Abstract
The bacterium Caulobacter crescentus and related stalk bacterial species are known for their distinctive ability to live in low-nutrient environments, a characteristic of most heavy metal-contaminated sites. Caulobacter crescentus is a model organism for studying cell cycle regulation with well-developed genetics. We have identified the pathways responding to heavy-metal toxicity in C. crescentus to provide insights for the possible application of Caulobacter to environmental restoration. We exposed C. crescentus cells to four heavy metals (chromium, cadmium, selenium, and uranium) and analyzed genome-wide transcriptional activities postexposure using an Affymetrix GeneChip microarray. C. crescentus showed surprisingly high tolerance to uranium, a possible mechanism for which may be the formation of extracellular calcium-uranium-phosphate precipitates. The principal response to these metals was protection against oxidative stress (up-regulation of manganese-dependent superoxide dismutase sodA). Glutathione S-transferase, thioredoxin, glutaredoxins, and DNA repair enzymes responded most strongly to cadmium and chromate. The cadmium and chromium stress response also focused on reducing the intracellular metal concentration, with multiple efflux pumps employed to remove cadmium, while a sulfate transporter was down-regulated to reduce nonspecific uptake of chromium. Membrane proteins were also up-regulated in response to most of the metals tested. A two-component signal transduction system involved in the uranium response was identified. Several differentially regulated transcripts from regions previously not known to encode proteins were identified, demonstrating the advantage of evaluating the transcriptome by using whole-genome microarrays.
Potentially hazardous levels of heavy metals have dispersed into subsurface sediment and groundwater in a number of metal-contaminated sites and represent a challenge for environmental restoration. Effective bioremediation of these sites requires knowledge of genetic pathways for resistance and biotransformation by component organisms within a microbial community. However, a comprehensive understanding of bacterial mechanisms of heavy metal toxicity and resistance has yet to be achieved. While many metals are essential to microbial function, heavy metals, i.e., most of those with a density above 5 g/cm3, have toxic effects on cellular metabolism (46). The majority of heavy metals are transition elements with incompletely filled d orbitals providing heavy metal cations which can form complex compounds with redox activity (46, 70). Therefore, it is important to the health of the organism that the intracellular concentrations of heavy metal ions are tightly controlled. However, due to their structural and valence similarities to nontoxic metals, heavy metals are often transported into the cytoplasm through constitutively expressed nonspecific transport systems (46). As such, heavy metals invariably find their way into the cell. Once inside the cell, toxic effects of heavy metals include nonspecific intracellular complexation, with thiol groups being particularly vulnerable (53). Interactions of these nonspecific complexes with molecular oxygen leads to the formation of reactive oxygen species such as H2O2, resulting in oxidative stress within the cell (23). In addition to oxidative stress, complexation of sulfhydryl groups with heavy metal cations results in reduced activity of sensitive enzymes (16). Previous studies identified several examples in which heavy metals were toxic to cellular processes, including the following. (i) Heavy metal cations (for example, cadmium) tend to bind to SH groups (46). In gram-negative bacteria, heavy metal cations can bind to glutathione. The resulting products (bisglutathione complex) tend to react with molecular oxygen to form oxidized bisglutathione, releasing the metal cation and hydrogen peroxide. The bisglutathione must be reduced in an NADPH-dependent reaction, with the released metal ion beginning another cycle of binding and bisglutathione oxidation (23, 46), resulting in considerable oxidative stress. (ii) Some metal ions structurally mimic physiologically important molecules (for example, chromate resembles sulfate, arsenate resembles phosphate) and thus interfere with physiological processes in which those molecules are required (16). (iii) Some metals, for example, chromate, are reduced intracellularly by both enzymatic and nonenzymatic mechanisms, with the reduction process producing reactive oxygen species. This process may inadvertently be a major factor in causing damage to many cellular components, including DNA and proteins (12, 58, 65).
Resistant bacteria possess a number of strategies to withstand elevated concentrations of heavy metals. Many resistance mechanisms revolve around removing the heavy metal or decreasing its toxicity (70). Alternatively, the concentration of metal entering the cytoplasm may be decreased through active (extracellular precipitation) and passive (native biosorption) processes (30, 35). Metal-chelating proteins have been reported as a means of resistance mainly in eukaryotes and also in some limited examples of prokaryotes (70). The major bacterial resistance mechanisms include (i) active efflux, (ii) transformation of the heavy metal ion to a less toxic form, for example, Cr(VI) can be reduced to Cr(III) (12), and (iii) precipitation, either intercellular or extracellular (35, 64, 66, 67).
Caulobacter spp. are extremely ubiquitous and are able to survive in low-nutrient environments (51). They have been found in freshwater, seawater, soil (51), ground water (37), wastewater (36), deep-sea sediment (38), and a deep subsurface gold mine (19) and have been noted for their ability to survive in broad environmental habitats where contamination may be present (8, 48). In addition, Caulobacter crescentus has been shown to form high-density biofilms with the potential for use in bioreactors for bioremediation (60) and has been used as a model organism to study cell cycle control (32, 40, 41). Previous knowledge of this organism, including its genome sequence (45), has provided new and extremely valuable tools to study genome-wide response to heavy metal stress. Both oligonucleotide cDNA microarrays and Affymetrix GeneChip microarrays have been used to study cell cycle regulation (31) (McGrath and McAdams, unpublished data). In this paper, we use a Caulobacter Affymetrix GeneChip microarray (Caulobacter chip) to study the transcriptional response of C. crescentus to heavy metal stress. This chip was designed by the McAdams laboratory at Stanford University in collaboration with Affymetrix. A complete description of all features of the chip will be published separately. In our work reported here, only the gene expression assay features, which are based on eight optimally selected 25-mer match/mismatch probe pairs per predicted open reading frame (ORF), were used.
MATERIALS AND METHODS
Bacterial strains, media, and growth.
The Caulobacter crescentus strain CB15N was used in this study. Cultures were maintained on PYE (peptone-yeast extract) agar plates containing 0.2% (wt/vol) Bacto peptone (Difco), 0.1% (wt/vol) yeast extract (Difco), 1 mM MgSO4, 0.5 mM CaCl2, and 1.5% (wt/vol) agar (Difco). The knockout mutants were maintained with 10 μg/ml tetracycline on solid medium (PYE agar), with tetracycline (5 μg/ml) being used in the overnight culture but not in the culture for transcriptional analysis. The liquid culture used for transcriptional response studies was M2 minimal salts medium (6.1 mM Na2HPO4, 3.9 mM KH2PO4, 9.3 mM NH4Cl, 0.5 mM MgSO4, 10 μM FeSO4 [EDTA chelate; Sigma Chemical Co.], 0.5 mM CaCl2), with 0.2% (wt/vol) glucose as the sole carbon source (M2G). Overnight culture was inoculated with colonies from PYE plates, grown at 30°C, and shaken continuously at 225 rpm. Escherichia coli K12 and Pseudomonas putida KT2440 were used as reference bacteria to uranium tolerance. Both were grown in M2G medium (identical conditions to that of C. crescentus) described above.
