Abstract
The bacterial quorum-sensing autoinducer 2 (AI-2) has received intense interest because the gene for its synthase, luxS, is common among a large number of bacterial species. We have identified luxS-controlled genes in Escherichia coli under two different growth conditions using DNA microarrays. Twenty-three genes were affected by luxS deletion in the presence of glucose, and 63 genes were influenced by luxS deletion in the absence of glucose. Minimal overlap among these gene sets suggests the role of luxS is condition dependent. Under the latter condition, the metE gene, the lsrACDBFG operon, and the flanking genes of the lsr operon (lsrR, lsrK, tam, and yneE) were among the most significantly induced genes by luxS. The E. coli lsr operon includes an additional gene, tam, encoding an S-adenosyl-l-methionine-dependent methyltransferase. Also, lsrR and lsrK belong to the same operon, lsrRK, which is positively regulated by the cyclic AMP receptor protein and negatively regulated by LsrR. lsrK is additionally transcribed by a promoter between lsrR and lsrK. Deletion of luxS was also shown to affect genes involved in methionine biosynthesis, methyl transfer reactions, iron uptake, and utilization of carbon. It was surprising, however, that so few genes were affected by luxS deletion in this E. coli K-12 strain under these conditions. Most of the highly induced genes are related to AI-2 production and transport. These data are consistent with the function of LuxS as an important metabolic enzyme but appear not to support the role of AI-2 as a true signal molecule for E. coli W3110 under the investigated conditions.
Bacteria can respond to a variety of chemical and physical changes in their environment by regulating gene expression. Changes such as heat shock, nutrient limitation, and high osmolarity can cause multigenic cellular responses in transcription and translation. Some changes which cause similar responses are traced to the bacteria themselves. For example, some bacteria produce metabolites that are released into the environment as the cell density increases. These molecules could be metabolic wastes, which are toxic to the normal physiological activities of the cells and are therefore secreted. However, some metabolic products may serve as a signaling molecule, which can be perceived by the cells to control the expression of specific genes as the population increases. It has been pointed out that such a chemical molecule can only be considered as truly signaling if the cellular response extends beyond physiological activities required to catabolize the signal molecule (52). This type of signaling molecule-dependent regulation confers upon bacteria the capability to communicate with each other and coordinate their activities and has been termed “quorum sensing.”
For example, many bacteria produce and secrete a freely diffusible signaling molecule, acyl-homoserine lactone (AHL). With an increase in cell density, the concentration of AHL can increase and reach a threshold stimulatory level, at which the signal molecule binds to a LuxR-like protein, the transcriptional regulator, to control gene expression and cell activity. AHL-mediated quorum sensing is well documented in gram-negative bacteria (28, 53).
A large number of gram-negative and gram-positive bacteria have been found to produce and release another type of signaling molecule, autoinducer 2 (AI-2), which can act via a phosphorelay cascade to stimulate production of bioluminescence in Vibrio harveyi. Schauder et al. showed that AI-2 is produced from S-adenosylmethionine in three enzymatic steps, wherein LuxS is the enzyme most directly linked to AI-2 production (38). More than 55 bacterial species possess a gene homologous to luxS (22, 43, 49, 54), and many have been shown to produce AI-2-like activities by using a V. harveyi BB170 reporter strain (44, 45). Recent advances have indicated that the AI-2 molecules from various bacterial species may differ in their structure, although all of them are derived from the product of the LuxS reaction, 4,5-dihydroxy-2,3-pentanedione (DPD) (29). DPD is a highly reactive molecule which likely undergoes cyclization and further arrangements to form a mixture of varied chemical molecules (38). The AI-2 molecules from Vibrio harveyi and Salmonella enterica serovar Typhimurium have been reported to be (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (S-THMF-borate) and (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF), respectively (6, 29). It was also suggested that DPD, R-THMF, and S-THMF-borate are in an equilibrium which is affected by the presence of borate (29).
Evidence accumulated during the last several years suggests that AI-2/luxS-mediated regulation may be important in controlling different cell activities in a variety of bacterial species (54). Some of these include biofilm production in Streptococcus mutans, S. enterica serovar Typhimurium, and Vibrio cholerae (17, 26, 35, 56); motility in Campylobacter jejuni, enterohemorrhagic Escherichia coli (EHEC), and enteropathogenic E. coli (12, 15, 42); iron acquisition in Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans, and V. harveyi (8, 13, 23); and expression of virulence factors in A. actinomycetemcomitans, E. coli EHEC, P. gingivalis, V. cholerae, and Clostridium perfringens (8, 13, 30, 41, 57). These studies have contributed to our understanding of the AI-2/luxS-mediated regulation of gene expression and cell activity but questions remain, as few genes appear to be directly influenced by AI-2. That is, many studies have depended upon comparison of a luxS mutant and its parent strain, and so there are questions regarding whether luxS-dependent phenotypes are caused by an AI-2 signaling defect, by metabolic perturbation, or by both (49, 52, 54).
In a search for luxS-regulated genes in S. enterica serovar Typhimurium, the lsrACDBFGE operon and the methionine synthase gene metE were identified by Taga et al. (47). They found that the lsrACDBFGE operon encodes an AI-2 uptake and modification system. In E. coli, there exists a similar lsr operon (b1513 operon), except that it does not have the lsrE homolog. It was shown recently that the functions of the E. coli lsr operon and its regulators, LsrR and LsrK, are similar to those in S. enterica serovar Typhimurium, and cyclic AMP (cAMP)-cAMP receptor protein (CRP) are involved in regulation of the lsr operon (50, 55).
In this study, we have attempted to identify the luxS-controlled genes by comparing the wild type and ΔluxS mutant under two different growth conditions using DNA microarrays. In the first case, we examined cells in the presence of glucose at low cell density (late exponential phase). Then, we examined cells in the absence of glucose at high density (early stationary phase). Profiles of gene regulation were very different under these two conditions, and many more genes were significantly affected by luxS deletion in the latter case. Importantly, we have shown new regulatory and structural characteristics for the E. coli lsr and lsrRK operons and demonstrated that both lsrRK and lsr operons are subject to two controllers: a repressor, LsrR, and an activator, cAMP-CRP. This study serves to enhance our understanding of the regulation of AI-2 transport and the growth conditions by which AI-2/luxS modulates gene expression in E. coli K-12.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. E. coli K-12 strain W3110 [F− λ− in(rrnD-rrnE)] was obtained from the Genetic Stock Center (New Haven, Conn.). ΔluxS::kan was moved into W3110 from LW7 (ZK126; ΔluxS::kan) (50) via P1vir transduction. Luria-Bertani broth (LB) contains 5 g liter−1 yeast extract (Difco), 10 g liter−1 Bacto tryptone (Difco), and 10 g liter−1 NaCl. Minimal media have been described previously (32, 39). Cultures of E. coli (wild type and the ΔluxS mutant) that had been grown overnight in LB or LB plus 0.8% glucose were diluted to an optical density at 600 nm (OD600) of about 0.02 in LB or LB plus 0.8% glucose. The cultures were then incubated at 30°C with shaking at 250 rpm in 50-ml flasks. When the cultures reached the appropriate OD600 (1.0 or 2.4), the cells were harvested for RNA extraction.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and property | Source or reference |
|---|---|---|
| E. coli strains | ||
| W3110 | Wild type | E. coli Stock Center |
| LW12 | W3110 ΔluxS::Kan | This study |
| ZK126 | W3110 ΔlacU169 tna-2 | 9 |
| ZK1000 | ZK126 ΔrpoS::Kan | 2 |
| LW2 | ZK126 Δcrp::Kan | 50 |
| LW7 | ZK126 ΔluxS::Kan | 50 |
| LW8 | ZK126 ΔlsrR::Kan | 50 |
| LW9 | ZK126 Δ(lsrACDBFG)::Kan | 50 |
| LW11 | ZK126 ΔlsrK::Kan | 50 |
| Plasmids | ||
| pFZY1 | galK′-lacZYA transcriptional fusion vector, Apr | 21 |
| pJLlsrR | pFZY1 derivative, containing lsrR promoter region, Apr | This study |
| pJLlsrK | pFZY1 derivative, containing lsrK promoter region, Apr | This study |
Plasmid construction.
