fig. 1).
TraFF, TrhFH, TrbBF, DsbCH, and HtdTH are predicted to be soluble periplasmic proteins.
TraFF and TrbBF are known to contain leader peptides (24), and SignalP predictions (56) detected leader peptides in all but one of the TrhFH, HtdTH, and DsbCH proteins. The exception is HtdTR27 (accession number, AAF69865), for which a leader sequence could not be found; however, when the sequence of htdTR27 was compared to the sequence of the nearly identical htdTpHCM1 (accession number, CAD09683), it became apparent that htdTR27 should begin 35 codons upstream from its current predicted start, at the rare TTG start codon. Analysis of this longer sequence revealed a 22-residue leader peptide in this protein. No other putative transmembrane regions were predicted in the mature proteins. Full-length TraFF and TrbBF can be expressed as soluble proteins (6; L. S. Frost, unpublished data), suggesting that TrhFR27, DsbCR27, and HtdTR27 are also soluble periplasmic proteins.
Transfer efficiency of pOX38-Tc is affected by DTT or dsbA null mutations.
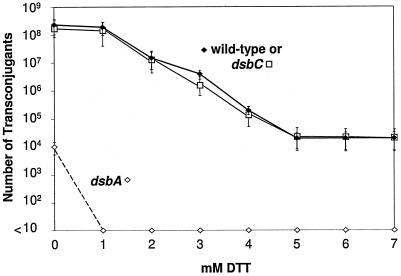
DsbA null mutants have been reported to affect assembly of the flagellar motor; pilus assembly, as determined by monitoring by M13 infection; and hypersensitivity to DTT (10, 21). We therefore determined the mating efficiencies of wild-type and dsbA and dsbC mutant donor cells containing the F derivative pOX38-Tc. The mating assays were subsequently repeated following incubation of the donor cells with various concentrations of DTT for 1 hour. DTT is a small reducing agent that can diffuse into the periplasm and maintain sulfhydryl groups in the reduced state in vivo. Wild-type bacteria have been shown to tolerate DTT concentrations up to 7 mM (52). The mating efficiency of wild-type and dsbC donor cells was not affected by 1 mM DTT, but higher concentrations reduced mating efficiency up to 4 orders of magnitude in a dose-dependent manner before a plateau was reached at 5 mM DTT (Fig. 2). Donor cells with a chromosomal dsbA null mutation had transfer efficiencies 4 logs lower than that of wild-type cells; addition of 1 mM DTT to these cells completely abolished DNA transfer, indicating a DTT-hypersensitive phenotype for this strain (Fig. 2).
FIG. 2.
Mating efficiency of pOX38-Tc is greatly decreased by the reducing agent DTT. The mating efficiencies of E. coli dsbA and dsbC mutants containing pOX38-Tc were plotted against increasing concentrations of DTT.
Construction of traF and trbB null mutants of pOX38-Tc.
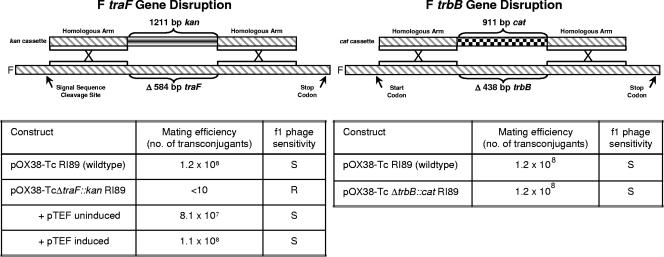
The essential role of TraFF in pilus formation was initially determined using a traF insertion mutant that could produce a C-terminally truncated TraF protein (24). To rule out any effect of the N-terminal region of TraF, we used the λ Red recombination system to replace codons 34 to 229 with a kan cassette, thus creating pOX38-Tc ΔtraF::kan (Fig. 3; Table 1). This mutation completely abolished DNA transfer and phage f1 infection, an indication that no F pili were assembled. The effects could be fully complemented by traF supplied in trans on pFTraF (Fig. 3).
FIG. 3.
Gene disruption of F traF and F trbB. Above are schematic diagrams of a kan and cat cassette insertion into F traF and F trbB, respectively, through homologous recombination. The native kan and cat promoters are present in both gene disruptions. Below the diagram are the mating efficiencies and f1 phage sensitivities of the traF and trbB null mutants.