Toxic metal effect on growth, survival, and morphology.
Metal stock solutions were prepared by dissolving the compounds (Sigma-Aldrich) in water to 10,000 ppm, with the exception of the uranyl nitrate stock solution, which was dissolved to 100 mM (23,800 ppm). All metal stocks were sterilized by filtration through a 0.2-μm membrane. Overnight cultures were diluted in fresh M2G medium with various concentrations of cadmium sulfate (CdSO4), sodium selenite (Na2SeO3), potassium chromate (K2CrO4), potassium dichromate (K2Cr2O7), and uranyl nitrate [(UO2)(NO3)2 · 6H2O]. Growth was followed spectrophotometrically (optical density at 600 nm). One metal concentration was selected to be used for each set of microarray experiments, based on the following requirements for each metal. (i) The stressed conditions only slightly affected growth (increased doubling time by 15 to 30 min). (ii) The addition of the heavy metal compound did not result in precipitation with salts or cause other obvious changes in the medium. (iii) The metal concentration was above a level considered toxic or close to those conditions used commonly in other studies. While not all these criteria were fully satisfied, the final concentrations used in this study were a compromise of the factors described above and are reported in the results.
For morphology and motility observations, a sample was taken from mid-log-phase culture (OD600 was 0.3 to 0.4) at 0 hours after end log-phase growth (T0). Metal stock was added to the final required concentration to be used in microarray experiments. After 30 min, another sample was taken as T1. A final sample was taken 3 to 4 h poststress as T2. Ten microliters of each sample was examined under 100× phase-contrast light microscopy. Fifty microliters of each sample was used to determine CFU. Bacterial membrane integrity was assessed using a Live/Dead BacLight bacterial viability kit (Molecular Probes, OR) according to the manufacturer's recommended protocol.
RNA extraction.
An overnight culture of C. crescentus CB15N was diluted into fresh M2G medium. When the culture reached exponential growth (OD600 was just over 0.3), 10 ml culture was removed (as nonstressed control). Heavy metal stock was added, and incubation was continued for another 30 min. After this period, a further 10 ml of sample was removed (as the stressed sample). Immediately after the samples were collected, they were centrifuged at 10,000 × g for 5 min and supernatant was removed. The cell pellets were frozen with liquid nitrogen and stored at −80°C. The RNA extraction protocol was described previously (18). Briefly, total RNA was extracted with Trizol reagent (Invitrogen) and any contaminating DNA was digested with DNase I. The RNA samples were further purified with acid phenol-chloroform-isoamyl alcohol (125:24:1, pH 4.5) (Ambion) extraction followed by salt-ethanol precipitation. RNA quantity was determined by OD260, and quality was determined by 2% (wt/vol) agarose gel electrophoresis and by a OD260/OD280 ratio.
Affymetrix GeneChip RNA expression analysis.
Procedures for sample preparation and array processing are described fully in the Affymetrix GeneChip Expression Analysis Technical Manual and briefly described here. Transcripts of three genes (Bacillus subtilis dab, phe, and thr) were added to the total RNA as spike-in controls to monitor labeling, hybridization, and staining efficiency. To generate the spike-in control RNA, the plasmids containing B. subtilis phe, thr, and dap genes were purified from strains ATCC 87483, ATCC 87484, and ATCC 87486, respectively. Linear template DNA was generated by digesting the plasmid with restriction enzyme NotI and sense RNA produced subsequently by in vitro transcription using T3 RNA polymerase (MEGAscript T3 kit; Ambion).
Total RNA (12 μg) was primed with random primer (Invitrogen), and cDNA was synthesized with reverse transcriptase (superscript II, Invitrogen). The resulting cDNA was fragmented with DNase I (Amersham) and biotin labeled using the Enzo BioArray terminal labeling kit (Affymetrix). Biotin-labeled samples were hybridized onto the Caulobacter microarray at 50°C overnight, and chip washing and staining followed standard Affymetrix GeneChip protocols (with stringent washing at 50°C). The high-density chip was scanned using an Affymetrix Scan3000 scanner.
Microarray data analysis and identifying differentially expressed transcripts under heavy metal stress.
The Caulobacter Affymetrix chip was used to assay gene expression levels for all 3,767 genes (45). For analysis of the protein-coding region, probe sets consisting of multiple (typically eight) 25-mer oligonucleotide probe pairs covering the gene were used for transcriptional interrogation. These probe sets were analyzed using the MAS5 statistical algorithm (3) for background adjustment and scaling in GCOS software (Affymetrix). Briefly, data from a minimum of three independent experiments were included as biological replicates in each comparison. Global scaling of all probe sets to a target signal intensity of 500 was applied to each chip (all microarray data are available at http://greengenes.lbl.gov/Download/Caulobacter_metal_stress_supplemental_microarray_data/). The data set was normalized using the spike-in controls mentioned above. For each comparison, the t test was performed on the data. For a probe set (gene) to be considered up-regulated under metal stress in these studies, it had to meet the following criteria. (i) The gene had to be called “present” by GCOS software for every experiment which was under metal stress. (ii) The average difference score (signal) for the gene had to be equal to or greater than 200, eliminating very low expression levels requiring more sophisticated analyses. (iii) The P value of the Student t test had to be less than 0.01, ensuring that the difference between the two conditions (nonstressed and stressed) was significant at a 99% confidence interval. (iv) The ratio of the average signals from stressed culture versus the average signals from nonstressed culture had to show at least twofold differences in expression.
The probes for predicted small protein regions were tiled at every 15 nucleotides on both strands. Since the boundary of these predicted proteins may be inaccurate, it is possible that some of the probes in the default probe set do not belong to a single transcript. It is inappropriate to use any of the existing software to obtain probe set values. At present, available microarray analysis programs do not allow dynamic definition of probe sets; thus, identification of transcripts driven by experimental data is not possible. To analyze these regions, we opted to examine their expression on a probe-by-probe basis. The background adjustment and normalization were performed using custom scripts developed by our lab. Probe-level data were examined in which individual probes meeting the following criteria were selected: (i) The average difference score (signal) for a probe was equal to or greater than 200. (ii) The P value of the Student t test was less than 0.015. (iii) The average signals from stressed culture versus the average signals from nonstressed culture were different by at least twofold to be considered for further analysis. Probes selected using these criteria were assembled in order to find continuous regions of up-regulation. The criteria defining a probe set for the small protein regions were as follows: (i) The maximum number of nonpassing probes (i.e., those not meeting the individual probe criteria listed above) within a probe set could not exceed three. (ii) The minimum length of assembled transcript was >100 bp. Probe set boundaries were also corrected according to probe behavior. For probe sets of interest, manual inspections were performed for final evaluation. This approach permitted monitoring of the transcriptional activities without bias.
