Abstract
Esterase (EST) from Pseudomonas putida IFO12996 catalyzes the stereoselective hydrolysis of methyl dl-β-acetylthioisobutyrate (dl-MATI) to produce d-β-acetylthioisobutyric acid (DAT), serving as a key intermediate for the synthesis of angiotensin-converting enzyme inhibitors. The EST gene was cloned and expressed in Escherichia coli; the recombinant protein is a non-disulfide-linked homotrimer with a monomer molecular weight of 33,000 in both solution and crystalline states, indicating that these ESTs function as trimers. EST hydrolyzed dl-MATI to produce DAT with a degree of conversion of 49.5% and an enantiomeric excess value of 97.2% at an optimum pH of about 8 to 10 and an optimum temperature of about 57 to 67°C. The crystal structure of EST has been determined by X-ray diffraction to a resolution of 1.6 Å, confirming that EST is a member of the α/β hydrolase fold superfamily of enzymes and includes a catalytic triad of Ser97, Asp227, and His256. The active site is located approximately in the middle of the molecule at the end of a pocket ∼12 Å deep. EST can hydrolyze the methyl ester group without affecting the acetylthiol ester moiety in dl-MATI. The examination of substrate specificity of EST toward other linear esters revealed that the enzyme showed specific activity toward methyl esters and that it recognized the configuration at C-2.
Esterases belong to the hydrolase family and are ubiquitous enzymes identified in various species (24). These enzymes exist as either monomers or oligomers with subunit molecular weights (MW) that range from 25,000 to 60,000 (13), with a conserved pentapeptide (Gly-X1-Ser-X2-Gly) sequence around the catalytic serine residue (3), despite sharing a low sequence homology. Unlike lipases, esterases preferentially hydrolyze water-soluble esters or short-chain fatty acid triglycerides. The proposed mechanism of catalysis for esterases is similar to the serine protease mechanism, which involves nucleophilic attack by the catalytic serine hydroxyl on the carbonyl carbon of the scissile bond. This hydroxyl group becomes more nucleophilic and the reaction is stabilized through hydrogen bonding to the imidazole group of the catalytic histidine that is stabilized by the carboxyl group of the acidic member of the catalytic triad (9, 12). The importance of esterases for application in pharmaceutical and agrochemical industries is increasing because their properties as stereo-selective catalysts in the synthesis of optically pure molecules are diverse. On the basis of their protein structures, the α/β hydrolase architecture of lipases possesses an additional movable helical lid that controls access to the active site, providing a molecular explanation for lipase-enhanced activity upon contact with a lipase-water interface (41).
d-β-acetylthioisobutyric acid (DAT), also known as S-β-acetylthio-2-methylpropionic acid, is a key intermediate for the synthesis of angiotensin-converting enzyme inhibitors, such as captopril and alacepril, which are used to treat hypertension and congestive heart failure. For DAT production, several processes, such as optical resolution (35) and chemical synthesis using dl-β-acetylthioisobutyric acid as a starting material (38), are reported, but these processes require expensive reagents. Sakimae et al. developed an enzymatic process for optical resolution to produce DAT by using microbial cells (32-34). A study of the hydrolytic reaction of methyl dl-β-acetylthioisobutyrate (dl-MATI) with an esterase from Pseudomonas putida strain MR-2068 revealed that DAT had an optical purity of more than 98% when the reaction was done below pH 7.5 and below 50°C (34). Honda et al. reported that 3,4-dihydrocoumarin hydrolase hydrolyzed dl-MATI to produce DAT with a high optical purity (>99.9% enantiomeric excess) (15).
Esterases are widely distributed in Pseudomonas, and several have been identified, cloned, and sequenced, such as esterases of Pseudomonas fluorescens (6, 7, 16, 20), Pseudomonas putida (27, 28), and other Pseudomonas spp. (22, 37, 39, 40). Esterase (EST) from Pseudomonas putida IFO12996 is capable of catalyzing the stereoselective hydrolysis of dl-MATI to produce DAT. We have cloned the EST gene, expressed the protein in Escherichia coli, and characterized and examined its kinetic constants and stabilities (36). Analysis of the nucleotide sequence of 854-bp EST revealed the gene coding for a protein of 276 amino acids, and the deduced amino acid sequence exhibited a high similarity to esterases from Pseudomonas putida MR-2068 (96% identity) (29) and 3,4-dihydrocoumarin hydrolase from Acinetobacter calcoaceticus F46 (73% identity) (15). Sequence alignments showed that EST is a member of the α/β hydrolase superfamily and indicated the presence of a functional catalytic triad of serine 97, aspartate 227, and histidine 256. To understand the biological and biochemical functions of EST in conjunction with its structure, we have determined the X-ray crystal structure at a 1.6-Å resolution, and we discuss here the substrate specificity, which can also provide us with an opportunity to engineer the enzyme for broader ester substrates.
MATERIALS AND METHODS
Chemicals.
Methyl methacrylate (MMA) and ethyl methacrylate (Janssen, Geel, Belgium), 2-hydroxyethyl methacrylate (Across, Geel, Belgium), dl-methyl 2-chloropropionate (dl-MCP) and dl-methyl 2-bromopropionate (dl-MBP) (TCI, Tokyo, Japan), and dl-methyl 2-methylbutyrate (dl-MMB) (Lancaster, Lancashire, United Kingdom) were obtained from the indicated suppliers.
Synthesis of substrates. (i) Synthesis of dl-MATI.