Similarly, the previous insertion of a kan cassette between codons 66 and 67 of trbB might express a truncated version of TrbB with a functional thioredoxin motif (37). To ensure against this, codons 14 to 160 of trbB were replaced with the chloramphenicol acetyltransferase gene (cat) from Tn9 to give pOX38-Tc ΔtrbB::cat (Fig. 3; Table 1). This null mutation affected neither plasmid transfer nor f1 phage infection (Fig. 3).
Mating efficiency is reduced in an E. coli dsbC trbBF double null mutant.
The trbBF null mutant has no obvious phenotype, similar to strains encoding several E. coli periplasmic thiol-redox proteins (35). Thus, significant functional redundancy could exist among this group of enzymes (62). To investigate whether a trbB dsbC double mutation affects F conjugation, pOX38-Tc or pOX38-Tc ΔtrbB::Cm was introduced into RI189 (wild-type) or RI179 (dsbC) cells. Mating assays revealed that transfer efficiency decreased 10-fold in the dsbC strain containing pOX38-Tc ΔtrbB::Cm compared to donor/plasmid combinations that contained either trbB or dsbC single mutations or completely wild-type donor cells (Table 4). Furthermore, complementation of RI179/pOX38-Tc ΔtrbB::Cm with TrbB supplied in trans from pFTrbB resulted in the restoration of wild-type mating efficiency (Table 4).
TABLE 4.
Effect of dsbC and trbB null mutations on pOX38-Tc transfer
| Construct | No. of transconjugants | Mating efficiency (±10) (no. of transconjugants/100 donor cells) |
|---|---|---|
| pOX38-Tc/RI89 (wild type) | 1.2 × 108 | 80 |
| pOX38-Tc/RI179 (dsbC) | 1.1 × 108 | 73 |
| pOX38-TcΔtrbB::cat/RI89 (wild type) | 1.0 × 108 | 67 |
| pOX38-TcΔtrbB::cat/RI179 (dsbC) | 1.1 × 107 | 7 |
| pOX38-TcΔtrbB::cat/RI179 (dsbC) + pFTrbB | 1.0 × 108 | 67 |
| pOX38-TcΔtrbB::cat/RI179 (dsbC) + pBAD30 | 1.0 × 107 | 7 |
Overproduced TrbBF, DsbCR27, or HtdTR27 complements the DTT hypersensitivity of a dsbA null mutant.
The overexpression of E. coli dsbC from a multicopy plasmid has been shown to alleviate the general hypersensitivity to DTT exhibited by dsbA null mutants (53, 66). Although pFTrbB (Table 1) could not complement the effect of the dsbA mutation on the mating ability of RI190/pOX38-Tc (dsbA) cells (data not shown), it could overcome the hypersensitivity to DTT as demonstrated by a mating assay (Table 5). This complementation required that pFTrbB be first induced with 0.1% arabinose, as uninduced samples showed no effect (data not shown). Similarly, overexpression of dsbCR27 or htdTR27 (pHDsbC or pHHtdT, respectively; Table 1) yielded comparable results (Table 5).
TABLE 5.
Effect of overproduction of F- and H-like transfer proteins in a dsbA mutanta
| E. coli strain | DTT concn (mM) | Plasmid construct | No. of transconjugants |
|---|---|---|---|
| pOX38-Tc/RI89 (wild type) | 1.2 × 108 | ||
| pOX38-Tc/RI90 (dsbA) | 1.1 × 104 | ||
| pOX38-Tc/RI90 (dsbA) | 1 | <10 | |
| pOX38-Tc/RI90 (dsbA) | 1 | pBAD30 | <10 |
| pOX38-Tc/RI90 (dsbA) | 1 | pFTraFb | <10 |
| pOX38-Tc/RI90 (dsbA) | 1 | pFTraF(CPYC)b | <10 |
| pOX38-Tc/RI90 (dsbA) | 1 | pFTrbBb | 9.0 × 103 |
| pOX38-Tc/RI90 (dsbA) | 1 | pFTrbB(Y63H)b | 3.0 × 103 |
| pOX38-Tc/RI90 (dsbA) | 1 | pFTrbB(SPYS)b | <10 |
| pOX38-Tc/RI90 (dsbA) | 1 | pHTrhFc | <10 |
| pOX38-Tc/RI90 (dsbA) | 1 | pHHtdTa | 8.0 × 103 |
| pOX38-Tc/RI90 (dsbA) | 1 | pHDsbCc | 1.0 × 104 |
| pOX38-Tc/RI90 (dsbA) | 1 | pHDsbC(SGFS)c | <10 |
All vector constructs were induced with the addition of 0.1% arabinose 2 hours prior to adding the recipient cells for the mating assay.