Gene annotation and metabolic pathway analysis.
Initial Caulobacter crescentus open reading frame annotations were taken from GenBank accession number NC_002696 (45). Clusters of orthologous gene descriptions (69) were used if they described the functions more clearly. If a clearly definitive annotation of function was not found, BLAST was performed against all bacterial genomes and Pfam analysis was run to identify any domains. The identification of pathways involved in metabolism was aided by the use of BioCyc (http://biocyc.org). Other analyses, such as operon prediction and gene ontology were improved through the use of the VIMSS (Virtual Institute for Microbial Stress and Survival) database (http://www.microbesonline.org/).
TEM observation.
The samples were fixed in 4% paraformaldehyde in 0.1 M cacodylate buffer at pH 7.4 (Electron Microscopy Sciences) for 1 h at room temperature. The fixed samples were dehydrated with a graded series of ethanol and t-butyl alcohol. The samples were freeze-dried and mounted on 200-mesh copper grids for characterization with an HF2000 field-emission transmission electron microscope (Hitachi, Inc.) which was equipped with energy-dispersive X-ray spectroscopy (EDX). Transmission electron microscopy (TEM) was operated at an accelerating voltage of 200 kV. The abiotic sample was prepared by adding uranium stock solution to the sterile growth medium followed by centrifugation at 10,000 × g for 1 min.
RESULTS AND DISCUSSION
Heavy metal affect on growth and survival.
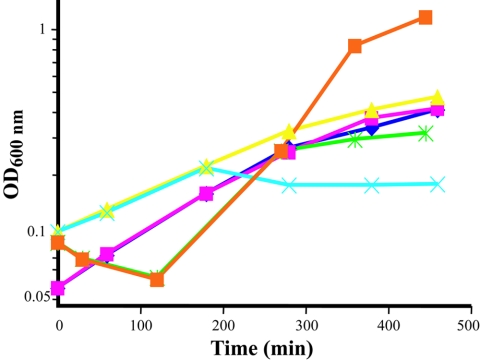
Relatively low concentrations of cadmium sulfate, potassium chromate, and potassium dichromate were sufficient to slow C. crescentus growth (see Fig. S1, S2, and S3 in the supplemental material). For microarray experiments, we defined the stressed condition as 6 μM cadmium sulfate, 40 μM potassium chromate, and 27 μM potassium dichromate. Under these conditions, the doubling times for the cells were 2.5 h under cadmium stress and over 3 h under chromium stress relative to nonstressed cells, which had doubling times of 2 h. Selenite stress was apparent at a concentration of 300 μM sodium selenite (doubling time increased by approximately 10 to 15 min). For uranium stress, a concentration 200 μM uranyl nitrate was used. The relevance of these metal concentrations to those in contaminated and uncontaminated groundwater is summarized in Table 1. Surprisingly, at the 200 μM uranium concentration used in this study (a concentration close to the highest observed at the NABIR Field Research Center, Oak Ridge, TN) (Table 1), Caulobacter growth rate was not significantly affected (see Fig. S4 in the supplemental material) and it was not until a concentration of 1 mM uranium was reached that Caulobacter growth slowed (doubling time slowed from 120 min to 180 min). Under these conditions (1 mM), visible precipitation was noted in the medium, hence this concentration was not used for microarray analysis. Under the same conditions (1 mM uranyl nitrate), growth of E. coli K-12 was completely stopped and the growth of Pseudomonas putida KT2440 was drastically reduced (Fig. 1). Growth of E. coli was significantly affected at 500 μM uranium, while growth of P. putida was initially inhibited at 800 μM uranium, although the P. putida culture did grow to higher density after more than 20 h. We believe this is the first study to identify C. crescentus as a uranium-tolerant bacterium. However, due to precipitation at higher concentrations of uranium, it was not possible to definitively determine the absolute concentration of Caulobacter uranium tolerance. The comparison to the reference bacteria (E. coli K12 and Pseudomonas putida KT2440), which was performed using the identical medium preparation and growth protocol and which demonstrated that C. crescentus is relatively more tolerant, is valid, but relative. A previous study showed that E. coli was killed at approximately 333 μM of uranium at pH 4 (66); however, it is not immediately comparable to our data, as uranium is known to be more soluble and more toxic at an acidic pH.
TABLE 1.
Comparison of metal concentrations used for stress response experiments with concentrations found in uncontaminated and contaminated groundwatera
| Metal | Concentration from:
|
Stress used (μM) | |
|---|---|---|---|
| US EPA (μM)b | NABIR FRC (μM)c | ||
| Cadmium | 0.04 | 39.59 | 6 |
| Chromium | 1.92 | 96.15 | 40-54d |
| Selenium | 0.63 | 632.91 | 300 |
| Uranium | 0.13 | 289.92 | 200-1000e |
Uncontaminated data was determined as per U.S. EPA Maximum Contaminant Level Goals, contaminated data was determined at NABIR Field Research Center (FRC).
Data are from the U.S. EPA website, http://www.epa.gov/safewater/hfacts.html.
Data are from the NABIR FRC website, http://www.esd.ornl.gov/nabirfrc/. The highest value of recently sampled wells is reported.
Twenty seven micromolars of potassium dichromate contains 54 μM chromium.
A concentration of 200 μM uranyl nitrate was used for transcriptional analysis, as significant precipitation was observed in the 1,000 μM treatment.
FIG. 1.
Growth of C. crescentus, E. coli, and P. putida at 1 mM uranyl nitrate. Bacteria were grown in M2G medium. Uranyl nitrate was added at early growth phase as indicated. Diamonds, C. crescentus control, not exposed to uranyl nitrate; pink squares, C. crescentus, uranium was added at 135 min to a final concentration of 1 mM uranyl nitrate; triangles, E. coli control, not exposed to uranium; X, E. coli, uranium was added at 135 min to a final concentration of 1 mM uranyl nitrate; orange squares, P. putida control, not exposed to uranium; star, P. putida, uranium was added at 275 min to a final concentration of 1 mM uranyl nitrate.
Microscopic examination after exposure to the various metals did not indicate significant changes in cell morphology, with the exception of some loss in motility after 3 to 4 h of exposure to chromate or dichromate. Membrane integrity test (by BacLight kit) at 30 min after chromium exposure did not reveal any obvious membrane damage at this point. No significant change in viability (CFU) was noted in stressed cells compared to that of nonstressed cells (data not shown).