Reacting MMA (22 g, 0.22 mol) with thioacetic acid (25 g, 0.33 mol) at 95°C for 6 h produced dl-MATI, which was purified by distillation under reduced pressure. After distillation at 78 to 81°C to remove thioacetic acid, the reaction mixture gave pure dl-MATI (37 g, 95.5% yield).
(ii) Synthesis of dl-EATI.
Reacting ethyl methacrylate (10 g, 0.088 mol) with thioacetic acid (10 g, 0.13 mol) at 95°C for 6 h produced ethyl dl-β-acetylthioisobutyrate (dl-EATI), which was purified by distillation under reduced pressure. After distillation at 78 to 81°C to remove thioacetic acid, the reaction mixture gave pure dl-EATI (16.2 g, 96.9% yield).
(iii) Synthesis of dl-HEATI.
Reacting 2-hydroxyethyl methacrylate (11.5 g, 0.08 mol) with thioacetic acid (10 g, 0.13 mol) at 95°C for 6 h produced 2-hydroxyethyl dl-β-acetylthioisobutyrate (dl-HEATI), which was purified by distillation under reduced pressure. After distillation at 78 to 81°C to remove thioacetic acid, the reaction mixture gave pure dl-HEATI (16.2 g, 98.3% yield).
(iv) Synthesis of MDMB.
2,2-Dimethylbutyic acid (11.6 g, 0.1 mol) was suspended in dry methanol (25 ml, 0.6 mol), and dry HCl gas (0.3 mol) was added. The solution was allowed to stand for 6 h in a sealed flask at 65°C; the solvent was then evaporated under vacuum below 30°C. The hydrochloride salt was shaken with water (50 ml) and CH2C12 (50 ml), and the pH of the solution was adjusted to 10 with aqueous NaOH. The organic phase was dried (MgSO4) and evaporated to give methyl 2,2-dimethylbutyrate (MDMB) (12 g, 92.3%).
Protein expression and purification.
The recombinant esterase from Pseudomonas putida IFO12996 was overexpressed in E. coli BL21(DE3) and purified using an ion-exchange column (Resource Q-6), as reported by Shaw et al. (36).
Enzymatic analysis.
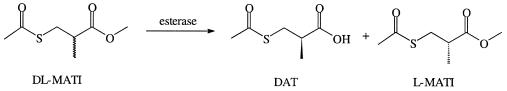
dl-MATI (2 g, 11 mmol) and pure Pseudomonas putida esterase (500 μl, 225 U) were mixed with Tris-HCl buffer (100 ml, 0.5 M, pH 7.0), followed by reaction at 37°C for 2 h (Fig. 1). The pH of the reaction mixture was maintained at 7.0 by adding NaOH solution (0.1 N) with a pH stat (Suntex pH controller PC-310). The concentration of hydrolyzed product from dl-ester in the reaction mixture was determined with a high-performance liquid chromatography system (Shimadzu LC-10AT) with a reverse-phase C18 column (Hypersil BDS). Injected samples were eluted with isocratic 50% solvent A (25 mM phosphate buffer, pH 6.5) and solvent B (acetonitrile) and monitored by UV at 229 nm. The concentration of eluted sample was quantified through comparison with a linear calibration curve for each sample. A unit of EST activity was defined as using enzyme (1 mg) to produce DAT (1 mM/min at 37°C and pH 7.0).
FIG. 1.
EST-catalyzed optical resolution of dl-MATI.
Substrate specificity assay.
The substrate specificity of EST was detected as acid production from hydrolysis of each substrate, and it was monitored by a pH indicator (bromocresol purple). As the acid was produced, the pH of the solution decreased and the color of the solution changed from blue to yellow. Tris-HCl (0.1 ml, pH 7.0, 0.5 M), 0.3 ml pH indicator standard (0.025 g bromocresol purple in 50 ml double-distilled H2O), and 0.6 ml double-distilled H2O were mixed in a microtube. Each substrate was added to the solution to final concentration as indicated, and then 2.1 μg EST was added and incubated at 37°C. The time until the color changed to yellow was counted. The activity toward each substrate was compared to its activity toward dl-MATI.
Gel filtration chromatography.
The molecular weight of native EST was determined by gel filtration chromatography (Superose 6 HR 10/30 column and ΔKTAexplorer fast-protein liquid chromatography system; Amersham Biosciences). The column was equilibrated with Tris-HCl (20 mM, pH 7.5) containing NaCl (0.15 M) and calibrated with these markers: thyroglobulin (670 kDa), bovine gamma globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa), and vitamin B12 (1.35 kDa). Purified EST protein (50 μg) was applied to the column per run, and the elution was detected by UV absorption spectrum at 280 nm.
Protein crystallization.
Crystallization was achieved by the hanging-drop vapor diffusion method at 18°C using screen kits (Hampton Research Co.). A shower of small crystals was obtained from one condition, using ammonium sulfate as precipitant after the initial screening. This condition was further refined to produce larger EST crystals by using hanging drops (2 μl) containing equal volumes of protein solution (10 mg/ml) and a reservoir solution containing ammonium sulfate (1.8 M) and citrate buffer (0.1 M) at pH 4.5. Crystals appeared after 3 days and reached a maximum size in 3 weeks. Crystals of diffraction quality were used for X-ray data collection.
X-ray data collection.