F-like.
H-like.
The C-X-X-C motif of TrbBF and DsbCR27 is required to complement DTT-hypersensitive dsbA mutants.
To determine whether the C-X-X-C motifs of TrbBF and DsbCR27 were important in the rescue of dsbA donor cells in the presence of DTT, site-directed mutagenesis (QuikChange; Stratagene) was used to convert the putative active-site cysteines to serines to give pFTrbB(SPYS) and pHDsbC(SGFS), respectively. Mating was undetectable using RI190/pOX38-Tc (dsbA) donor cells containing either pFTrbB(SPYS) or pHDsbC(SGFS), even after induction with 0.1% arabinose. Thus, the C-X-X-C motif appears to be essential for plasmid-encoded TrbB and DsbC function, and it acts as an active redox site in these proteins. Because TraFF lacks the putative active-site cysteines, site-directed mutagenesis was used to restore a C-P-Y-C motif at amino acids 135 to 138 of the mature protein. This mutant plasmid, pFTraF(CPYC), when supplied in trans, encoded a protein that behaved like wild-type TraF and complemented the transfer deficiency of RI89/pOX38-Tc ΔtraF::kan donor cells (data not shown) without induction with arabinose, suggesting that basal expression of TraF was sufficient for complementation. However, mating was undetectable when either the wild-type (pFTraF) or mutated [pFTraF(CPYC)] TraFF was provided in trans in RI90 (dsbA) donor cells treated with 1 mM DTT (Table 5). Wild-type TrhFR27 (pHTrhF) was also unable to complement DTT hypersensitivity when provided in trans even though TrhFR27 contains a C-X-X-C motif (Table 5). Thus TraF and TrhF do not appear to have DsbC-like activity and the presence or absence of the C-X-X-C motif does not affect their function in pilus formation or conjugation.
Periplasmic secretion of the C-terminal regions containing thioredoxin folds of TrbBF and DsbCR27, but not TrhFR27, partially complements a dsbA mutation.
E. coli thioredoxin, a disulfide oxidoreductase, can be secreted to the periplasm via the DsbA signal sequence. Although periplasmic wild-type thioredoxin cannot replace DsbA, thioredoxin variants containing the Grx-type (C-P-Y-C) or DsbA-type (C-P-H-C) active-site sequences could complement a DsbA deficiency by approximately 40 to 60% as measured by motility on a swarm plate (34). Furthermore, a DsbC variant with a 76-amino-acid deletion in its dimerization domain is able to complement the effect of a dsbA null mutation by 44% as measured by alkaline phosphatase activity (9). To determine whether the C-terminal regions containing putative thioredoxin folds of TrbBF, DsbCR27, or TrhFR27 could complement a DsbA deficiency during conjugation, we constructed fusions between the DsbA signal sequence and our truncated proteins using the method of Jonda et al. (34) (Table 1). Mating was assayed using RI190/pOX38-Tc (dsbA) donor cells in the absence of DTT. As a control, the expression of E. coli dsbA from uninduced pKDsbA was shown to nearly fully complement a dsbA mutation as measured by mating efficiency (Table 6). The expression of genes encoding the C-terminal putative thioredoxin domains of TrbBF and DsbCR27 (pFBASS and pHCASS, respectively) was also shown to increase mating efficiency by 11- to 20-fold in the dsbA mutant (Table 6). To test whether having a DsbA-type (C-P-H-C) active site rather than the wild-type (C-P-Y-C) active site of truncated TrbBF would be more effective incomplementing the dsbA mutation, we constructed pFBASS(Y63H) (Table 1), which gave results similar to pFBASS and pHCASS (Table 6), implying that DsbA has attributes other than the thioredoxin domain that contribute to its activity. In contrast, the mating efficiency of pOX38-Tc in RI90 (dsbA) did not increase when the putative thioredoxin domain of TrhFR27 was expressed from pHFASS despite the presence of a C-X-X-C motif (Table 6).
TABLE 6.