Differentially regulated genes common to multiple heavy metals.
Transcriptional response to cadmium, chromate, dichromate, and uranium shared four up-regulated transcripts (Table 2); however, the only gene with known function was the superoxide dismutase with Mn as its cofactor (sodA). The greatest induction of this gene occurred under cadmium and chromium stress, with induction under uranium stress being lower. Superoxide dismutases are known to remove superoxide radicals which may be generated upon exposure to heavy metals (22, 23, 46, 63). While Caulobacter crescentus has three superoxide dismutase genes with different cofactors, sodA (CC1777, Mn), sodB (CC3557, Fe) and sodC (CC1579, Cu-Zn), the major gene involved in heavy metal response appears to be sodA. Under nonstressed conditions, it was at background levels but was up-regulated 19-, 14-, 9-, and 3-fold in cadmium, chromate, dichromate, and uranium stress, respectively. Transcription of sodB, on the other hand, was constitutive and was only twofold up-regulated under cadmium stress. Since sodB was induced under only cadmium stress, when sodA was already substantially up-regulated (19-fold), it is possible that sodB was up-regulated as a compensation mechanism under substantial oxidative stress. Transcription of sodC was also constitutive under nonstressed conditions but did not increase above the twofold threshold under any of the metal stress conditions. The difference in the regulation of the three enzymes may reflect their subtle differences in response to various oxidative stressors. It is known that C. crescentus sodA(Mn) and sodB(Fe) are cytosolic and sodC(Cu-Zn) is periplasmic (55). The induction of sodA and sodB but not sodC suggests that the oxidative stress imposed by the metals is intracellular rather than extracellular, which is consistent with the view that the oxidative stress originated from the reactive oxygen species generated by the interaction of metals and cellular components. The different scale of induction of sodA with various metals probably reflects the variable oxidative potentials of the metals inside the cells. Chromium and cadmium are well known for inducing oxidative stress in cells, while uranium, in our system, seemed to provoke a smaller response, although the precise intracellular quantity of metals is not directly measured in our system. As such, it is not possible to determine whether the variable oxidative response is a function of metal properties or whether it is a dose response.
TABLE 2.
Genes (and descriptions) up-regulated under cadmium, chromium, and uranium stress
| Gene | Fold change
|
Annotation | |||
|---|---|---|---|---|---|
| Cadmium | Chromate | Dichromate | Uranium | ||
| CC1777 | 18.9 | 14.1 | 8.6 | 2.9 | Superoxide dismutase (cofactor, Mn2+) (sodA) |
| CC3500 | 2.9 | 8.5 | 6.7 | 2.3 | TonB-dependent outer membrane receptor |
| CC1532 | 3.2 | 3.8 | 3.6 | 2 | Conserved hypothetical protein |
| CC3291 | 6.6 | 3.9 | 2.8 | 2.2 | Hypothetical protein |
Four genes (CC3254, CC3255, CC3256, and CC3257) consecutively located on the chromosome were commonly up-regulated under cadmium, chromate, dichromate, and selenite stress (see the supplemental data). The last three of the genes were suggested to be in an operon. Since two (CC3254 and CC3257) out of the four genes were predicted to carry membrane proteins, this general response is likely to be associated with membrane protein function; however, protein sequences did not show any similarity to typical efflux pumps or transporters.
We identified a total of 59 genes which were down-regulated at least fivefold under cadmium, chromium, or uranium stress, although we did not observe any genes commonly down-regulated more than fivefold. The most significant finding was the down-regulation of a sulfate transporter under chromium stress (see the supplemental data). Since chromate structurally resembles sulfate, and chromate typically enters the cell by the sulfate uptake system (47), we believe the response intended to reduce chromium entering into the cell. Similarly, dichromate stress provoked the down-regulation of the same transporter despite its structural differences. The reason may be rooted in chromate and dichromate equilibrium. Chromate and dichromate equilibrium are concentration and pH dependent (K = 10−2.2). Under our experimental conditions (pH 6 to 7 and concentration below 0.05 mM), a significant amount of chromate will be present (44) in the dichromate solution, thus eliciting a transcriptional response to the chromate stress.
Differential gene expression under cadmium stress.
A total of 144 genes were up-regulated by at least twofold under cadmium stress (see the supplemental data). Several groups of annotated genes (Table 3) are discussed here.
TABLE 3.
Selected genes (and descriptions) up-regulated under cadmium stress
| Gene | Fold change | Annotation |
|---|---|---|
| Efflux pumps (Cluster I) | ||
| CC2721 | 21.3 | Outer membrane efflux protein |
| CC2722 | 36 | Metal ion efflux membrane fusion protein, contains HlyD domain |
| CC2723 | 20 | Hypothetical protein |
| CC2724 | 22.8 | Homologous to nccA and czcA |
| CC2725 | 8 | Conserved hypothetical protein |
| CC2726 | 12.4 | Cation transporting P-type ATPase |
| CC2727 | 6 | Conserved hypothetical protein |
| Efflux pumps (Cluster II) | ||
| CC3195 | 3.4 | Outer membrane efflux protein |
| CC3196 | 2.6 | Contains HlyD domain |
| CC3197 | 3 | Cation/multidrug efflux pump, with AcrB/AcrD/AcrF domain |
| Protect against oxidative stress | ||
| CC1777 | 18.9 | Superoxide dismutase (cofactor, Mn2+) (sodA) |
| CC3557 | 2.2 | Superoxide dismutase (cofactor, Fe2+) (sodB) |
| CC1316 | 3.2 | Glutathione S-transferase |
| CC2434 | 2.2 | Glutathione S-transferase |
| CC0062 | 2.4 | Thioredoxin-like protein |
| CC0110 | 2 | Thioredoxin |
| CC3539 | 2.3 | Thioredoxin |
| CC2505 | 2.3 | Glutaredoxin-related protein |
| CC0994 | 2.5 | Peptide methionine sulfoxide reductase |
| CC1039 | 2.3 | Peptide methionine sulfoxide reductase |
| CC0141 | 2.3 | Glutathione synthetase |
| CC0885 | 3.6 | Riboflavin biosynthesis protein (ribD) |
| CC0886 | 4.3 | Riboflavin synthase, alpha subunit (ribE) |
| CC0887 | 4 | GTP cyclohydrolase II (ribAB) |
| CC0888 | 3.6 | Riboflavin synthase, beta subunit (ribH) |
| CC0459 | 4.1 | GTP cyclohydrolase I (tetrahydrofolate biosythesis pathway) |
| Arsenic resistance pathway | ||
| CC1503 | 4.8 | Arsenic reductase (arsC) |
| CC1504 | 4.4 | Transmembrane channel protein |
| CC1505 | 4.4 | Transcriptional regulator (arsR) |
| CC1506 | 9.9 | Arsenic resistant protein (arsH) |
| DNA repair | ||
| CC1428 | 6 | Deoxyribodipyrimidine photolyase, removes cyclobutane-type pyrimidine dimers in DNA |
| CC2590 | 2 | Excinuclease ABC, subunit A |
| Others | ||
| CC0260 | 2.7 | Ribonucleotide reductase, alpha subunit |
| CC3492 | 2.2 | Ribonucleotide reductase, beta subunit |
| CC2129 | 4.5 | NADH:flavin oxidoreductase |
The expression data demonstrated that one major detoxification mechanism appears to be an active efflux employing a translocation P-type ATPase to lower the intracellular cadmium ion concentration. This has previously been recognized as a detoxification mechanism in other bacteria (46, 57). There were two groups of efflux proteins involved that were physically clustered at different chromosomal locations. The main cluster (cluster I) was from CC2721 to CC2727, and expression of all the genes in this cluster were at background levels under nonstressed conditions but specifically induced under cadmium stress. In fact, the majority of the genes were up-regulated over 10-fold. CC2721 is an outer membrane efflux protein and CC2722 overlaps CC2721, presumably in the same operon. CC2722 contains a HlyD family domain, which is involved in the export of proteins not requiring cleavage of N-terminal signal peptides. CC2726 is a cation transporting P-type ATPase, which has previously been reported to be primarily responsible for translocating cadmium ions (and other closely related divalent heavy metals such as cobalt, mercury, lead, and zinc) across membranes (6). CC2724 is highly homologous to nccA (nickel-cobalt-cadmium resistance protein) and czcA (heavy metal efflux pump for divalent cations—cadmium, zinc, and cobalt). While CC2723, CC2725, and CC2727 were also up-regulated over 10-fold on the same chromosomal cluster, their functions are not yet known.