The plasmids used in this study are listed in Table 1 and were generated using standard procedures (37). Restriction enzymes, T4 DNA ligase, and Vent DNA polymerase were used as specified by the manufacturer (New England BioLabs, Beverly, MA). The E. coli W3110 chromosomal DNA preparation was performed using the QIAGEN DNeasy tissue kit (QIAGEN, Valencia, CA). Extractions of DNA from agarose gels were performed using the QIAGEN QIAEX II gel extraction kit. Oligonucleotides were from Integrated DNA Technologies (Coralville, IA). DNA sequencing was performed at the DNA core facility of the Center of Biosystems Research (University of Maryland Biotechnology Institute). All constructs made by PCR were sequenced to verify their integrity.
Plasmid pFZY1 is a mini-F derivative (average copy number, 1 to 2 per cell) with a polycloning site upstream of a promoterless galK′-lacZYA reporter segment (21). To create pJLlsrR, the lsrR promoter region [−340 to +59 relative to the start codon of lsrR (b1512)] was amplified by PCR using primers lsrRpF (CCGGAATTCTCGATGCCTTTCAGGACATTG) and lsrRpR (CTCGGATCCGCGACCTGTTCTTCTTCACACATT). The purified PCR product was digested with EcoRI-BamHI and was inserted into EcoRI-BamHI-digested pFZY1. To create pJLlsrK, the lsrK promoter region [−367 to +53 relative to the start codon of lsrK (b1511)] was amplified by PCR using primers lsrKpF (CCGGAATTCTCGCTCCGGTTATATCAGCCAGGGCGAACA) and lsrKpR (CTCGGATCCTCCAGCGCCATCAGGTAGTACTTT). The purified PCR product was digested with EcoRI-BamHI and was inserted into EcoRI-BamHI-digested pFZY1.
Total RNA isolation.
Total RNA was isolated from the cultures using an RNeasy Mini kit (QIAGEN, Inc., Valencia, CA) according to the manufacturer's instructions. RNAprotect bacteria reagent (QIAGEN, Inc., Valencia, CA) was added to the cultures to stabilize RNA before isolation. The RNase-free DNase set (QIAGEN, Inc., Valencia, CA) was used for on-column DNase digestion to remove residual DNA; removal of contaminant DNA was confirmed by PCR. RNA quality was examined spectrophotometrically and with gel electrophoresis.
cDNA synthesis and labeling.
cDNA was synthesized and labeled according to the manufacturer's suggestions for the Affymetrix E. coli antisense genome array (Affymetrix, Inc., Santa Clara, CA). Briefly, in 60 μl of reaction mixture, 10 μg of total RNA was used for cDNA synthesis by random primers (12.5 ng/μl) and SuperScript II reverse transcriptase (25 U/μl) (both from Invitrogen Corp., Carlsbad, CA). RNA was removed by addition of 20 μl of 1 N NaOH and incubation at 65°C for 30 min. cDNA was purified with a Qiaquick PCR purification kit (QIAGEN, Inc., Valencia, CA) and then fragmented using DNase I (0.6 U/μg of DNA; Amersham Pharmacia Biotech, Piscataway, NJ) at 37°C for 10 min. The Enzo BioArray terminal labeling kit with biotin-ddUTP (Affymetrix, Inc., Santa Clara, CA) was used to label the 3′ termini of the fragmented cDNA. A gel shift assay with NeutrAvidin (Pierce Biotechnology, Inc., Rockford, IL) was performed to estimate the labeling efficiency based on the instructions from Affymetrix.
Microarray hybridization, washing, and scanning.
Hybridization solution mix was made with the labeled cDNA according to the manufacturer's instructions (Affymetrix, Inc., Santa Clara, CA), and the mixture was hybridized to the E. coli antisense genome arrays at 45°C for 16 h. A GeneChip fluidics station (Affymetrix, Inc., Santa Clara, CA) was used to automate the washing and staining of the arrays. Sequentially, the arrays were stained with ImmunoPure streptavidin (Pierce Biotechnology, Inc., Rockford, IL), antistreptavidin goat antibody (Vector Laboratories, Inc., Burlingame, CA), and R-phycoerythrin streptavidin (Molecular Probes, Inc., Eugene, OR). Finally, the probe arrays were scanned using the Affymetrix GeneArray scanner.
Data analysis.
Microarray data were analyzed with the Affymetrix Microarray Suite software version 5.1 (Affymetrix, Inc., Santa Clara, CA) and the four-comparison survival method (7). The fluorescence of each array was normalized by scaling total chip fluorescence intensities to a common value of 500. For each growth condition, two independent experimental cell cultures (wild type) were compared with two independent control groups (ΔluxS mutant), and four comparisons were made. The fold change for each gene was calculated as the ratio of signal intensity for the wild type to the signal intensity for the ΔluxS mutant. The reported value for the fold change is the average of the four comparisons. Genes with a consistent increase or decrease in all comparisons were determined and used for the analysis. However, the induced genes with absent calls of the array signal in the experimental groups (wild type) and the repressed genes with absent calls of the array signal in the control groups (ΔluxS mutant) were eliminated from the analysis. Genes were considered to be statistically significantly over- or underexpressed based on the following criteria: average change of at least 1.8-fold and P values of <0.05 (t test). Our full microarray data are available at our website (http://www.umbi.umd.edu/%7Ecbr/lab_web/home.htm).
RT-PCR and real-time RT-PCR.
cDNA was synthesized from total RNA and random hexamers using the SuperScript III first-strand synthesis system for reverse transcription-PCR (RT-PCR; Invitrogen) according to the manufacturer's instructions. Real-time RT-PCR was performed in 50 μl of reaction mixture containing the Platinum SYBR Green qPCR Supermix UDG (Invitrogen), 0.2 μM of primers, and cDNA (50°C for 2 min, 95°C for 2 min, 95°C for 15 s, and 60°C for 30 s). The dye-labeled PCR products were detected with a GeneAmp 5700 sequence detection system (Applied Biosystems). Regular RT-PCR was used to check for the existence of the lsrRK and lsr-tam operons, and data presented are from reactions using 22 amplification cycles. Primers were designed and purchased from Integrated DNA Technologies (Coralville, IA) (primer sequences are available upon request). Controls were always used to ensure absence of genomic DNA in the DNase I-treated RNA samples. clpB was used as the normalizing gene for all reactions, since its transcript levels were not significantly different between the wild type and the luxS mutant (data not shown).