Effect of periplasmic secretion of C-terminal putative thioredoxin folds of F- and H-like transfer proteins in a dsbA mutanta
| E. coli strain | Plasmid construct | No. of transconjugants |
|---|---|---|
| pOX38-Tc/RI89 (wild type) | 1.2 × 108 | |
| pOX38-Tc/RI90 (dsbA) | 1.1 × 104 | |
| pOX38-Tc/RI90 (dsbA) | pBAD30 | 1.0 × 104 |
| pOX38-Tc/RI90 (dsbA) | pKDsbA uninduced | 8.0 × 107 |
| pOX38-Tc/RI90 (dsbA) | pKDsbA induced | 5.5 × 104 |
| pOX38-Tc/RI90 (dsbA) | pFBASS uninduced | 1.2 × 105 |
| pOX38-Tc/RI90 (dsbA) | pFBASS induced | 1.3 × 103 |
| pOX38-Tc/RI90 (dsbA) | pFBASS(Y63H) uninduced | 1.6 × 105 |
| pOX38-Tc/RI90 (dsbA) | pFBASS(Y63H) induced | 1.1 × 103 |
| pOX38-Tc/RI90 (dsbA) | pHFASS uninduced | 3.0 × 104 |
| pOX38-Tc/RI90 (dsbA) | pHFASS induced | 2.1 × 104 |
| pOX38-Tc/RI90 (dsbA) | pHCASS uninduced | 2.0 × 105 |
| pOX38-Tc/RI90 (dsbA) | pHCASS induced | 1.1 × 103 |
Induced refers to the addition of 1% arabinose 2 hours prior to adding the recipient cells for the mating assay; uninduced refers to the continued growth for 2 hours without the addition of arabinose.
Interestingly, induction of these constructs (pKDsbA, pFBASS, and pHCASS) with 1% arabinose gave a decrease in mating efficiency. Perhaps this overexpression caused an excessive increase of oxidizing potential in the periplasm that interferes with the disulfide bond isomerization. For instance, overproduction of DsbB, but not DsbA, results in a defect in periplasmic disulfide bond formation, supporting the idea that isomerization has an optimal upper limit (32).
DISCUSSION
Sequence homology searches detected a putative thioredoxin fold within the C-terminal regions of TraFF and TrbBF encoded by F-like T4SS and TrhFH, DsbCH, and HtdTH within or near regions encoded by the T4SS in IncH plasmids. TrbBF and homologues encoded by the three H plasmids contain the thioredoxin C-X-X-C active-site motif, as well as the essential proline residue that is adjacent to the active-site loop. They also contain a hydrophobic or aromatic residue at the third position, which is consistent with a possible role in disulfide bond isomerization (13). The second position can vary and affects interactions with the substrate (43), suggesting substrate specificity in each system. DsbCH and HtdTH also conserve the extra helical domain that is present in E. coli DsbC, including the disulfide bond between residues Cys141 and Cys163. This disulfide bond is required for the chaperone activity of E. coli DsbC (44), further suggesting that DsbCH and HtdTH are homologues of DsbC.
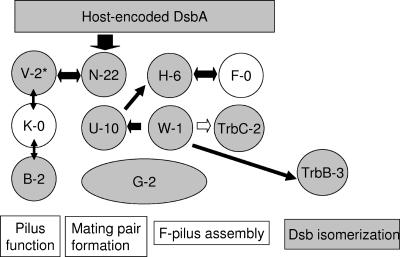
Proteins that promote disulfide bond formation have previously been shown to be essential for the proper assembly of a number of periplasmic protein complexes, including the E. coli flagellar apparatus, the enteropathogenic E. coli bundle-forming pilus, the E. coli pap pili, the Klebsiella oxytoca type II secreton, and Salmonella enterica serovar Typhimurium plasmid-encoded fimbriae (15, 58). In addition to previous studies on the effect of a dsbA mutation on F piliation as measured by filamentous phage sensitivity and electron microscopy (10), we observed that the mating efficiency of the F plasmid is also affected by the presence of a dsbA mutation in the donor cell and that the addition of DTT further decreases mating ability to undetectable levels. Interestingly, the F-like T4SS-specific proteins TraN, -U, -H, and -W and TrbC have 22, 11, 6, 1, and 2 conserved cysteines, respectively. The F-like universal T4SS envelope proteins TraB, -V, and -G also have 2, 3, and 2 cysteines, respectively, suggesting there are many candidates for disulfide bond formation and isomerization (Fig. 4). Nonreducing gel electrophoresis has demonstrated that wild-type TraV and a TraVC18S mutant can form mixed disulfides with many cell envelope proteins (30). Similar experiments based on protein mobility under nonreducing conditions have determined the presence of intra- and intermolecular disulfide bonds in TraN (39). In addition, the T4SS of the Ti plasmid, which has no apparent thioredoxin-like homologues, relies on disulfide bond formation for the stability of its transfer protein complex. In this case, VirB7 stabilizes VirB9 by a disulfide cross-link and the B7-B9 dimer in turn stabilizes other VirB proteins (16). Since the dsbA mutation affects F piliation, TraB, -G, -H, -V, and -W and TrbC, which are involved in pilus assembly, are the best candidates. The effect of a dsbA mutation on other proteins that are specific to F-like T4SS, which are involved in mating pair stabilization and possibly pilus retraction, cannot be readily assessed in the absence of the pilus.