Another cluster (cluster two) of efflux pumps, CC3195, CC3196, and CC3197 were also specifically up-regulated under cadmium stress; however, they were transcribed at basal but clearly above background levels under nonstressed conditions. The increase of these transcripts was moderate, about threefold under cadmium stress. It is possible that this group of proteins acted as “patrol” agents, responding to stresses if needed. On the other hand, proteins in cluster I were truly specifically induced by cadmium (expression was at background level under nonstressed conditions). It is not been experimentally demonstrated whether the two groups of transporters have subtle differences in functions. If the differential transcriptional patterns cannot be explained by functional requirement, this may be another example of supplementary induction similar to that observed with superoxide dismutase. In this scenario, it appears that the main system is specifically induced, and if it alone is not sufficient to manage the effects of intracellular stressors, a supplementary mechanism is also induced. At this point, it is not clear whether any of these efflux pumps work synergistically as essential components of one system or simultaneously to increase total output.
Cadmium is known to cause oxidative stress by depleting glutathione and protein-bound sulfhydryl groups, resulting in the production of reactive oxygen species. Consequently, it leads to enhanced lipid peroxidation, DNA damage, and altered calcium and sulfhydryl homeostasis (63). We observed many groups of genes which were up-regulated to deal with this type of stress. As described above (“Differentially regulated genes common to multiple heavy metals”), the enzyme directly involved in response to oxygen stress is the superoxide dismutase (Mn). Other proteins involved in removing toxic compounds and protecting thio groups were also up-regulated. Glutathione S-transferase has been shown to be induced by heavy metals in plants (39). Glutathione has two general functions: to remove toxic metabolites from the cell and to maintain cellular sulfhydryl groups in their reduced form. Glutathione S-transferase detoxifies xenobiotic compounds or products of oxidative stress by the covalent linking of glutathione to hydrophobic substrates (13, 17). C. crescentus glutathione S-transferases CC1316 and CC2434 were up-regulated in response to cadmium stress. Thioredoxin is a general protein disulfide reductase, believed to serve as a cellular antioxidant by reducing protein disulfide bonds produced by various oxidants. In this study, we noted three thioredoxin-coding transcripts (CC0062, CC0110, and CC3539) that were up-regulated under cadmium stress. Glutaredoxin is also known as thioltransferase; it is a small protein of approximately 100 amino acid residues which functions as an electron carrier in the glutathione-dependent synthesis of deoxyribonucleotides by the enzyme ribonucleotide reductase. Like thioredoxin, which functions in a similar way, glutaredoxin possesses an active center disulphide bond and exists in either a reduced or an oxidized form where the two cysteine residues are linked in an intramolecular disulphide bond. It is not surprising that C. crescentus glutaredoxin (CC2505) was up-regulated under cadmium stress, while the other glutaredoxin (CC0829) was not, since CC0829 has more sequence homology to E. coli glutaredoxin 3 (grx3), which has been shown to have narrower substrate specificity in vivo (54). Peptide methionine sulfoxide reductase (CC0994 and CC1039) was also up-regulated, and, together with thioredoxin, it can reverse the effects of oxidative damage on methionine residues in proteins (34, 42). Under cadmium stress, both were up-regulated slightly over twofold.
Both the riboflavin biosynthesis pathway and GTP cyclohydrolase I (folE) were up-regulated (3.5- to 4-fold) under cadmium stress (Table 3). Their up-regulation may be due to oxidative stress. Previous studies have shown that H2O2-induced expression of GTP cyclohydrolase I mRNA in vascular endothelial cells (56) is extremely sensitive to oxidative stress and is also one of the major targets of H2O2 in E. coli (33). There are several possibilities for the induction of the riboflavin biosynthesis pathway; for example, studies in E. coli have demonstrated that GTP cyclohydrolase II is induced by redox-cycling agents and is positively regulated by soxR and soxS (29), the global regulators for oxidative stress. Riboflavin is a precursor of both flavin mononucleotide (FMN) and flavin adenine dinucleotide, which are important coenzymes of several oxidoreductases (1, 52). In addition, ribonucleotide reductase was up-regulated, and, in E. coli at least, this enzyme is activated by flavins (15). It provides precursors necessary for DNA synthesis by catalyzing the reductive synthesis of deoxyribonucleotides from their corresponding ribonucleotides. This reaction also replenishes the pool of reduced thioredoxin, which also has a role in protection against oxidative stress (54). Another example of an up-regulated enzyme with FAD and FMN as cofactors is CC2129, which belongs to the NADH:flavin oxidoreductase family, members of which are capable of reducing a range of alternative electron acceptors. It is also possible that GTP cyclohydrolase II was up-regulated to protect genetic material against damage from oxygen radicals. For example E. coli GTP cyclohydrolase II can hydrolyze 8-oxo-dGTP, which is an oxidized form of dGTP; overproduction of this enzyme has been shown to reduce mutation frequency of the mutT strain (mutT protein prevents A:T to C:G transversion by hydrolyzing 8-oxo-dGTP) to almost normal levels (28).