β-Galactosidase assays.
Cultures of E. coli were grown overnight in LB, diluted 100-fold into fresh LB, and grown to mid-exponential phase and then diluted into different media with the OD600 below 0.03. The cultures were incubated at 30°C with shaking at 250 rpm in flasks. Samples were removed at regular intervals for determination of the OD600 and β-galactosidase activity. Specific activity of β-galactosidase is expressed in Miller units (27).
Gel mobility shift assay.
The 46-bp DNA fragments containing the wild-type or mutated lsrR promoter regions were synthesized by Integrated DNA Technologies (Coralville, IA). A digoxigenin gel shift kit (Boehringer Mannheim) was used for labeling of DNA fragments and detection of signals according to the manufacturer's instructions. Binding reactions were performed by incubating the labeled DNA fragments with various amounts of purified CRP (generously provided by Fred Schwarz, University of Maryland Biotechnology Institute) in 20 μl of binding buffer (10 mM Tris-HCl [pH 8.0], 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 50 μg ml−1 bovine serum albumin, 15 μg ml−1 sonicated salmon sperm DNA, 100 μM cAMP). Following incubation at 37°C for 10 min, 5 μl of gel loading buffer (0.25× Tris-borate-EDTA [TBE], 60%; glycerol, 40%; bromphenol, 0.2% [wt/vol]) was added, and mixtures were electrophoresed in a 6% native polyacrylamide gel in 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8.0) containing 100 μM cAMP. DNA bands were detected according to the manufacturer's instructions.
Motility assays.
The medium used for the motility swimming assay was tryptone broth (10 g/liter tryptone [Difco], 5 g/liter NaCl) containing 0.3% Difco agar. Cultures of E. coli were grown overnight in liquid tryptone broth, diluted 100-fold into the same fresh medium, and grown to mid-exponential phase. Swim plates were inoculated at the center with 5 μl of cell culture and incubated at 30°C in a humid environment for 11 h.
Biofilm assays.
Biofilm assays were performed as described previously (34) with modifications. E. coli cells were grown in polypropylene tubes in LB at room temperature without shaking for 24 h and subcultured at a 1:100 dilution into different media: LB, LB and glucose, glycerol minimal, glucose minimal, glycerol minimal with Casamino Acids (CAA), glucose minimal with CAA, or minimal medium with CAA. CAA was used at 5%. These cultures were grown for 48 h at room temperature without shaking and then rinsed with distilled water and stained with 1.0% crystal violet. After 20 min, the tubes were rinsed. The biofilm-associated crystal violet was solubilized by dimethyl sulfoxide, and the OD570 of the suspension was measured.
RESULTS
Deletion of the E. coli W3110 luxS gene does not affect growth, motility, and biofilm formation.
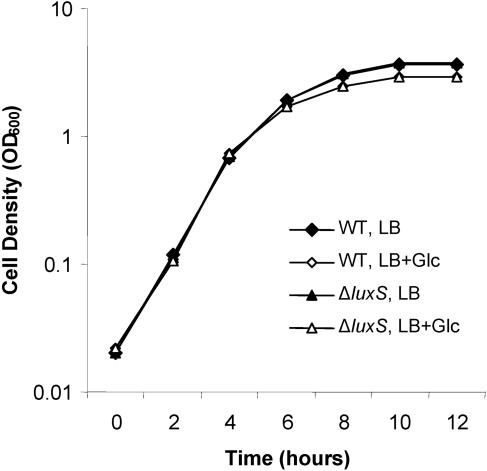
Previous reports showed that deletion of luxS resulted in increased growth rate and reduced motility in EHEC (42). Similarly, a luxS mutant of Campylobacter jejuni had reduced motility (12). In addition, it was shown that the luxS mutant of S. enterica serovar Typhimurium and Streptococcus mutans has a defective ability to form biofilms (26, 35, 56). We investigated whether the mutation of luxS of E. coli K-12 strain W3110 has similar effects on these phenotypes. Figure 1 shows that the ΔluxS mutant grows as well as its isogenic parent when the cells are grown in LB or in LB plus glucose. We further tested the motility of the ΔluxS mutant. On the 0.3% agar swim plate of tryptone broth, there was no apparent difference of the swimming, as measured by the ability to form halos, between the mutant and the wild type (data not shown). Finally, we tested biofilm formation of both strains in various growth media (see Materials and Methods). No significant differences were observed between the wild type and the ΔluxS mutant; biofilm formation was supported in both strains in LB (or with glucose) or various minimal media containing Casamino Acids but not in minimal media without Casamino Acids (see Materials and Methods) (data not shown), consistent with a previous study with E. coli 2K1056 (34). In summary, there were no apparent phenotypic differences between the wild-type and the ΔluxS mutant cells under the investigated conditions, although we did not rule out the possibility that some specific conditions for these assays may affect such differences. To investigate how luxS deletion affects cellular activities, we further performed microarray analysis of these two strains (see below).
FIG. 1.
Growth of the wild type and ΔluxS mutant of E. coli W3110. Overnight cultures of E. coli W3110 and the ΔluxS mutant were diluted in LB or LB plus 0.8% glucose to an OD600 of about 0.02. At different time points during cell growth, aliquots were collected for measurement of the OD600.
Genomic transcriptional analyses of the luxS deletion.
Using DNA microarrays, we compared genomic transcript levels of the wild type and ΔluxS mutant of E. coli W3110 under two different growth conditions. One condition (I) was in LB medium when the cells reached an OD600 of 2.4 (early stationary phase), while the other (II) was in LB plus 0.8% glucose when the cells reached an OD600 of 1.0 (late exponential phase). In the first case, the wild type had low levels of AI-2 and the mutant had none. In addition, the expression level of the lsrACDBFG operon is much higher in wild-type cells than in the ΔluxS mutant, as CRP is required for its activation (50). It is possible that there exist additional genes regulated by luxS under this condition. The second case is characterized by high extracellular AI-2 activity in cultures of the wild type (50) and no AI-2 activity in cultures of the ΔluxS mutant. This is another condition under which AI-2 signaling may be important; the wide disparity in AI-2 for these cultures should be revealing (11).
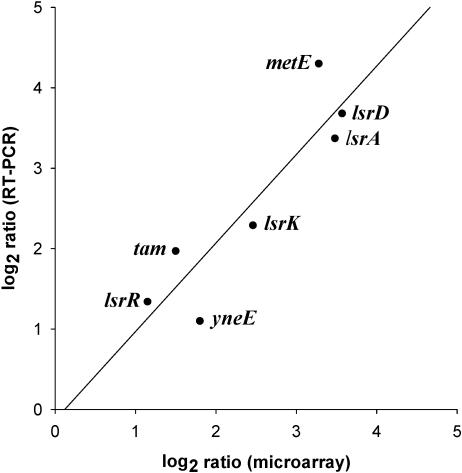
To report the number of genes differentially expressed, we used a 1.8-fold induction ratio as a cutoff limit. Although a 2-fold cutoff is commonly used for analysis of microarray data (20), we have used a slightly less stringent cutoff, as previous studies indicated that even a 1.5-fold difference in transcript level can be biologically significant (3, 19, 40). Table 2 shows that, under condition I (no glucose, OD of 2.4), there were 17 and 46 genes that were induced and repressed at least 1.8-fold by luxS, respectively. To verify our microarray data, we further performed real-time RT-PCR on a selected number of the identified luxS-regulated genes. Figure 2 shows that there was a strong positive correlation (r2 = 0.90) between the two techniques, validating our microarray profiles.