FIG. 4.
A representation of possible interactions in the F pilus assembly and mating pair formation system. The proteins for F pilus assembly and mating pair formation are divided into three groups: those that are highly conserved among T4SS and are involved in pilus assembly and function (TraV, -K, and -B), F-T4SS proteins involved in mating pair stabilization (TraN, -U, and -G), and F-T4SS proteins involved in pilus assembly (TraF,-G [N-terminal region], -H, and -W and TrbC). DsbA is known to affect pilus assembly (10) and TraN stability (39). Host-encoded DsbC and -G may be involved in disulfide bond isomerization (not shown) as might be F TrbB. For the R27 plasmid, DsbC and HtdT might also be involved (not shown). The number of cysteines is shown after each protein (single letter for Tra proteins), indicating a potential for disulfide bond formation. Proteins with no cysteines are emphasized in white. The possible interactions between the proteins are summarized according to Harris and Silverman (31). White arrows indicate inferred interactions from sequence data for TrbC and TraW, which are fused into one protein in R27 (42).
In addition to TraFH, the H-like T4SS also encode homologues (DsbCH and HtdTH) that closely resemble the host DsbC protein, which is known to be involved in disulfide bond isomerization (49). DsbCR27 and HtdTR27 along with TrbBF could act as disulfide bond isomerases, rather than disulfide bond-forming proteins, because (i) TrbBF cannot complement a host dsbA mutant; (ii) TrbBF, DsbCR27, and HtdTR27 can alleviate the DTT hypersensitivity of a dsbA null mutant, similar to the E. coli DsbC disulfide bond isomerase (53, 66); (iii) truncated TrbBF and DsbCR27 proteins, containing the predicted thioredoxin fold, can partially complement a dsbA null mutation; (iv) single trbB and E. coli dsbC null mutants have no phenotype whereas a trbB dsbC double mutant has a 10-fold decrease in mating efficiency; and (v) DsbCR27 and HtdTR27 both conserve the extra helical domain of E. coli DsbC, including the important disulfide bond between Cys141 and Cys163, which determines specificity for chaperone activity.
TraFF is essential for pilus assembly even though it does not contain a C-X-X-C active-site motif or any other cysteines. When a C-P-Y-C motif was introduced by mutagenesis, TraFF remained able to complement a traF mutation in trans but could not alleviate the DTT hypersensitivity of a dsbA donor cell. Similarly, TrhFR27, which has the conserved C-X-X-C motif, is unable to counteract the hypersensitivity of a dsbA mutant to DTT. Furthermore, the cloned C-terminal domain of TrhFR27, containing the thioredoxin fold element, was unable to complement the dsbA mutation. TrhFR27 might have substrate specificity or redox potential requirements that do not allow it to affect general periplasmic disulfide bond formation. Liu and Wang (44) have shown that the active-site cysteine residues of E. coli DsbC are necessary for enzyme activity but are not required for substrate binding and chaperone function. Therefore, TraFF and TrhFR27 may act as chaperones during pilus assembly. Yeast two-hybrid studies have indicated that TraFF interacts with TraHF (six cysteines) (31), which has a C-terminal coiled-coil domain (42), making it an excellent candidate for chaperone-assisted assembly in the periplasm.
It should be noted that thioredoxin has been found to play a role in processes unrelated to disulfide bond chemistry. It has a role in processivity of the T7 DNA polymerase (29) and in filamentous phage assembly (22), suggesting that TraF and TrhF might also have an unexpected role in pilus biogenesis.