Interestingly four genes resembling arsenic resistance system components were induced by cadmium (Table 3). These genes have homologues in Pseudomonas putida KT2440 (11), in Thiobacillus ferrooxidans (10), and in virulence plasmid pYV of Yersinia enterocolitica (43). CC1503 is most similar to arsC, an arsenic reductase that is responsible for arsenate reduction to arsenite which is then translocated out of the cell by arsB using proton motive force. However, while CC1504 would be the primary candidate for arsB according to its location and the fact that it also contains a transmembrane domain, its sequence and the domain structure are different from those of arsB. As such, its capacity to function in a similar manner to arsB or indeed its involvement in response to cadmium stress is not clear. CC1505 is most similar to arsR, which is a transcriptional repressor with a helix-turn-helix DNA-binding domain that is thought to dissociate from DNA in the presence of metal ions (74). Sequence comparison shows that the metal-binding site of arsR resembles that of cadC from Staphylococcus aureas plasmid pI258 (76) and from Bacillus firmus OF4 (7). Cadmium ion is thought to relieve repression by cadC (20, 74). Therefore, it is possible that the arsRBC operon in C. crescentus was up-regulated because cadmium binds to arsR and releases the repression. CC1506 is most similar to arsH and was highly up-regulated in C. crescentus cells under cadmium stress. Despite the fact that CC1506 contains a NADPH-dependent FMN reductase domain, its direct involvement in metal resistance is not definitive. Likewise the detailed function of other arsH genes is unclear.
Response to uranium stress.
Bacteria are known to possess several mechanisms for resistance to uranium that frequently involve precipitation to reduce toxicity. For example, uranyl ions may be sequestered intracellularly by complexation with phosphate granules, as in the case of Arthrobacter spp (66) and Pseudomonas spp (64). In Citrobacter spp., inorganic phosphate is liberated from organic forms, resulting in precipitation of various uranyl phosphate crystal complexes outside the cell (35). To investigate if such mechanisms were utilized as a uranium resistance mechanism by Caulobacter crescentus, we performed TEM and EDX analysis and demonstrated that C. crescentus did not form any uranium-containing phosphate granules intracellularly. However, TEM images of whole cells of C. crescentus revealed extracellular precipitates associated with the cells (Fig. 2A). EDX spectra from cells and extracellular precipitates show that while uranium is almost absent within cells (Fig. 2B), extracellular precipitates contain high concentrations of uranium, phosphorus, and calcium (Fig. 2C), suggesting that the extracellular precipitates are composed mainly of these elements. The selected-area electron diffraction patterns of the extracellular precipitates indicated that the extracellular precipitates are amorphous, although uranyl phosphate minerals are known to readily become amorphous under the high vacuum of TEM (68). In order to examine whether the formation of the extracellular precipitates was catalyzed by C. crescentus or was simply an abiotic phenomenon, uranium-bearing precipitates formed in the same medium in the absence of C. crescentus were also characterized (Fig. 2D). EDX analysis revealed that abiotic precipitates were mainly composed of uranium and phosphorus with substantially lower calcium content than the biological precipitates (Fig. 2E), indicating either direct or indirect biological involvement in the formation of the calcium-uranium-phosphate complex.
FIG. 2.
TEM image of uranium precipitates formed in the presence and absence of C. crescentus CB15N. (A) Extracellular precipitates associated with the cells. EDX spectra were taken in the areas indicated by arrows. (B) The EDX spectrum shows uranium (U) is almost absent in cells. (C) Cell-associated extracellular precipitates are composed of uranium, calcium (Ca), and phosphorus (P). (D) Abiotic precipitates that formed between uranium and culture medium are mostly composed of uranium and phosphorus. (E) The EDX spectrum from the abiotic precipitates shows calcium is nearly depleted in the uranyl-phosphate precipitates.
Based on the chemical composition, the precipitates are thought to be the uranyl phosphate mineral autunite {Ca[(UO2)2(PO4)]2 · 11H2O}. Autunite is a major source of naturally occurring secondary uranium ore and is known to persist under oxidizing conditions on a geological time scale (21). In contrast, the uranium-bearing precipitates formed abiotically in the uninoculated control experiment appear to be chernikovite [H(UO2)(PO4) · 4H2O]. Interestingly, transcriptional analysis did show a candidate, CC1295, which may involved in the uranium precipitation process. CC1295 has a phytase domain. The protein with this domain was found to bind to Ca2+ and organic phosphate (myo-inositol hexakisphosphate) (49). Further investigations are needed to prove or disprove the hypothesis that the active site of this enzyme may facilitate the precipitation of calcium-uranium-phosphate complexes by providing a nucleation site.
Uranium stress also induced transcription of 48 transcripts which were up-regulated at least twofold (see the supplemental data); however, the response to uranium does not appear to overlap substantially with other heavy metal stresses evaluated in this study. It is believed that uranium, unlike cadmium and chromate, imposes less direct oxidative damage to cells, thus it is not surprising that most of the commonly up-regulated genes noted under cadmium and chromium stress were not up-regulated under uranium stress.
A large portion of the up-regulated genes were difficult to classify into pathways due to mainly a lack of functional annotation (Table 4), although even with clearly annotated proteins, such as two-component systems, their targets have not previously been identified. Two up-regulated two-component systems, CC1293-CC1294 and CC1304-CC1305, were identified. They are DNA-binding response regulators consisting of a CheY-like receiver domain and were up-regulated (seven- to ninefold) under only uranium stress. Both CC1293 and CC1304 have signal-sensing domains at the N-terminal, and a DNA-binding domain at the C-terminal but no sigma factor interaction domains were detected. CC1294 and CC1305 have histidine kinase domains at the C-terminal, and a HAMP membrane domain was also detected at the N-terminal of CC1294, indicating it could be a membrane protein. The E value of Pfam analysis for a membrane domain in CC1305 is too low (0.27, compared to 6.6e−12 for CC1294) to believe it has a membrane component; therefore, it may be cytoplasmic. The domain structure of the proteins suggests that CC1294 and CC1305 receive their respective metabolic/environmental signals in the following manner. The histidine kinase catalyzes ATP and transfers a phosphoryl group to the response regulator (CC1293 or CC1304), resulting in activation of the DNA-binding domain that elicits the specific response—activation or repression of the transcription of their targeted gene(s).
TABLE 4.