The crystal was transferred into a cryoprotectant solution containing glycerol (20%), mounted on a glass fiber loop (0.2 to 0.3 mm; Hampton Research Co.), and then flash cooled in liquid nitrogen at 100 K. X-ray diffraction data were collected from frozen crystals with a synchrotron X-ray at the Taiwan contract beamline BL12B2 equipped with a Quantum 4R charge-coupled-device (ADSC) detector at SPring-8 in Japan. For complete collection of high-resolution data, 360° of data with 1° oscillations was measured at an X-ray wavelength of 1.00 Å with an exposure time of 40 s and a distance of 180 mm from crystal to detector in a nitrogen stream (110 K) (produced with an X-Stream cryosystem [Rigaku/MSC]). The data were indexed, integrated, and scaled using the HKL2000 programs (26). The crystals belonged to the orthorhombic space group P212121, with unit cell dimensions of a = 50.56 Å, b = 98.11 Å, and c = 153.63 Å. The data set was 95.5% complete, with an internal agreement (Rsym) of 5.6%. Details of the data statistics appear in Table 1.
TABLE 1.
Crystal diffraction and structural statistics of EST monomer
| Parameter | Valuea |
|---|---|
| Crystal data | |
| Wavelength (Å) | 1.00 |
| Temp (K) | 110 |
| Resolution range (Å) | 25-1.6 |
| Space group | P212121 |
| No. of unique reflections | 97,023 |
| Completeness (%) (outermost shell) | 95.5 (82.3) |
| I/σI (outermost shell) | 8.6 (4.8) |
| Average redundancy | 6.30 |
| Rsymb (%) | 5.6 (21.2) |
| Mosaicity | 0.463 |
| Unit cell parameters (Å) | a = 50.56, b = 98.11, c = 153.63 |
| No. of molecules per asymmetric unit | 3 |
| Solvent content (%) | 40.1 |
| Refinement results (R)c | |
| Initial model (%) (25-2.5 Å) | 43.5 |
| After refinement (%) (25-2.5 Å) | 21.9/23.9d |
| RMSD bond length (Å) | 0.005 |
| RMSD bond angles (degrees) | 1.300 |
| No. of amino acid residues (built/total) | 275/276 |
| Avg no. of atoms (nonhydrogen) | 2,140 |
| Avg no. of waters per monomer | 106 |
Values in parentheses are for the highest-resolution shell (1.66-1.60 Å). Values were determined by use of beamline BL12B2 at SPring-8.
Rsym = ΣhΣi [|Ii(h) − <I(h)>|/ΣhΣi Ii(h)], where Ii is the ith measurement and <I(h)> is the weighted mean of all measurements of I(h). Reflection cutoffs (I/σI > 0) were applied in generating the data statistics.
R = Σh|Fo − Fc|/ΣhFo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h.
Second value is Rfree value.
Determination and refinement of the crystal structure.
The crystal structure of EST was solved by the molecular replacement method (31), using the cofactor-free chloroperoxidase structure (RCSB Protein Data Bank [PDB] code 1A88) (14), which shows a 61% sequence identity by the Crystallographic and NMR System (CNS) v1.1 (5), as a search model. The self-rotation function with data resolution of 15 to 4 Å showed a noncrystallographic threefold symmetry, indicating that the EST structure comprises three homologous subunits, i.e., trimers, in an asymmetric unit. We thus successfully obtained the complete molecular replacement solution and confirmed three molecules in one asymmetric unit.
Further crystallographic refinement was carried out using CNS v1.1 (5). Throughout the refinement, a randomly selected fraction (10%) of the data was set aside as a “free data set” for Rfree calculation, and the model was refined against the data with an F of ≥0 (working data set) (4). The composite omit electron density maps with coefficients |2Fo−Fc| were calculated and visualized using O v7.0 (18), and the model was rebuilt and adjusted iteratively as required. The trimeric protein model obtained through molecular replacement was initially treated to rigid-body refinement using data from resolution 25.0 to 3.0 Å for which the group B values were first restrained to 20.0 Å2. The R factor decreased to 36.1% after 120 cycles of conventional energy-restrained positional refinement. The refinement was followed by a simulated annealing step with the slow-cooling protocol provided by CNS, applied to the observed data between 25.0 and 1.6 Å. A starting temperature of 3,000 K was gradually decreased to 300 K in steps of 25 K, using time steps of 0.5 fs between energy calculations. For this resolution range, the R factor and Rfree values were reduced to 28.6% and 32.2%, respectively. In subsequent refinement stages, a bulk solvent correction was applied and group B factors were adjusted. Another simulated annealing refinement, with a starting temperature of 1,000 K, was then performed without noncrystallographic symmetry (NCS) restraint, which brought the R factor and Rfree values to 22.3% and 27.4%, respectively. To locate water molecules, we employed the PICKWATER subroutine from CNS to define peaks in the difference maps (3σ cutoff level). A water molecule was accepted if the identified feature correlated with a separate peak in the corresponding |Fo−Fc| electron density map and if one or more hydrogen bonds (3.3 to 2.3 Å) could be identified. Based on these criteria, 317 water molecules were located automatically. The protein model and water molecules were then subjected to another iteration of positional, simulated annealing and B factor refinement.