TABLE 2.
Genes regulated by luxS at OD 2.4 in LB
| B no. | Gene | Gene producta | Fold change (WT/ΔluxS) |
|---|---|---|---|
| b1518 | lsrG | ORF, hypothetical protein | 12.64 |
| b1515 | lsrD | Putative transport system permease protein | 11.9 |
| b1513 | lsrA | Putative ATP-binding component of a transport system | 11.16 |
| b1516 | lsrB | Putative LACI-type transcriptional regulator | 10.14 |
| b3829 | metE | Tetrahydropteroyltriglutamate methyltransferase | 9.74 |
| b1517 | lsrF | Putative aldolase | 6.54 |
| b1511 | lsrK | Putative kinase | 5.52 |
| b1520 | yneE | ORF, hypothetical protein | 3.49 |
| b2236 | yfaE | ORF, hypothetical protein | 3.19 |
| b1519 | tam | trans-aconitate 2-methyltransferase | 2.82 |
| b4308 | yjhR | Putative frameshift suppressor | 2.46 |
| b1512 | lsrR | Putative transcriptional regulator, SorC family | 2.22 |
| b1514 | lsrC | Putative transport system permease protein | 2 |
| b4395 | gpmB | Phosphoglyceromutase 2 | 1.99 |
| b3796 | argX | Arginine tRNA3 | 1.97 |
| b3852 | ileT | Isoleucine tRNA1, triplicate | 1.91 |
| b4017 | arpA | Putative regulator of acetyl CoA synthetase | 1.89 |
| b2087 | gatR | Split galactitol utilization operon repressor, interrupted | −1.81 |
| b0974 | hyaC | Probable Ni/Fe-hydrogenase 1 b-type cytochrome subunit | −1.83 |
| b1127 | pepT | Putative peptidase T | −1.85 |
| b1019 | ycdB | ORF, hypothetical protein | −1.85 |
| b1550 | gnsB | ORF, hypothetical protein, GnsB protein | −1.86 |
| b4196 | sgaH | Probable hexulose-6-phosphate synthase | −1.87 |
| b0630 | lipB | Protein of lipoate biosynthesis | −1.87 |
| b1022 | ycdQ | ORF, hypothetical protein | −1.88 |
| b1437 | ORF, hypothetical protein | −1.88 | |
| b4186 | yjfC | Putative synthetase/amidase | −1.92 |
| b2406 | xapB | Xanthosine permease | −1.93 |
| b3939 | metB | Cystathionine gamma-synthase | −1.93 |
| b3945 | gldA | Glycerol dehydrogenase (NAD) | −1.95 |
| b0648 | ybeU | Putative tRNA ligase | −1.98 |
| b1680 | sufS | ORF, hypothetical protein, selenocysteine lyase, PLP dependent | −1.98 |
| b0823 | ybiW | Putative formate acetyltransferase | −2 |
| b1112 | ycfR | ORF, hypothetical protein | −2 |
| b4288 | fecD | Citrate-dependent iron transport, membrane-bound protein | −2.03 |
| b4310 | yjhT | ORF, hypothetical protein | −2.05 |
| b3103 | yhaH | Putative cytochrome | −2.07 |
| b0621 | dcuC | Transport of dicarboxylates | −2.09 |
| b1407 | ydbD | ORF, hypothetical protein | −2.11 |
| b0076 | leuO | Probable transcriptional activator for leuABCD operon | −2.14 |
| b2723 | hycC | Membrane-spanning protein of hydrogenase 3 (part of FHL complex) | −2.14 |
| b3683 | glvC | PTS system, arbutin-like IIC component | −2.2 |
| b3028 | mdaB | NADPH-quinone reductase (modulator of drug activity B) | −2.22 |
| b2968 | yghD | Putative secretion pathway protein | −2.33 |
| b0579 | ybdF | ORF, hypothetical protein | −2.34 |
| b3220 | yhcG | ORF, hypothetical protein | −2.35 |
| b0260 | ykfD | Putative amino acid/amine transport protein | −2.38 |
| b3046 | yqiG | Putative membrane protein | −2.39 |
| b2919 | ygfG | Putative enzyme | −2.4 |
| b3906 | rhaR | Positive regulator for rhaRS operon | −2.47 |
| b2060 | wzc | ORF, hypothetical protein, tyrosine-protein kinase | −2.49 |
| b0790 | ybhP | ORF, hypothetical protein | −2.51 |
| b1720 | ORF, hypothetical protein | −2.52 | |
| b1010 | ycdK | ORF, hypothetical protein | −2.53 |
| b2797 | sdaB | l-serine dehydratase (deaminase), L-SD2 | −2.57 |
| b2993 | hybD | Probable processing element for hydrogenase-2 | −2.59 |
| b0042 | fixB | Probable flavoprotein subunit, carnitine metabolism | −2.7 |
| b1001 | yccE | ORF, hypothetical protein | −2.87 |
| b2549 | yphG | ORF, hypothetical protein | −2.97 |
| b1311 | ycjO | Putative binding protein-dependent transport protein | −3.06 |
| b3141 | agaI | Putative galactosamine-6-phosphate isomerase | −3.15 |
| b1012 | ycdM | ORF, hypothetical protein | −3.24 |
| b1571 | ydfA | ORF, hypothetical protein | −6.85 |
Abbreviations: ORF, open reading frame; CoA, coenzyme A.
FIG. 2.
Correlation of microarray and real-time RT-PCR results. The differences in expression of seven luxS-controlled genes (in LB at high cell density) were log2 transformed and plotted against each other, microarray versus real-time RT-PCR.
Under condition II (with glucose, OD of 1.0), there were fewer genes significantly regulated by luxS (Table 3). With the 1.8-fold cutoff, only 15 and 8 genes were up- and down-regulated, respectively, by the presence of the luxS gene, indicating expression of most of the genes were not affected markedly by the luxS deletion. In addition, the genes regulated by luxS were different from those observed under the first condition (I). These results indicate that luxS-controlled gene expression varies with conditions, suggesting that both (i) careful experimental designs are important in identifying the luxS-controlled genes among various bacteria, and (ii) the role of luxS may vary with conditions.
TABLE 3.