There are more proteins with disulfide bonds than there are disulfide bond isomerases, suggesting there may be overlapping specificities that can explain the lack of a phenotype in trbB and E. coli dsbC null mutants (35). Interestingly, many large F- and H-like conjugative plasmids encode at least one Dsb homologue containing the C-X-X-C motif. H-like transfer regions such as those of pNL1 (63), SXT (11), R391 (14), Rts1 (55), and pCAR1 (46) encode either DsbCH or HtdTH but not both, whereas R27 (40, 41, 65), R478 (26), and pHCM1 (57) have two transfer regions, each encoding either HtdTH or DsbCH. Both F- and H-like transfer regions encode TraF or TrhF; however, trbB is found only in plasmids encoding TraF lacking the C-X-X-C motif. The strict conservation of at least one functional Dsb homologue in each plasmid suggests a need for assisted disulfide bond formation. This conserved presence of plasmid-derived Dsb homologues, in spite of the chromosomally expressed DsbC, might be due to the relatively low isomerase activity of E. coli DsbC compared to eukaryotic protein disulfide isomerase, for instance (20). Since very few periplasmic and outer membrane proteins require more than two disulfide bonds per subunit, a high isomerase activity might not be necessary (20) whereas a resident plasmid producing several cell envelope proteins containing multiple disulfide bonds might need auxiliary isomerase activities. Unfortunately, we cannot determine definitively at this time whether these plasmid-encoded proteins are disulfide bond isomerases for their respective DNA transfer systems since there are no assays for their activity beyond pilus formation and conjugation. We are currently examining which of the many candidate proteins encoded by the F- and H-like transfer regions besides TraN (39) require DsbA for pilus assembly and transfer.
Acknowledgments
This work was supported by grants from NSERC and CIHR toL.S.F. and the Alberta Heritage Foundation for Medical Research toB.H.
We thank Diane Taylor and members of her laboratory for useful discussions and providing plasmid R27.
REFERENCES
- 1.Achtman, M., N. Kennedy, and R. Skurray. 1977. Cell-cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc. Natl. Acad. Sci. USA 74:5104-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anfinsen, C. B., E. Haber, M. Sela, and F. H. White, Jr. 1961. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc. Natl. Acad. Sci. USA 47:1309-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony, K. G., C. Sherburne, R. Sherburne, and L. S. Frost. 1994. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol. Microbiol. 13:939-953. [DOI] [PubMed] [Google Scholar]
- 5.Anthony, K. G., W. A. Klimke, J. Manchak, and L. S. Frost. 1999. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J. Bacteriol. 181:5149-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Audette, G. F., S. J. Holland, T. C. Elton, J. Manchak, K. Hayakawa, L. S. Frost, and B. Hazes. 2004. Crystallization and preliminary diffraction studies of TraF, a component of the Escherichia coli type IV secretory system. Acta Crystallogr. Sect. D Biol. Crystallogr. 60:2025-2027. [DOI] [PubMed] [Google Scholar]
- 7.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 8.Bader, M., W. Muse, D. P. Ballou, C. Gassner, and J. C. Bardwell. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217-227. [DOI] [PubMed] [Google Scholar]
- 9.Bader, M. W., A. Hinniker, J. Regeimbal, D. Goldstone, P. W. Haebel, J. Riemer, P. Metcalf, and J. C. Bardwell. 2001. Turning a disulfide isomerase into an oxidase: DsbC mutants that imitate DsbA. EMBO J. 20:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 11.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analysis of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bessette, P. H., J. J. Cotto, H. F. Gilbert, and G. Georgiou. 1999. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J. Biol. Chem. 274:7784-7792. [DOI] [PubMed] [Google Scholar]
- 13.Bessette, P. H., J. Qiu, J. C. Bardwell, J. R. Swartz, and G. Georgiou. 2001. Effect of sequences of the active-site dipeptides of DsbA and DsbC on in vivo folding of multidisulfide proteins in Escherichia coli. J. Bacteriol. 183:980-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Böltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouwman, C. W., M. Kohli, A. Killoran, G. A. Touchie, R. J. Kadner, and N. L. Martin. 2003. Characterization of SrgA, a Salmonella enterica serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J. Bacteriol. 185:991-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charbonnier, J. B., P. Belin, M. Moutiez, E. A. Stura, and E. Quemeneur. 1999. On the role of the cis-proline residue in the active site of DsbA. Protein Sci. 8:96-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. T. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, J., J. L. Song, S. Zhang, Y. Wang, D. F. Cui, and C. C. Wang. 1999. Chaperone activity of DsbC. J. Biol. Chem. 274:19601-19605. [DOI] [PubMed] [Google Scholar]