Selected genes (and descriptions) up-regulated under uranium stress
| Gene | Fold change | Annotation |
|---|---|---|
| Protect against oxidative stress | ||
| CC1777 | 2.9 | Superoxide dismutase (cofactor, Mn2+) (sodA) |
| Two-component signal transduction systems | ||
| CC1293 | 9.5 | DNA-binding response regulator consisting of a CheY-like receiver domain |
| CC1294 | 6.6 | Signal transduction histidine kinase |
| CC1304 | 9.7 | DNA-binding response regulator consisting of a CheY-like receiver domain |
| CC1305 | 7.7 | Signal transduction histidine kinase |
| ABC transporter | ||
| CC2090 | 3.8 | Predicted permease component |
| CC2091 | 4.3 | ABC transporter, ATPase component |
| CC2092 | 3.8 | HlyD family secretion protein |
| Possible extracellular activities | ||
| CC1295 | 5.4 | Possible phosphatase |
| CC0696 | 3.7 | Similar to gumN (biosynthesis of extracellular polysaccharide) |
| CC0697 | 2.2 | Similar to gumN (biosynthesis of extracellular polysaccharide) |
| Others | ||
| CC0411 | 3.1 | Conserved hyperthetical protein |
| CC0412 | 3.8 | Conserved hyperthetical protein |
| CC0815 | 7.9 | TonB-dependent outer membrane receptor |
| CC3500 | 2.3 | TonB-dependent outer membrane receptor |
| CC1303 | 7.4 | Hypothetical protein |
| CC1306 | 3.3 | Unknown function. Contains a dihydrouridine synthase domain |
| CC1638 | 9.8 | GMC (glucose-methanol-choline) family oxidoreductase |
| CC1891 | 8.6 | Unknown function. Contains beta-helical repeats |
| CC2111 | 3.8 | Conserved hyperthetical protein |
| CC2312 | 9.8 | Predicted transcriptional regulator |
ΔCC1293 and ΔCC1294 are knockout mutants in which CC1293 and CC1294 were replaced in-frame by tetracycline-resistant cassettes (59). Both mutants were generated using strain CB15N, thus the results of the mutants should be directly comparable with other data in this study. Sequencing of PCR amplification products confirmed the deletion and correct chromosomal location; furthermore, transcription of CC1293 or CC1294 was not detected in their respective mutants. Compared to that of the wild type, the growth of ΔCC1294 was unaffected under 200 μM uranyl nitrate stress; however, growth did slow after 120 min of uranium stress at 1 mM. Growth of ΔCC1293 showed no significant difference from the wild type at any uranium concentration tested. Expression of CC1293 decreased in the ΔCC1294 background, possibly because CC1294 is closer to the transcriptional start of the operon and replacing it with a tetracycline cassette negatively impacted the overall quantity of the CC1293 message; despite this, the regulation was not lost (Table 5). With the exception of four transcripts (Table 5) whose functions are unknown, transcripts responding to uranium maintained the same regulation patterns in the knockout mutants as in the wild type. Therefore it appears from growth and microarray data that CC1293-CC1294 is not a master regulator of uranium response despite its specific up-regulation under uranium stress.
TABLE 5.
Altered regulation to uranium in wild type and ΔCC1293 and ΔCC1294 mutantsa
| Gene | Regulation of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
ΔCC1293
|
ΔCC1294
|
|||||||
| −U | +U | P value | −U | +U | P value | −U | +U | P value | |
| CC3291 | 995 | 3,296 | 0.02 | 2,500 | 5,000 | 0.06 | 2,300 | 2,585 | 0.76 |
| CC0139 | 460 | 1,390 | 0.02 | 989 | 813 | 0.45 | 1,013 | 1,458 | 0.03 |
| CC3446 | 1,190 | 304 | 0.01 | 852 | 561 | 0.3 | 803 | 1,107 | 0.1 |
| CC2334 | 1,481 | 229 | 0.004 | 1,097 | 729 | 0.3 | 977 | 614 | 0.05 |
| CC1293 | 482 | 6,303 | 0.006 | 40 | 10 | 0.43 | 150 | 864 | 0.008 |
| CC1294 | 946 | 8,220 | 0.006 | 600 | 6,500 | 0.004 | 91 | 20 | 0.38 |
Numbers in the −U or +U columns are expression levels (average difference scores); each was an average of three independent experiments. Numbers in the P value columns represent results from a t test. Genes were differentially regulated under uranium stress in a wild-type (CB15N) background, but the regulation was lost or altered when either CC1293 or CC1294 was deleted, except CC1293 and CC1294 were included to demonstrate the loss of the respective genes in the mutants. Evidently the regulation of the CC1293-CC1294 operon is still maintained.
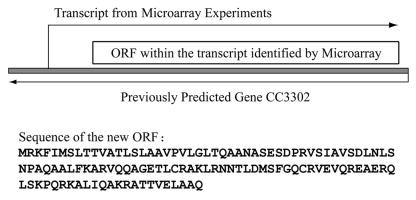
In plant and human studies, many antisense transcripts have been identified (24, 75) and, in this study, the transcript that had the highest induction (27.5-fold) specific to uranium stress came from an antisense transcript of CC3302 (transcribed from the opposite strand of the predicted CC3302). CC3302 was annotated as in the minus strand. The up-regulated probe set interrogates the plus strand of the same region. Since the boundaries of this probe set were arbitrary, we examined the data at the probe level and discovered that the first three probes of this probe set and the upstream region immediately adjacent to this probe set were not transcribed. The total length of the regulated transcript is about 420 bp, with an open reading frame containing two possible start codons (ATG) of five amino acids apart (Fig. 3). The size of the possible protein would be 112 or 117 amino acids. Further examples were detected of such regulated anti-sense transcripts responding to uranium or other heavy metals, particularly chromium and cadmium (Table 6). While the function of most anti-sense transcripts is not currently understood, these may play important and previously overlooked roles in response to heavy metal stress. Detection of such regulated transcripts is possible only by using microarrays designed to cover the whole genome without automatic assumption of previous (possibly incomplete) predictions.
FIG. 3.
A possible ORF differentially expressed under uranium stress. This transcript was from the opposite strand of predicted hypothetical protein CC3302. The antisense transcript is up-regulated 27.5-fold under uranium stress. An ORF encoding 112 or 117 amino acids was predicted within the transcript.
TABLE 6.
Up-regulated antisense transcripts
| Corresponding genes | Metal stressor | Estimated fold changea | Possible ORF | Comments |
|---|---|---|---|---|
| CC1040 and CC1041 | Chromate/dichromate | 5.3/5.7 | No | There is no gap between the probes covering the two genes, thus it is likely to be one transcript. CC1040 and CC1041 were in one operon |
| CC1127 | Uranium | 7.8 | Yes (116 aa) | |
| CC1416 | Uranium | 8.4 | Maybe | A small ORF of 52 aab may exist |
| CC2602 | Cadmium | 4.8 | Yes (135 aa) | |
| CC3553 | Chromate/dichromate | 3.1/3.7 | No | Eight consecutive probes were differentially regulated. The transcript is predicted to be 200 bp |
| CC3302 | Uranium | 27.5 | Yes (112 or 117 aa) | First three probes were not part of the transcript |
The fold change was estimated from the responding probes. It is marginally different from the algorithms used to obtain probe set values from all probes.
aa, amino acids.