Genes regulated by luxS at OD 1.0 in LB plus glucose
| B no. | Gene | Gene producta | Fold change (WT/ΔluxS) |
|---|---|---|---|
| b1561 | rem | ORF, hypothetical protein | 2.67 |
| b1700 | ydiT | Putative ferredoxin | 2.58 |
| b4186 | yjfC | Putative synthetase/amidase | 2.51 |
| b3711 | yidZ | Putative transcriptional regulator, LYSR type | 2.42 |
| b3580 | lyxK | l-xylulose kinase, cryptic | 2.32 |
| b1567 | ydfW | ORF, hypothetical protein | 2.25 |
| b3004 | ORF, hypothetical protein | 2.19 | |
| b4002 | zraP | ORF, hypothetical protein, Zn-binding periplasmic protein | 2.04 |
| b0805 | ybiL | Putative outer membrane receptor for iron transport | 1.97 |
| b0667 | Putative RNA | 1.93 | |
| b1834 | yebT | ORF, hypothetical protein, putative membrane protein | 1.92 |
| b0671 | Putative RNA | 1.84 | |
| b4367 | fhuF | ORF, hypothetical protein, ferric hydroxamate transport protein | 1.82 |
| b3829 | metE | Tetrahydropteroyltriglutamate methyltransferase | 1.81 |
| b0872 | hcr | Putative enzyme, NADH oxidoreductase for HCP | 1.8 |
| b2597 | yfiA | Putative YhbH sigma 54 modulator | −1.83 |
| b1482 | osmC | Osmotically inducible protein | −1.91 |
| b1461 | ydcE | ORF, hypothetical protein | −1.92 |
| b3267 | yhdV | ORF, hypothetical protein | −1.92 |
| b3110 | yhaO | Putative transport system permease protein | −1.98 |
| b2715 | ascF | PTS system enzyme II ABC (asc), cryptic, transports specific beta-glucosides | −2.02 |
| b3108 | yhaM | ORF, hypothetical protein | −2.14 |
| b3109 | yhaN | ORF, hypothetical protein | −2.87 |
ORF, open reading frame.
Genes controlled by luxS in the absence of glucose at OD of 2.4.
There were more genes down-regulated than up-regulated by luxS when cells were grown to an OD of 2.4 in the absence of glucose. Table 2 shows that 17 and 46 genes were induced and repressed at least 1.8-fold by luxS, respectively. The most significantly induced genes belong to the lsrACDBFG operon. This result is consistent with the previous lsr-lacZ fusion studies performed in E. coli (50, 55), which showed the lsr operon was differentially expressed between the wild type and the luxS mutant mainly in stationary phase. A relatively lower fold change of lsrC compared to the other genes in the lsr operon might have resulted from interfering effects of certain cDNA fragments, which masked the hybridization of lsrC to its probes. Further analysis of lsrC expression by RT-PCR indicated a similar induction level by the luxS gene (∼12-fold change) (data not shown). Surprisingly, expression of lsrR, lsrK, tam, and yneE, which flank the lsrACDBFG operon, were significantly induced by luxS (2.2-, 5.5-, 2.8-, and 3.5-fold, respectively). The lsrR and lsrK genes encode the lsr regulator and the AI-2 kinase, respectively. We investigated their regulation in more detail later. The tam gene encodes an S-adenosyl-l-methionine-dependent methyltransferase, which catalyzes the methyl esterification of trans-aconitate (4). The trans-aconitate appears to be formed spontaneously from the citric acid cycle intermediate cis-aconitate (4). The benefit of methylation of the trans-aconitate to the E. coli cells is not clear. The other luxS-dependent gene, yneE, is within close proximity to the lsr operon and encodes a protein with unknown function. This gene is transcribed in the opposite direction to the tam gene and the lsr operon, and so its up-regulation is potentially important and worthy of further investigation.
metE, which encodes methionine synthase, has much lower expression in the ΔluxS mutant than in the wild-type strain (9.7-fold decrease). This gene was identified before by Taga etal. in their search for luxS-controlled genes in S. enterica serovar Typhimurium (47). MetE catalyzes the last step of methionine synthesis in a vitamin B12-independent pathway from homocysteine which, in turn, can be recycled from the LuxS-catalyzed reaction with S-ribosylhomocysteine. It was previously reported that homocysteine is required for the full induction of metE expression (5, 25, 48). Lack of homocysteine in the ΔluxS mutant was suggested to result in lower transcription of metE (47). On the other hand, homocysteine was shown to play an inhibitory role in the expression of MetA, which catalyzes the first reaction unique to the homocysteine synthetic pathway from homoserine (24). Consistent with this, we found that metA expression was slightly higher in the ΔluxS mutant (1.43-fold), which does not synthesize homocysteine via SAH detoxification. In addition, the expression level of MetB, the enzyme immediately downstream of MetA in homocysteine synthesis, was also increased by luxS deletion (1.93-fold).
Our results also showed that deletion of the luxS gene results in induction of several genes involved in utilization of various carbohydrates. The rhaBAD operon, which encodes enzymes responsible for utilization of l-rhamnose, and its regulatory gene rhaSR (51) all have increased expression in the luxS mutant (1.44-, 1.96-, 1.65-, 1.58-, and 2.47-fold, respectively). Expression of the glvCBG operon, which encodes putative proteins involved in utilization of arbutin, is also induced by 2.2-, 1.68-, and 1.78-fold, respectively. In addition, the luxS mutant has higher expression of dcuC and xapB, which encode proteins involved in transport of dicarboxylates and xanthosine (2.09- and 1.93-fold, respectively). It is not clear why the luxS deletion increases expression of these carbohydrate utilization genes.
In addition to the genes mentioned above, many of the luxS-regulated genes have unknown functions, which are worthy of further investigation.
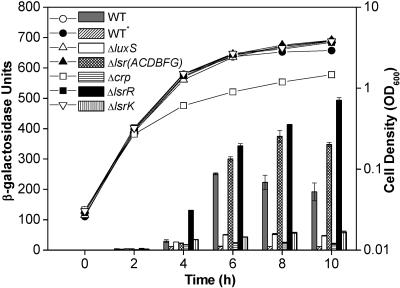
Transcriptional regulation of lsrR by LsrR and CRP.
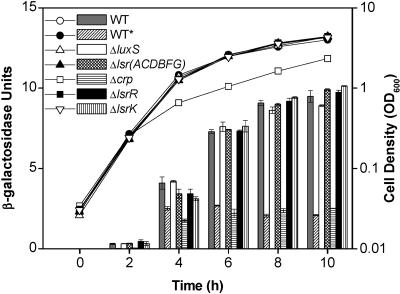
It is interesting that lsrR expression is induced by luxS, as shown by the microarray experiment and quantitative RT-PCR. This was initially unexpected, because the produced LsrR cannot repress the lsr operon (or lsr regulon) in the presence of the inducer phospho-AI-2 under this condition. To investigate the control of lsrR transcription in greater detail, we constructed a lacZ fusion plasmid containing the lsrR promoter region and checked its expression levels in different mutant strains and under different growth conditions (Fig. 3). The overall regulatory pattern of the lsrR gene is similar to that of the lsr operon (50). When the wild-type ZK126 cells (lsrR-lacZ) were grown in LB medium, transcription from the lsrR promoter remained low until the cells entered the stationary phase (Fig. 3). Deletion of lsrR significantly increased lsrR expression, indicating that the lsrR transcription was autorepressed. Since phospho-AI-2 is an inducer that releases LsrR repression (46), it is reasonable to see that deletion of either luxS or lsrK reduced the lsrR expression, because there is no phospho-AI-2 available in these two deletion strains. Figure 3 also showed that deletion of the lsrACDBFG operon increased the lsrR transcription. The presence of an alternative AI-2 transport mechanism, and the absence of phospho-AI-2 degradation enzymes (LsrF and LsrG), may account for this increase, as suggested for lsr expression in the same mutant (50).
FIG. 3.
Transcriptional regulation of lsrR expression. E. coli ZK126 (wild type) and strains containing deletion of crp, luxS, lsrK, lsrR, and lsrACDBFG carry plasmid pJLlsrR (lsrR-lacZ). All strains were grown in LB medium except for ZK126 (WT*), which was grown in LB plus 0.8% glucose. At different time points during cell growth, aliquots were collected for measurement of the OD600 (circles, triangles, and squares) and β-galactosidase activity (bars).