The final EST model contained three protein molecules (residues 2 to 276) as a trimer and an average of 2,126 protein nonhydrogen atoms and 106 solvent molecules for each monomer of three protein molecules in an asymmetric unit. For data between resolutions of 25.0 and 1.6 Å, the resulting model had a final R factor of 21.9% and Rfree of 23.9% (Table 1). The correctness of the stereochemistry of the model was verified with PROCHECK (21), and calculations of root mean square deviations (RMSD) from ideal values for bonds, angles, and dihedral and improper angles were performed with CNS. For all criteria applied with PROCHECK, the model was flagged as being either better than or within good regions. The calculations showed also a satisfactory stereochemistry with RMSD from ideality (10) in bond length of 0.008 Å and angle of 1.356°. In a Ramachandran plot (30), 99.2% of residue dihedral angles were in the most favored and additionally allowed regions, and 0.6% of main-chain dihedral angles, which all arise from catalytic residue Ser97 of three monomers, were observed in disallowed regions. This result seemed satisfactory for a model at a resolution of 1.6 Å.
Protein structure accession number.
The atomic coordinates have been deposited in the RCSB Protein Data Bank under accession code 1ZOI.
Nucleotide sequence accession number.
The nucleotide and protein sequence data of EST have been deposited in GenBank under accession number DQ187948.
RESULTS AND DISCUSSION
Enzymatic activity.
A cell extract of E. coli BL21(DE3) transformed with pET-EST exhibited specific dl-MATI hydrolyzing activity at 35 U/mg. With a purification procedure involving precipitation with ammonium sulfate and ion-exchange chromatography, we obtained homogeneous EST for enzyme activity analysis and crystallization. EST hydrolyzed the methyl ester bond of dl-MATI to produce DAT with a degree of conversion of 49.5% and an enantiomeric excess value of 97.2% at pH 7 and 37°C. The enantiomeric excess value is equal to enantiomeric purity. It is calculated from the absolute number of the percentage of d form compound minus the percentage of l form compound (|d%−l%|) (19). In contrast, the hydrolysis of the thioester bond could not be detected (36). ESTs before and after purification were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reduced and nonreduced conditions. A major band was detected at 33 kDa in all samples, which suggested the molecular mass of the monomeric subunit of EST to be 33 kDa.
EST monomer structure.
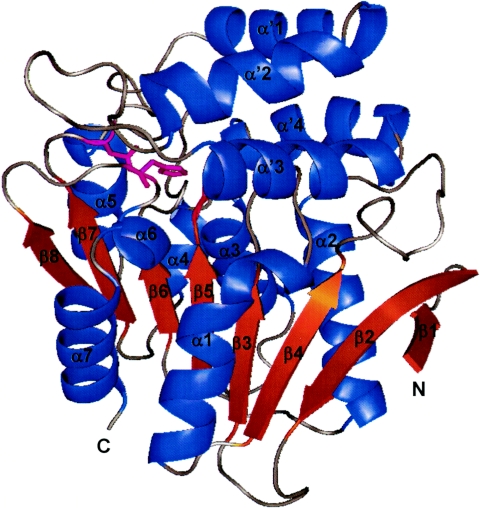
Superimposing three monomers in the asymmetric unit shows that the foldings of three EST molecules are essentially identical. EST has the α/β canonical hydrolase fold, which is composed of repeating α-β-α motifs that form a central eight-strand β-sheet, all parallel with the exception of the second strand. The topology of this arrangement corresponds to the general α/β hydrolase fold (strand order of β1, β2, β4, β3, β5, β6, β7, and β8) (25). In total, 11 helices are flanked on both sides of the β-sheets. The loop regions connecting the C termini of β-strands and the N termini of α-helices constitute the substrate-binding site. Except for the hairpin loop connecting strands 1 to 3, strands 3 to 6 and strands 7 and 8 are connected by helices α1, α2, and α3, which “cross” the central sheet. A large loop is formed between strands 6 and 7 by five helices, labeled α4 and α′1 to α′4 (Fig. 2). The variability in the loop regions contributes the versatile functionalities of α/β hydrolase superfamilies.
FIG. 2.
Ribbon drawing of EST monomer structure, made with PyMOL (http://www.pymol.org). The catalytic residues Ser97, Asp227, and His256 are shown as thin sticks in magenta.
EST active site.
In the Ser-His-Asp catalytic triad, the three residues, which occur far apart in the amino acid sequence of the enzyme, come together in a specific conformation in the active site to perform the hydrolytic cleavage of the appropriate bond in the substrate. This triad was first identified in the serine proteases (2, 43), which cleave peptides at the amide bond. In EST, the active catalytic triad is located approximately in the middle of the molecule at the end of a pocket 10 to 12 Å deep. The nucleophilic serine residue, Ser97, is located in a sharply turning, nucleophilic elbow, between strand β5 and helix α3 (Fig. 2). The main-chain conformation of Ser97 is strained (φ = 54.2°, ϕ = −91°). This is an energetically unfavorable conformation also observed with other α/β hydrolases. The sequence surrounding Ser97 is G-H-S-T-G, which is a conserved sequence, G-X1-Ser-X2-G, in most members of the α/β hydrolase fold superfamily (17, 23, 25). This sharp turn is stabilized with two strong backbone hydrogen bonds between Ser97-CO · · · HN-Gly100 (R = 2.84 Å) and Ser97-NH · · · OC-Ile121 (R = 2.85 Å). A moderate hydrogen bond can form between the carboxylic group of Ser97 and the amide hydrogen atom of Gly99 (R = 3.05 Å). The carboxylate oxygen OG also forms an additional strong hydrogen bond to the water molecule (W42) that can be connected to the nitrogen of Trp31.