- 21.Dailey, F. E., and H. C. Berg. 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:1043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng, J. N., P. Model, and M. Russel. 1999. A trans-envelope protein complex needed for filamentous phage assembly and export. Mol. Microbiol. 34:745-755. [DOI] [PubMed] [Google Scholar]
- 23.Foster, G. C., G. C. McGhee, A. L. Jones, and G. W. Sundin. 2004. Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 70:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuya, N., and T. Komano. 1996. Nucleotide sequence and characterization of the trbABC region of the IncI1 plasmid R64: existence of the pnd gene for plasmid maintenance within the transfer region. J. Bacteriol. 178:1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmour, M. W., N. R. Thomson, M. Sanders, J. Parkhill, and D. E. Taylor. 2004. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52:182-202. [DOI] [PubMed] [Google Scholar]
- 27.Gubbins, M. J., W. R. Will, and L. S. Frost. 2005. The F-plasmid, a paradigm for bacterial conjugation, p. 151-206. In P. Mullany (ed.), The dynamic bacterial genome. Cambridge University Press, Cambridge, United Kingdom.
- 28.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamdan, S. M., B. Marintcheva, T. Cook, S.-J. Lee, S. Tabor, and C. C. Richardson. 2005. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc. Natl. Acad. Sci. USA 102:5096-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 30.Harris, R. L., and P. M. Silverman. 2002. Roles of internal cysteines in the function, localization, and reactivity of the TraV outer membrane lipoprotein encoded by the F plasmid. J. Bacteriol. 184:3126-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris, R. L., and P. M. Silverman. 2004. Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J. Bacteriol. 186:5480-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jander, G., N. L. Martin, and J. Beckwith. 1994. Two cysteines in each periplasmic domain of the membrane protein DsbB are required for its function in protein disulfide bond formation. EMBO J. 13:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeng, M. F., A. P. Campbell, T. Begley, A. Holmgren, D. A. Case, P. E. Wright, and H. J. Dyson. 1994. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure 2:853-868. [DOI] [PubMed] [Google Scholar]
- 34.Jonda, S., M. Huber-Wunderlich, R. Glockshuber, and E. Mössner. 1999. Complementation of DsbA deficiency with secreted thioredoxin variants reveals the crucial role of an efficient dithiol oxidant for catalyzed protein folding in the bacterial periplasm. EMBO J. 18:3271-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadokura, H., F. Katzen, and J. Beckwith. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72:111-135. [DOI] [PubMed] [Google Scholar]
- 36.Kadokura, H., H. Tian, T. Zander, J. C. Bardwell, and J. Beckwith. 2004. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 303:534-537. [DOI] [PubMed] [Google Scholar]
- 37.Kathir, P., and K. Ippen-Ihler. 1991. Construction and characterization of derivatives carrying insertion mutations in F plasmid transfer region genes, trbA, artA, traQ, and trbB. Plasmid 26:40-54. [DOI] [PubMed] [Google Scholar]
- 38.Klimke, W. A., and L. S. Frost. 1998. Genetic analysis of the role of the transfer gene, traN, of the F and R100-1 plasmids in mating pair stabilization during conjugation. J. Bacteriol. 180:4036-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klimke, W. A., C. D. Rypien, B. A. Klinger, R. A. Kennedy, J. M. Rodriguez-Maillard, and L. S. Frost. The mating pair stabilization protein, TraN, of the F plasmid, is an outer membrane protein with two regions that are important for its function in conjugation. Microbiology, in press. [DOI] [PubMed]