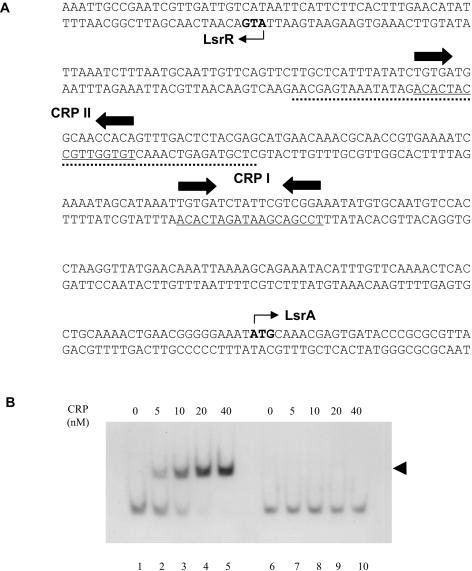
Finally, we found that addition of glucose to the growth medium significantly reduced lsrR transcription and that deletion of the crp gene had effects on lsrR expression similar to the addition of glucose (Fig. 3). These results suggested that lsrR expression was subject to catabolite repression, and CRP was needed for stimulation of lsrR transcription. In our previous paper (50), we identified one CRP binding site (CRP I) located upstream of the lsr promoter region which is necessary for activation of the lsr operon (Fig. 4A). Examination of the intergenic region between lsrR and the lsr operon, which are divergently transcribed, revealed another CRP binding site (CRP II), which has a typical 6-bp spacer between two conserved motifs (Fig. 4A). Gel mobility shift assay results (Fig. 4B) demonstrated that cAMP-CRP binds to a 46-bp DNA fragment in the intergenic region containing this site. CRP did not bind the identical DNA fragment with substitutions in 4 bp of one of the CRP-binding motifs. These findings positively confirm the CRP binding capability to the lsrR regulatory region. Whether the two CRP binding sites are independent or cooperate in stimulation of transcription of lsrR and the lsr operon needs further investigation; however, our results clearly indicate that the promoters of lsrR and the lsr operon are both subject to LsrR repression and CRP activation.
FIG. 4.
CRP binding to regulatory regions of lsrR and the lsr operon. (A) Sequences of the promoter and regulatory regions of lsrR and the lsr operon. The underlined sequences indicate locations of the CRP binding site (CRP I and CRP II). The inverted arrows denote the conserved CRP binding motifs. The dotted DNA sequence was used in gel mobility shift assays. The translation start sites of LsrR and LsrA are shown by small arrows. (B) Gel mobility shift assays were performed as described in Materials and Methods. Digoxigenin-labeled DNA fragments which contained CRP II (the dotted sequence) or changed CRP II (with substitutions in 4 bp in the left CRP binding motif) (see Materials and Methods) were incubated with 0 to 40 nM of purified CRP, as indicated. cAMP was included in all reaction mixtures at a final concentration of 100 μM. The arrow denotes the CRP-DNA complex.
lsrK and lsrR belong to the same operon.
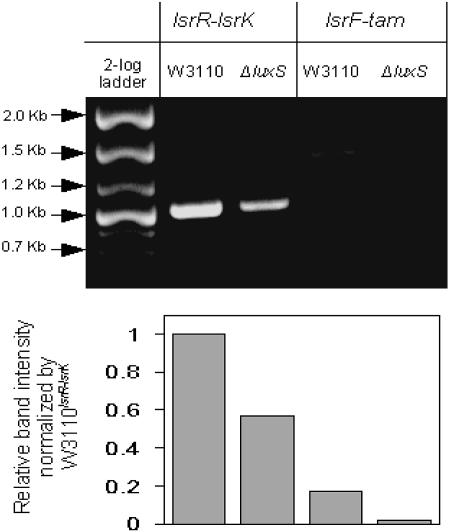
Microarray and quantitative RT-PCR results showed that lsrK expression was increased by the presence of luxS. This is consistent with its role as a kinase that phosphorylates AI-2 newly taken up. To confirm regulation of lsrK expression, we constructed another lacZ fusion plasmid with the lsrK promoter region (−367 to +53 relative to the start codon of lsrK). Surprisingly, deletion of luxS, lsrR, lsrK, or the lsr operon did not affect lsrK expression compared to the wild type (Fig. 5). In addition, the β-galactosidase activities for the lsrK-lacZ fusion were much lower than those of the lsrR-lacZ fusion (Fig. 3 and 5). However, we observed similar hybridization signals for lsrK and lsrR in the wild-type cells during the microarray experiments (data not shown). Further, we found that the luxS mutant cells had lsrK hybridization signals that were much lower than that for lsrR (data not shown). We speculated that lsrK could be transcribed together with lsrR under control of the lsrR promoter. To test this idea (lsrRK operon), we performed regular RT-PCR (differential display). Figure 6 shows that there exists a transcript spanning the coding sequences of both lsrR and lsrK. In addition, the level of this transcript was much lower in the luxS deletion mutant compared to the wild type. These results indicated that lsrR and lsrK belong to one operon. It is possible that weak transcription from the lsrR promoter in the luxS mutant, which is therefore repressed by LsrR, accounts for the polar effect of transcription.
FIG. 5.
Transcriptional regulation of lsrK expression. E. coli ZK126 (wild type) and strains containing deletions of crp, luxS, lsrK, lsrR, and lsrACDBFG carry plasmid pJLlsrK (lsrK-lacZ). All strains were grown in LB medium except ZK126 (WT*), which was grown in LB plus 0.8% glucose. At different time points during cell growth, aliquots were collected for measurement of the OD600 (circles, triangles, and squares) and β-galactosidase activity (bars).
FIG. 6.
Transcriptional analysis of lsr and the lsrRK operon. The agarose gel was run to show DNA fragments obtained from RT-PCR of total RNA prepared from the OD 2.4 cell cultures of the wild type (WT) and the ΔluxS mutant grown in LB. Specific primers were used to amplify the fragments that span coding sequences of the lsrR-lsrK and lsrF-tam genes. The 2-log DNA ladder (New England BioLabs) and intensity results (NIH Image J) are depicted.
Intriguingly, we also found that addition of glucose (0.8%) to the growth medium or deletion of the crp gene reduced transcription of the lsrK-lacZ fusion (Fig. 5), suggesting additional control of lsrK by catabolite repression and CRP, although there are no apparent CRP binding sequences in the lsrK promoter region. Whether CRP directly acts on this promoter awaits further study.
The E. coli lsr operon includes an additional gene, tam.
The E. coli lsrACDBFG operon is similar to the S. enterica serovar Typhimurium lsrACDBFGE operon, except that it does not contain an lsrE homolog. However, the tam gene is located immediately downstream of the lsrE.c operon. Although several programs predict the existence of a potential tam promoter, there appears to be no obvious transcription terminator between lsrG and tam. Both microarray and quantitative RT-PCR showed that tam expression was increased by luxS (about 3.4-fold). Although this induction was lower than those for the other lsr genes, additional transcriptional controls may exist that cause this difference. To check whether the tam gene belongs to the lsr operon, we again used RT-PCR to see whether we could amplify a region spanning coding sequences of lsrF, lsrG, and tam. The results in Fig. 6 support our hypothesis, showing amplification of this region from wild type, but not from ΔluxS (with limited PCR cycles). It should be noted that higher numbers of PCR cycles resulted in the appearance of lsrF-tam products from both strains (data not shown).