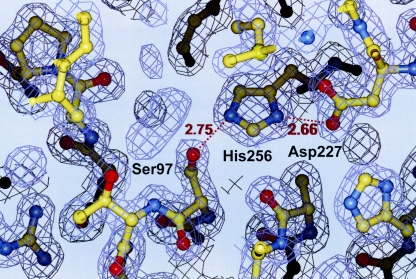
The second member of the triad, His256, is located at the C-terminal end of strand β8 (Fig. 3). His256 is present in an appropriate conformation corresponding to the active orientation for a catalytic triad histidine residue to form hydrogen bonds with both Ser97 and the third catalytic residue, Asp227. Instead, since the ɛ-1 carbon of His256 is in close proximity to the carbonyl oxygen of Ala122 located in the adjacent loop, the important function of this hydrogen bond is to facilitate a general acid-base catalytic role of the histidine residue during both the acylation and the deacylation step of hydrolysis (8).
FIG. 3.
Composite omit electron density maps with coefficients |2Fo−Fc| around activity site region. Distances (in Å) are shown with red numbers.
In addition to the catalytic triad, another essential component of the active center is the oxyanion hole. The architecture of the oxyanion hole is generated from a special arrangement of hydrogen bond donors that can stabilize the tetrahedral transition enzyme-substrate complex through accommodation of the negatively charged carbonyl oxygen of the scissile bond (11, 42).
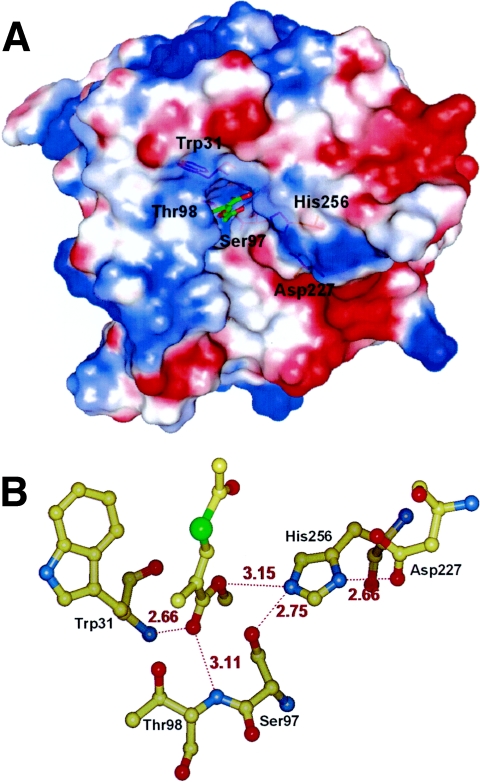
To achieve this objective, the position of the other substrate in the complex for which the enzymes possess an overall structural similarity with EST serves as a template. Chain B of the chloroperoxidase T/benzoate complex (PDB code 1A8U) and chain B of the chloroperoxidase F/propionate complex (PDB code 1A8S) were maximally superimposed to chain B of EST. In this way, the position of the substrate within the binding site is adjustable with increased precision. In the EST catalytic site, we have docked dl-MATI and performed energy minimization with CNS v1.1. Figure 4 shows that in the EST-dl-MATI complex the oxyanion hole can form by peptide NH groups of Thr98 and Trp31 that form hydrogen bonds with the carbonyl oxygen of the dl-MATI.
FIG. 4.
(A) The molecular electrostatic surface of the enzyme was calculated and displayed by PyMOL (http://www.pymol.org). The darkest blue denotes a potential of 15 kT/e, and the darkest red denotes a potential of −15 kT/e. The docking dl-MATI-binding site is in a slightly positive potential oxyanion hole. dl-MATI is shown as green and red sticks, and the important residues are shown as orange sticks. (B) In the modeled dl-MATI-EST complex, the oxyanion hole can form by peptide NH groups of Thr98 and Trp31, which form hydrogen bonds with the carbonyl oxygen of the dl-MATI. Distances (in Å) are shown with red numbers.
EST quaternary structure and crystal contact.
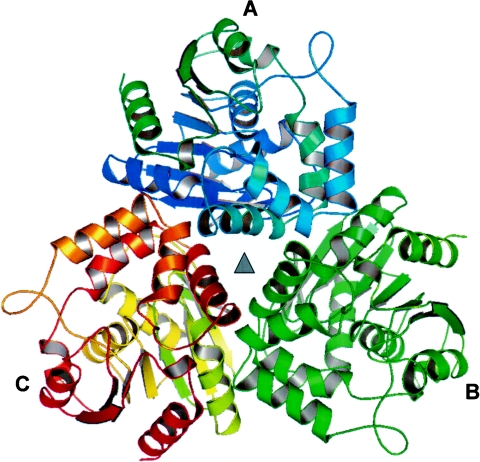
There are three EST monomers in one asymmetric unit, in which a noncrystallographic threefold axis is located in the center of the trimer (Fig. 5). Intermolecular interactions that form a trimer involve mainly the residues between β2 and the loop connecting β4 with α2, α1, and the sharp turn connecting α′1 with α′2 and α′3 with α1, which makes the outward directions of the active sites of substrate binding. This extensive interface between two monomers of the trimer accounts for a buried surface area of ∼1,445 Å2 for each monomer. The distance among their respective active sites of ∼38 Å indicates that the three subunits might act independently.
FIG. 5.