- 40.Lawley, T. D., M. W. Gilmour, J. E. Gunton, L. J. Standeven, and D. E. Taylor. 2002. Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J. Bacteriol. 184:2173-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawley, T. D., M. W. Gilmour, J. E. Gunton, D. M. Tracz, and D. E. Taylor. 2003. Functional and mutational analysis of conjugative transfer region 2 (Tra2) from the IncHI1 plasmid R27. J. Bacteriol. 185:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 43.Lin, T. Y., and T. S. Chen. 2004. A positive charge at position 33 of thioredoxin primarily affects its interaction with other proteins but not redox potential. Biochemistry 43:945-952. [DOI] [PubMed] [Google Scholar]
- 44.Liu, X., and C. C. Wang. 2001. Disulfide-dependent folding and export of Escherichia coli DsbC. J. Biol. Chem. 276:1146-1151. [DOI] [PubMed] [Google Scholar]
- 45.Lu, J., J. Manchak, W. Klimke, C. Davidson, N. Firth, R. A. Skurray, and L. S. Frost. 2002. Analysis and characterization of the IncFV plasmid pED208 transfer region. Plasmid 48:24-37. [DOI] [PubMed] [Google Scholar]
- 46.Maeda, K., H. Nojiri, M. Shintani, T. Yoshida, H. Habe, and T. Omori. 2003. Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J. Mol. Biol. 326:21-33. [DOI] [PubMed] [Google Scholar]
- 47.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin, J. L. 1995. Thioredoxin—a fold for all reasons. Structure 3:245-250. [DOI] [PubMed] [Google Scholar]
- 49.Maskos, K., M. Huber-Wunderlich, and R. Glockshuber. 2003. DsbA and DsbC-catalyzed oxidative folding of proteins with complex disulfide bridge patterns in vitro and in vivo. J. Mol. Biol. 325:495-513. [DOI] [PubMed] [Google Scholar]
- 50.McCarthy, A. A., P. W. Haebel, A. Torronen, V. Rybin, E. N. Baker, and P. Metcalf. 2000. Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat. Struct. Biol. 7:196-199. [DOI] [PubMed] [Google Scholar]
- 51.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 52.Missiakas, D., C. Georgopoulos, and S. Raina. 1993. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc. Natl. Acad. Sci. USA 90:7084-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Missiakas, D., C. Georgopoulos, and S. Raina. 1994. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 13:2013-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore, D., K. Maneewannakul, S. Maneewannakul, J. H. Wu, K. Ippen-Ihler, and D. E. Bradley. 1990. Characterization of the F-plasmid conjugative transfer gene traU. J. Bacteriol. 172:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murata, T., M Ohnishi, T. Ara, J. Kaneko, C. G. Han, Y. F. Li, K. Takashima, H. Nojima, K. Nakayama, A. Kaji, Y. Kamio, T. Miki, H. Mori, E. Ohtsubo, Y. Terawaki, and T. Hayashi. 2002. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J. Bacteriol. 184:3194-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 57.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 58.Pugsley, A. P., N. Bayan, and N. Sauvonnet. 2001. Disulfide bond formation in secreton component PulK provides a possible explanation for the role of DsbA in pullulanase secretion. J. Bacteriol. 183:1312-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raina, S., and D. Missiakas. 1997. Making and breaking disulfide bonds. Annu. Rev. Microbiol. 51:179-202. [DOI] [PubMed] [Google Scholar]
- 60.Rees, C. E. D., D. E. Bradley, and B. M. Wilkins. 1987. Organization and regulation of the conjugation genes of IncI1 plasmid ColIb-P9. Plasmid 18:223-236. [DOI] [PubMed] [Google Scholar]
- 60a.Rietsch, A., D. Belin, N. Martin, and J. Beckwith 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:13048-13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rietsch, A., P. Bessette, G. Georgiou, and J. Beckwith. 1997. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol. 179:6602-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ritz, D., and J. Beckwith. 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 55:21-48. [DOI] [PubMed] [Google Scholar]
- 63.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed]
- 64.Sambrook, J., J. E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 65.Sherburne, C. K., T. D. Lawley, M. W. Gilmour, F. R. Blattner, V. Burland, E. Grotbeck, D. J. Rose, and D. E. Taylor. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shevchik, V. E., I. Bortoli-German, J. Robert-Baudouy, S. Robinet, F. Barras, and G. Condemine. 1995. Differential effect of dsbA and dsbC mutations on extracellular enzyme secretion in Erwinia chrysanthemi. Mol. Microbiol. 16:745-753. [DOI] [PubMed] [Google Scholar]
- 67.Wilkins, B. M., and L. S. Frost. 2001. Mechanisms of gene exchange between bacteria, p. 355-400. In M. Sussman (ed.), Molecular medical microbiology. Academic Press, London, United Kingdom.
- 68.Wunderlich, M., and R. Glockshuber. 1993. Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli. Protein Sci. 2:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]