DISCUSSION
In our previous search for AI-2-regulated genes in E. coli (11), conditioned media (with or without AI-2) from the same wild-type cells (W3110) and a luxS mutant (MDAI2) were added to luxS mutant cultures, revealing 242 genes that were significantly affected by the resultant 300-fold differential in AI-2. Our study provided useful information on the global effects of AI-2 and luxS; however, because the addition of AI-2 was accompanied by conditioned media to the mutant, there remains ambiguity as to whether the observed effects were caused by AI-2, other compounds in the conditioned medium, or luxS. In the current study, we directly compared the transcriptional profiles of W3110 and the luxS mutant and identified the luxS-controlled genes under two different growth conditions. Some of these genes, such as metE and the lsrACDBFG operon, were identified previously in S. enterica serovar Typhimurium as luxS regulated by using a different method (47). The identified luxS-controlled genes in E. coli K-12 strain W3110 are different from those identified in EHEC O157:H7 (42); there are significantly fewer genes regulated by luxS in W3110. In addition, unlike the EHEC strain, there were no apparent phenotypic differences between the W3110 wild-type and the ΔluxS mutant cells under the investigated conditions.
Sperandio et al. (42) reported that 404 genes were regulated by luxS at least fivefold in the EHEC strain, in which the flagellum and motility genes were highly induced by luxS. In that study, the EHEC wild-type and luxS deletion cells were grown to an OD600 of 1.0 in Dulbecco's modified Eagle's medium at 37°C. One phenotypic difference between the luxS mutants of W3110 and EHEC is that the luxS mutant of EHEC grows much faster than its parent strain (42), while the growth of the W3110 luxS mutant and its parent strain are indistinguishable under the investigated conditions. Although we do not understand the reason for the luxS-mediated growth stimulation in EHEC, we suspect that the faster growth rate in the EHEC luxS mutant may have distorted the effects reported forthe luxS mutation. A second phenotypic difference between the two strains was that the EHEC luxS mutant had reduced motility relative to the wild type (42), while the W3110 luxS mutant does not.
Genomic comparisons of K-12 and EHEC O157:H7 strains revealed that they share a 4.1-Mb backbone sequence, which is punctuated by hundreds of strain-specific genomic regions (K-islands and O-islands) (14, 18, 33). These genetic differences as well as the difference in growth conditions between the studies may account for the divergence in identified genes controlled by luxS. That the genes identified under the two growth conditions in our work were mostly of orthogonal sets supports the latter of these hypotheses.
Both the current and previous studies (47) indicated that methionine metabolism and regulation were affected by deletion of luxS. At an OD of 2.4 without glucose, the expression of metE genes was most repressed in the ΔluxS mutant, while expression levels of other methionine synthesis and regulation genes were either unaffected or to a lesser degree. Consistent with this, the metJ gene, which encodes a major repressor of the met regulon (16), did not exhibit significant differences in expression level. It should be noted that the stable expression of MetH, the B12-dependent methionine synthase, might ensure that the reduced MetE level in the ΔluxS mutant did not decrease methionine synthesis significantly under the investigated conditions. That is, relative stability in the biosynthesis and utilization of methionine and S-adenosylmethionine might be important for the cells to function normally, i.e., to preserve peptide synthesis, methyl donation, and spermidine synthesis (16). We do not understand the reason for increased expression of metB in the ΔluxS mutant (1.93-fold), although it may suggest potential relatedness between luxS and metB. It is interesting that the two genes are transcribed in a 4.9-kb operon (ycgJ-metB-cysK-luxS) in the gram-positive pathogen Clostridium perfringens (30, 31), although they are transcribed separately in E. coli. Our results also indicated that the growth conditions influenced the degree of repression of metE. When grown in glucose at an OD of 1.0, the repression of metE expression in the ΔluxS mutant was much lower (1.81-fold). It is not clear whether the global effect of glucose on cell metabolism caused this difference.
Importantly, our microarray and RT-PCR results revealed an unexpected regulatory mode for the E. coli lsr operon (Fig. 7). It was shown that lsrR and lsrK belong to the same lsrRK operon, and the E. coli lsr operon includes an additional gene, tam. Although we do not understanding the advantages conferred upon E. coli cells by Tam-mediated methylation of trans-aconitate, it appears linked to the AI-2 biosynthesis pathway through the reaction of the S-adenosylmethionine-dependent methyl transfer. Transcriptional control of tam may be complicated due to the potential promoter/regulatory sequence between lsrG and tam in addition to the lsrA promoter. It is interesting that another luxS-induced gene, yneE, which is adjacent to tam and is transcribed in the opposite direction as the lsr operon, was significantly affected. We do not know the function of yneE, or whether it is involved in lsr regulation. It was shown earlier that the S. enterica serovar Typhimurium lsr operon does not contain tam or yneE (47). Instead, it includes the lsrE gene at the end of the lsr operon, which is homologous to the rpe gene that encodes a ribulose phosphate epimerase (47). Whether these genetic differences between these two organisms cause differences in luxS-regulated genes and related cellular activities awaits further investigation.
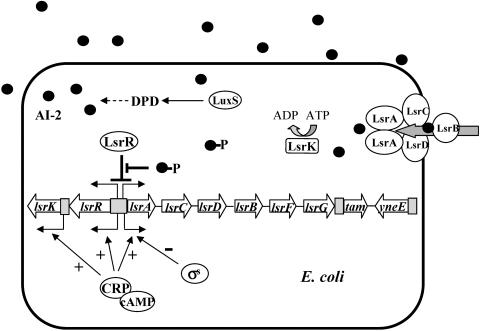
FIG. 7.
Schematic of lsr and lsrRK operon regulation. cAMP-CRP and LsrR are involved in transcriptional control of lsr and lsrRK operons. cAMP-CRP, a positive regulator, stimulates expression of both operons in the absence of glucose, while LsrR prevents their expression. The function of LsrR is inhibited when cell density increases and inducer phospho-AI-2 accumulates. In addition, cAMP-CRP stimulates transcription from the lsrK promoter, and σs negatively regulates lsr expression (50). Also included in the schematic is the yneE gene, whose expression is increased by luxS. Gray boxes denote promoter. Plus and minus signs indicate positive and negative transcriptional regulation. DPD, 4,5-dihydroxy-2,3-pentanedione.
It was unexpected that the lsr and lsrRK operons, which are divergently transcribed, are controlled by the same transcriptional regulators: CRP and LsrR (Fig. 7). CRP, as a global regulator, is needed for activation of these two operons through direct binding to two different sequences within the operator regions. LsrR, as a repressor protein for both operons, undergoes dynamic control of the transcription network through a negative autoregulation feedback loop, which has been shown to speed up the transcription response and reduce cell-to-cell fluctuations in the steady-state level of the transcription repressor (1, 10, 36). In the absence of active inducer phospho-AI-2, the LsrR proteins not only repress expression of the lsr operon but also repress transcription of the lsrRK operon. However, when inducers are available, repression by LsrR is released, and transcription of the two operons is increased rapidly. The effect of this autoregulation of LsrR is a more severe or amplified “switch” that responds to high levels of AI-2. Increased levels of LsrR provide a mechanism for quickly shutting down expression of the lsr and lsrRK operons when the inducer phospho-AI-2 is not available later due to the degradation by LsrF and LsrG (46).