Three EST monomers packing with a noncrystallographic threefold symmetry (▴) in an asymmetric unit.
Oligomeric state of EST in solution.
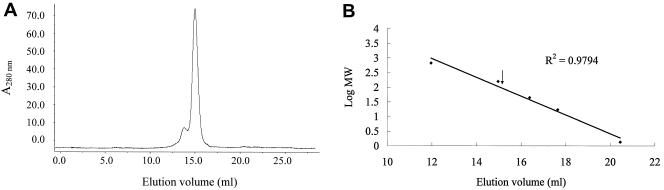
The oligomeric state and molecular weight of EST under a native condition were determined by gel filtration chromatography (Superose 6 HR 10/30) with a fast-protein liquid chromatography system. We observed a linear dependence of log MW on elution volume. The EST was eluted as a major peak at an elution volume of 15 ml (Fig. 6A), corresponding to the MW of 104,000 relative to the calibration curve (Fig. 6B). Together, sodium dodecyl sulfate-polyacrylamide gel electrophoresis and the gel filtration data indicate that EST exists as a homotrimer under a native condition. This result indicates that EST is a trimer in solutions of the physiology condition (pH 7.5), showing a similarity with an observation from the crystal structure (pH 4.0). The trimeric units of EST in both crystals and solution show the unique and important biological function of this enzyme. By means of an association and dissociation of multimers, it can eliminate some deficient or defective monomers to maintain overall activity ability.
FIG. 6.
(A) Gel filtration chromatogram of EST. (B) Calibration curve showing the dependence of log MW (in thousands) on elution volume. The position of EST is indicated by an arrow.
Substrate specificity.
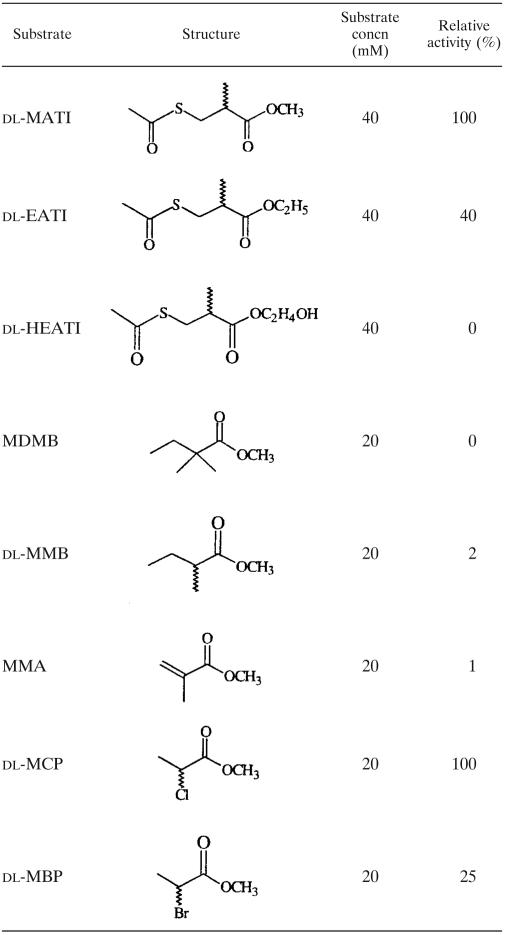
We examined linear ester compounds over a broad range to reveal the substrate specificity of EST (Table 2). The nomenclature of each compound and the location of each substituent are based on IUPAC. The data indicated that EST has three potential recognition sites for its substrates. (i) The ester moiety of substrates requires a small and neutral group: dl-MATI had 2.5 times the activity of dl-EATI, but dl-HEATI completely lost its activity. (ii) The substrates require a small and neutral group at C-2: the methyl (dl-MATI) and chloro (dl-MCP) groups have the highest activities, the bromo (dl-MBP) group shows less activity, and the dimethyl (MDMB) group completely loses its activity. The results support that a larger group at C-2 increases the steric effect. Although there is a methyl group at C-2 of MMA, the configuration of its 2-carbon shows trigonal coordination (sp2) instead of tetrahedral (sp3), by which MMA fails to be an effective substrate. Moreover, for the MMA compound the resonance effect effectively decreased the relative activity to 1%. (iii) Substitution at C-3 of substrates is also allowed. In a comparison of dl-MATI and dl-MMB, substitution of an acetylthiol group for a methyl group at C-3 increased the relative activity about 60 times.
TABLE 2.
Substrate specificity of recombinant EST toward linear estersa
Reactions were carried out as described in the text.
Although the acetylthiol ester moiety of dl-MATI is more labile than the methyl ester site for nucleophilic attack with EST, the fascinating nature of this enzymatic catalysis is not only governed by decrease in its activation energy but is also determined by its substrate specificity or affinity. Bianchi and Cesti reported that lipases can be used to prepare optically active thiols with varied thiol-containing substrates, such as dl-MATI (1). Both lipases and esterases are hydrolytic enzymes and can serve extensively as catalysts in enantioselective and regioselective synthesis, but the substrate specificities of lipases and esterases differ. In the case of dl-MATI as a substrate, lipase P from Pseudomonas cepacia and porcine pancreatic lipase can catalyze only the thiotransesterification, whereas EST can hydrolyze the methyl ester group without affecting the acetylthiol ester moiety in dl-MATI (1). This distinction reflects varied substrate specificities but not substrate reactivities. The knowledge of substrate specificity and structure of a substrate-binding site of EST is useful for enzymatic engineering for broader ester substrates.