It was shown in this study that deletion of luxS affects genes associated with different cell activities in E. coli K-12, such as AI-2 transport, biosynthesis of methionine, transfer of methyl groups, iron uptake, and the utilization of different carbon sources. Many of the differentially expressed proteins have unknown functions. For this study, we had expected to identify genes whose expression might respond to AI-2 signaling and to the metabolic effects of the luxS deletion, such as accumulation of S-ribosylhomocysteine and a reduced level of homocysteine. It was shown that the most highly induced genes (such as the lsr operon, lsrR, lsrK, tam, and metK) are related to AI-2 production and transport, while the genes involved in other activities are induced to a lesser degree. These data are consistent with the function of LuxS as an important metabolic enzyme in E. coli K-12 and suggest that its role as a signal molecule might require additional cellular factors not revealed in our study. In a previous study (50), we showed that the presence of glucose strongly inhibits expression of the lsr operon through control of cAMP-CRP. That the lsr operon is induced only in the absence of glucose is consistent with the notion that AI-2 is used here as a carbon source. Although Taga et al. reported that S. enterica serovar Typhimurium could not grow on AI-2 as the sole carbon source (47), additional conditions may be needed for AI-2 utilization as a carbon source, as suggested by Winzer et al. (52). It is possible that AI-2, which is synthesized by many species, is perceived as a true signal only by specific bacterial species or under specific conditions.
Acknowledgments
We thank F. P. Schwarz for the gift of CRP and R. Kolter, L. I. Rothfield, and M. Berlyn for generously providing strains and plasmids used in the study. We are grateful to S. W. Hutcheson, and R. C. Stewart, D. C. Straney, and S. K. Samal for helpful discussions.
This work was supported by the U.S. Army, SBCCOM, APG, Md. (DAAD 13-01-C-0036) and the National Science Foundation (BES −0222687).
REFERENCES
- 1.Becskei, A., and L. Serrano. 2000. Engineering stability in gene networks by autoregulation. Nature 405:590-593. [DOI] [PubMed] [Google Scholar]
- 2.Bohannon, D. E., N. Connell, J. Keener, A. Tormo, M. Espinosa-Urgel, M. M. Zambrano, and R. Kolter. 1991. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of sigma 70. J. Bacteriol. 173:4482-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2002. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect. Immun. 70:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, H., and S. Clarke. 1999. A novel methyltransferase catalyzes the methyl esterification of trans-aconitate in Escherichia coli. J. Biol. Chem. 274:13470-13479. [DOI] [PubMed] [Google Scholar]
- 5.Cai, X. Y., B. Redfield, M. Maxon, H. Weissbach, and N. Brot. 1989. The effect of homocysteine on MetR regulation of metE, metR and metH expression in vitro. Biochem. Biophys. Res. Commun. 163:79-83. [DOI] [PubMed] [Google Scholar]
- 6.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y. W., P. Zhao, R. Borup, and E. P. Hoffman. 2000. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J. Cell Biol. 151:1321-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, B. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connell, N., Z. Han, F. Moreno, and R. Kolter. 1987. An E. coli promoter induced by the cessation of growth. Mol. Microbiol. 1:195-201. [DOI] [PubMed] [Google Scholar]
- 10.De Keersmaecker, S. C., K. Marchal, T. L. Verhoeven, K. Engelen, J. Vanderleyden, and C. S. Detweiler. 2005. Microarray analysis and motif detection reveal new targets of the Salmonella enterica serovar Typhimurium HilA regulatory protein, including hilA itself. J. Bacteriol. 187:4381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elvers, K. T., and S. F. Park. 2002. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology 148:1475-1481. [DOI] [PubMed] [Google Scholar]
- 13.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukiya, S., H. Mizoguchi, T. Tobe, and H. Mori. 2004. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 186:3911-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 16.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 17.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 20.Ichikawa, J. K., A. Norris, M. G. Bangera, G. K. Geiss, A. B. van 't Wout, R. E. Bumgarner, and S. Lory. 2000. Interaction of pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc. Natl. Acad. Sci. USA 97:9659-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koop, A. H., M. E. Hartley, and S. Bourgeois. 1987. A low-copy-number vector utilizing beta-galactosidase for the analysis of gene control elements. Gene 52:245-256. [DOI] [PubMed] [Google Scholar]
- 22.Lerat, E., and N. A. Moran. 2004. The evolutionary history of quorum-sensing systems in bacteria. Mol. Biol. Evol. 21:903-913. [DOI] [PubMed] [Google Scholar]
- 23.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 24.Mares, R., M. L. Urbanowski, and G. V. Stauffer. 1992. Regulation of the Salmonella typhimurium metA gene by the MetR protein and homocysteine. J. Bacteriol. 174:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maxon, M. E., B. Redfield, X. Y. Cai, R. Shoeman, K. Fujita, W. Fisher, G. Stauffer, H. Weissbach, and N. Brot. 1989. Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc. Natl. Acad. Sci. USA 86:85-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 29.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 30.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171-179. [DOI] [PubMed] [Google Scholar]
- 31.Ohtani, K., H. Takamura, H. Yaguchi, H. Hayashi, and T. Shimizu. 2000. Genetic analysis of the ycgJ-metB-cysK-ygaG operon negatively regulated by the VirR/VirS system in Clostridium perfringens. Microbiol. Immunol. 44:525-528. [DOI] [PubMed] [Google Scholar]
- 32.Pardee, A. B., F. Jacob, and J. Monod. 1959. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase in E. coli. J. Mol. Biol. 1:165-178. [Google Scholar]
- 33.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 34.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 35.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenfeld, N., M. B. Elowitz, and U. Alon. 2002. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 323:785-793. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 39.Silhavy, T. J., M. L. Berman, L. W. Enquist, and Cold Spring Harbor Laboratory. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun, J., R. Daniel, I. Wagner-Dobler, and A. P. Zeng. 2004. Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 47.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 48.Urbanowski, M. L., and G. V. Stauffer. 1989. Genetic and biochemical analysis of the MetR activator-binding site in the metE metR control region of Salmonella typhimurium. J. Bacteriol. 171:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making “sense” of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 50.Wang, L., Y. Hashimoto, C. Y. Tsao, J. J. Valdes, and W. E. Bentley. 2005. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187:2066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickstrum, J. R., and S. M. Egan. 2004. Amino acid contacts between sigma 70 domain 4 and the transcription activators RhaS and RhaR. J. Bacteriol. 186:6277-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winzer, K., K. R. Hardie, and P. Williams. 2002. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 5:216-222. [DOI] [PubMed] [Google Scholar]
- 53.Withers, H., S. Swift, and P. Williams. 2001. Quorum sensing as an integral component of gene regulatory networks in gram-negative bacteria. Curr. Opin. Microbiol. 4:186-193. [DOI] [PubMed] [Google Scholar]
- 54.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 55.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida, A., T. Ansai, T. Takehara, and H. K. Kuramitsu. 2005. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl. Environ. Microbiol. 71:2372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]