Acknowledgments
We thank the support staff, Y.-C. Jean, Y.-S. Huang, C.-S. Chao, J.-H. Jiang, and C.-C. Tseng, for their technical assistance and discussion during data collection at BL17B2 of NSRRC in Taiwan and BL12B2 of SPring-8 in Japan.
This research was supported in part by the National Synchrotron Radiation Research Center (grant no. 934RSB02) and National Science Council (grant no. NSC 93-2321-B-213-001) in Taiwan to C.-J.C.
REFERENCES
- 1.Bianchi, D., and P. Cesti. 1990. Lipase-catalyzed stereoselective thiotransesterification of mercapto esters. J. Org. Chem. 55:5657-5659. [Google Scholar]
- 2.Blow, D. M., J. J. Birkoft, and B. S. Hartley. 1969. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature 221:337-340. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, S. 1988. The molecular evolution of genes and proteins: a tale of two serines. Nature 334:528-530. [DOI] [PubMed] [Google Scholar]
- 4.Brünger, A. T. 1992. The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-474. [DOI] [PubMed] [Google Scholar]
- 5.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallographic and NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Cheeseman, J. D., A. Tocilj, S. Park, J. D. Schrag, and R. J. Kazlauskas. 2004. Structure of an aryl esterase from Pseudomonas fluorescens. Acta Crystallogr. Sect. D 60:1237-1243. [DOI] [PubMed] [Google Scholar]
- 7.Choi, K. D., G. H. Jeohn, J. S. Rhee, and O. J. Yoo. 1990. Cloning and nucleotide sequence of an esterase gene from Pseudomonas fluorescens and expression of the gene in Escherichia coli. Agric. Biol. Chem. 54:2039-2045. [PubMed] [Google Scholar]
- 8.Derewenda, Z. S., U. Derewenda, and P. M. Kobos. 1994. (His)Cɛ-H · · · O=C< hydrogen bond in the active sites of serine hydrolases. J. Mol. Biol. 241:83-93. [DOI] [PubMed] [Google Scholar]
- 9.Dodson, G., and A. Woldawer. 1998. Catalytic triads and their relatives. Trends Biochem. Sci. 23:347-352. [DOI] [PubMed] [Google Scholar]
- 10.Engh, R. A., and R. Huber. 1991. Accurate bond and angles parameters for X-ray protein structure refinement. Acta Crystallogr. Sect. A 47:392-400. [Google Scholar]
- 11.Fuxreiter, M., and A. Warshel. 1998. Origin of the catalytic power of acetylcholinesterase: computer simulation studies. J. Am. Chem. Soc. 120:183-194. [Google Scholar]
- 12.Hedstrom, L. 2002. Serine proteases mechanism and specificity. Chem. Rev. 102:4501-4523. [DOI] [PubMed] [Google Scholar]
- 13.Heikinheimo, P., A. Goldman, C. Jeffries, and D. W. Ollis. 1999. Of barn owls and bankers: a lush variety of α/β hydrolases. Structure 7:141-146. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann, B., S. Tolzer, I. Pelletier, J. Altenbuchner, K. H. van Pée, and H. J. Hecht. 1998. Structural investigation of the cofactor-free chloroperoxidases. J. Mol. Biol. 279:889-900. [DOI] [PubMed] [Google Scholar]
- 15.Honda, K., M. Kataoka, and S. Shimizu. 2002. Enzymatic preparation of d-β-acetylthioisobutyric acid and cetraxate hydrochloride using a stereo- and/or regioselective hydrolase, 3,4-dihydrocoumarin hydrolase from Acinetobacter calcoaceticus. Appl. Microbiol. Biotechnol. 60:288-292. [DOI] [PubMed] [Google Scholar]
- 16.Hong, K. H., W. H. Jang, K. D. Choi, and O. J. Yoo. 1991. Characterization of Pseudomonas fluorescens carboxylesterase: cloning and expression of the esterase gene in Escherichia coli. Agric. Biol. Chem. 55:2839-2845. [PubMed] [Google Scholar]
- 17.Hotelier, T., L. Renault, X. Cousin, V. Negre, P. Marchot, and A. Chatonnet. 2004. Esther, the database of the α/β hydrolase fold superfamily of proteins. Nucleic Acids Res. 32:D145-D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 19.Juaristi, E. 1991. Introduction to stereochemistry and conformational analysis, p. 129. John Wiley & Sons, New York, N.Y.
- 20.Kim, K. K., H. K. Song, D. H. Shin, K. Y. Hwang, S. Choe, O. J. Yoo, and S. W. Suh. 1997. Crystal structure of carboxylesterase from Pseudomonas fluorescens, an α/β hydrolase with broad substrate specificity. Structure 5:1571-1584. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 22.McKay, D. B., M. P. Jennings, E. A. Godfrey, I. C. MacRae, P. J. Rogers, and I. R. Beacham. 1992. Molecular analysis of an esterase-encoding gene from a lipolytic psychrotrophic pseudomonad. J. Gen. Microbiol. 138:701-708. [DOI] [PubMed] [Google Scholar]
- 23.Nardini, M., and B. W. Dijkstra. 1999. α/β Hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9:732-737. [DOI] [PubMed] [Google Scholar]
- 24.Okuda, H. 1991. Esterases, p. 563-577. In S. A. Kuby (ed.), A study of enzymes. CRC Press, Boca Raton, Fla.
- 25.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, J. Schrag, J. L. Sussman, K. H. G. Verchueren, and A. Goldman. 1992. The α/β hydrolase fold. Protein Eng. 5:197-211. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode, p. 307-326. In C. W. Carter, Jr., and R. M. Sweet (ed.), Methods in enzymology, vol. 276. Macromolecular crystallography, part A. Academic Press Ltd., London, United Kingdom. [DOI] [PubMed] [Google Scholar]
- 27.Ozaki, E., and A. Sakimae. 1997. Purification and characterization of recombinant esterase from Pseudomonas putida MR-2068 and its application to the optical resolution of dimethyl methylsuccinate. J. Ferment. Bioeng. 83:535-539. [Google Scholar]
- 28.Ozaki, E., A. Sakimae, and R. Numazawa. 1994. Cloning and expression of Pseudomonas putida esterase gene in Escherichia coli and its use in enzymatic production of d-β-acetylthioisobutyric acid. Biosci. Biotech. Biochem. 58:1745-1746. [DOI] [PubMed] [Google Scholar]
- 29.Ozaki, E., A. Sakimae, and R. Numazawa. 1995. Nucleotide sequence of the gene for a thermostable esterase from Pseudomonas putida MR-2068. Biosci. Biotech. Biochem. 59:1204-1207. [DOI] [PubMed] [Google Scholar]
- 30.Ramachandran, G. N., and V. Sasisekharan. 1968. Conformation of polypeptides and proteins. Adv. Protein Chem. 23:283-437. [DOI] [PubMed] [Google Scholar]
- 31.Rossmann, M. G. 1990. The molecular replacement method. Acta Crystallogr. Sect. A 46:73-82. [DOI] [PubMed] [Google Scholar]
- 32.Sakimae, A., A. Hosoi, E. Kobayashi, N. Ohsuga, R. Numazawa, I. Watanabe, and H. Ohnishi. 1992. Screening of microorganisms producing d-β-acetylthioisobutyric acid from methyl dl-β-acetylthioisobutyrate. Biosci. Biotech. Biochem. 56:1252-1256. [Google Scholar]
- 33.Sakimae, A., R. Numazawa, and H. Ohinishi. 1992. A newly isolated microorganism producing d-β-acetylthioisobutyric acid from methyl dl-β-acetylthioisobutyrate. Biosci. Biotech. Biochem. 56:1341. [Google Scholar]
- 34.Sakimae, A., E. Ozaki, H. Toyama, N. Ohsuga, R. Numazawa, I. Muraoka, E. Hamada, and H. Ohnishi. 1993. Process conditions for production of d-β-acetylthioisobutyric acid from methyl dl-β-acetylthioisobutyrate with the cells of Pseudomonas putida MR-2068. Biosci. Biotech. Biochem. 57:782-786. [Google Scholar]
- 35.Samayama, T., M. Tsukamoto, T. Sasagawa, S. Naruto, J. Matsumoto, and H. Uno. 1989. An improved optical resolution of 3-acetylthio-2-methylpropionic acid by use of a new chiral amine, N-isopropyl (phenylalaninol). Chem. Pharm. Bull. 37:1382-1383. [Google Scholar]
- 36.Shaw, S.-Y., Y.-J. Chen, J.-J. Ou, and L. Ho. Enzymatic resolution of methyl dl-β-acetylthioisobutyrate and dl-β-acetylthioisobutyramide using a stereoselective esterase from Pseudomonas putida IFO12996. Submitted for publication.
- 37.Shimada, Y., T. Nagao, A. Sugihara, T. Iizumi, T. Yui, K. Nakamura, T. Fukase, and Y. Tominaga. 1993. Cloning and sequence analysis of an esterase gene from Pseudomonas sp. KWI-56. Biochim. Biophys. Acta 1174:79-82. [DOI] [PubMed] [Google Scholar]
- 38.Shimazaki, M., J. Hasegawa, K. Kan, K. Nomura, Y. Nose, H. Kondo, T. Ohashi, and K. Watanabe. 1982. Synthesis of captopril from an optically active β-hydroxy acid. Chem. Pharm. Bull. 30:3139-3146. [Google Scholar]
- 39.Sugihara, A., Y. Shimada, T. Nagao, T. Iizumi, K. Nakamura, T. Fukase, and Y. Tominaga. 1994. Purification and characterization of a carboxylesterase from Pseudomonas sp. KWI-56. Biosci. Biotechnol. Biochem. 58:752-755. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki, T., T. Nakayama, D. W. Choo, Y. Hirano, T. Kurihara, T. Nishino, and N. Esaki. 2003. Cloning, heterologous expression, renaturation, and characterization of a cold-adapted esterase with unique primary structure from a psychrotroph Pseudomonas sp. strain B11-1. Protein Expr. Purif. 30:171-178. [DOI] [PubMed] [Google Scholar]
- 41.Verger, R. 1997. Interfacial activation of lipases: fact and artifacts. Trends Biotechnol. 15:32-38. [Google Scholar]
- 42.Warshel, A., G. Naray-Szabo, F. Sussman, and J. K. Hwang. 1989. How do serine proteases really work? Biochemistry 28:3629-3637. [DOI] [PubMed] [Google Scholar]
- 43.Wright, C. S., R. A. Alden, and J. Kraut. 1969. Structure of subtilisin BPN′ at 25 Å resolution. Nature 221:235-242. [DOI] [PubMed] [Google Scholar]