Differential gene expression under chromium stress.
Hexavalent chromium, Cr(VI), is found together with a variety of aromatic compounds in a number of contaminated sites, including groundwater aquifers, lake and river sediments, and soils. Both chromate resistance and toxicity can be related to Cr(VI) reduction. The central mechanism of chromate toxicity is thought to be the reactive oxygen species that initial intracellular chromate reduction generates (27, 58, 65). A common pathway for Cr(VI) reduction to the less toxic Cr(III) is through an unstable Cr(V) intermediate (14) which is subject to redox cycling and, as such, can generate large amounts of reactive oxygen species which induce cellular damage. While Cr(VI) reduction to Cr(V) occurs spontaneously by cellular components (61, 62), only chromium-resistant bacteria are capable of reducing Cr(V) to Cr(III) intracellularly, thus minimizing the oxidative damage induced by Cr(V) as it occurs transiently within the cell (1, 2). Although we present no direct evidence to discount intracellular chromate reduction in C. crescentus, both the phenotypic data (the concentration of chromium severely reducing growth was relatively low [50 μM] compared to that in efficient chromium reducing bacteria, e.g., 400 μM in Pseudomonas putida [2]) and the transcriptional analysis in this study strongly suggest that there was significant intracellular damage induced by oxidative stress and, therefore, it is unlikely that significant chromium reduction occurred.
In C. crescentus, it appears that the majority of up-regulated genes were in response to chromium-induced oxidative stress (Table 7). Both chromate and dichromate contain Cr(VI) and, subsequently, a large portion of the up-regulated genes (214) are common to both metal stresses; however, their chemical structures differ and this may account for the number of genes up-regulated only twofold upon exposure to one form of chromium. Following chromate exposure, 84 genes were specifically up-regulated, while 23 were specifically up-regulated following dichromate exposure (see the supplemental data).
TABLE 7.
Selected genes (and descriptions) up-regulated under chromium stress
| Genes | Fold change
|
Annotation | |
|---|---|---|---|
| Chromate | Dichromate | ||
| Protect against oxidative stress | |||
| CC1777 | 14.1 | 8.6 | Superoxide dismutase (cofactor, Mn2+) (sodA) |
| CC1124 | 2 | 2.5 | Glutathione S-transferase |
| CC2311 | 5.6 | 6.1 | Glutathione S-transferase |
| CC0220 | 6.4 | 4.3 | Thioredoxin-like |
| CC1039 | 9.3 | 6.5 | Peptide methionine sulfoxide reductase |
| Outer membrane response | |||
| CC3500 | 8.5 | 6.7 | TonB-dependent outer membrane receptor |
| CC0201 | 3.9 | 4.3 | OmpA family protein, outer membrane protein and related peptidoglycan-associated lipoproteins |
| CC0747 | 3.9 | 4.5 | OmpA family protein, outer membrane protein and related peptidoglycan-associated lipoproteins |
| Two-component signal transduction system | |||
| CC0247 | 2.6 | 2.2 | DNA-binding response regulator consisting of a CheY-like receiver domain |
| CC0248 | 2.2 | <2 | Histidine kinase |
| DNA repair | |||
| CC2200 | 2.7 | 3 | HNH endonuclease |
| CC2272 | 3.3 | 2.8 | EndoIII-related endonuclease |
| CC1087 | 4.1 | 3.5 | recA protein |
| CC1902 | 3.2 | 2.9 | Repressor LexA SOS-response transcriptional |
| Repressors (RecA-mediated autopeptidases) | |||
| Electron transport process/cytochrome oxidases | |||
| CC1770 | 2.7 | 2 | Heme copper-type cytochrome c oxidase, subunit 4 |
| CC1771 | 2.8 | 2.3 | Heme copper-type cytochrome c oxidase, subunit 3 |
| CC1772 | 3.8 | 3.1 | Heme copper-type cytochrome c oxidase, subunit 1 |
| CC1773 | 5.8 | 4.1 | Heme copper-type cytochrome c oxidase, subunit 2 |
| CC0762 | 2.8 | 2.3 | Cytochrome d ubiquinol oxidase subunit 1 |
| CC0763 | 3.6 | 3.6 | Cytochrome d ubiquinol oxidase subunit 2 |
| CC2269 | 2.7 | 3 | Isoquinoline 1-oxidoreductase, beta subunit |
| CC2270 | 2.4 | 2.2 | Isoquinoline 1-oxidoreductase, alpha subunit |
| CC0946 | 3.1 | 2.5 | Cytochrome P450 family protein |
| Genes involved in metabolism | |||
| Ammonium transport | |||
| CC1338 | 5.1 | 3.6 | Ammonium transporter. Ammonia permeases |
| CC1339 | 5.7 | 4.3 | Nitrogen regulatory protein PII 2 |
| CC1740 | 2.6 | 2.2 | Nitrogen regulation protein NR(II) (ntrB). Signal transduction histidine kinase |
| CC1741 | 3.4 | 3 | Nitrogen regulation protein NR(I) (ntrC). DNA-binding response regulator |
| Glutamate sysnthesis | |||
| CC3606 | 4.1 | 4.1 | Glutamate synthase, small subunit |
| CC3607 | 3 | 2.7 | Glutamate synthase, large subunit |
| Phosphate starvation response | |||
| CC2644 | 24.4 | 24.9 | PhoH homolog. Phosphate starvation-inducible ATPase |
| PHB synthesis | |||
| CC0510 | 6 | 4.7 | Acetyl-CoA transferase (phbA) |
| CC0511 | 3.4 | 3.1 | NADPH-dependent acetoacetyl-CoA reductase (phbB) |
| CC0947 | 2.7 | 2.3 | Enoyl-CoA hydratase/isomerase (fatty acid oxidation) |
| CC2478 | 2.3 | <2 | Acyl-CoA dehydrogenase (fatty acid oxidation) |
| CC3087 | 2.5 | 2.3 | Acyl-CoA dehydrogenase (fatty acid oxidation) |
| PHB and carbon/energy utilization | |||
| CC0250 | 5.5 | 6.1 | PHB depolymerase |
| CC0797 | 2.7 | 2.6 | 1,4-beta-d-glucan glucohydrolase D hydrolysis of the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety |
| Histidine degradation | |||
| CC0957 | 2.7 | 2.2 | Urocanate hydratase (hutU) |
| CC0958 | 3.1 | 2.8 | Formiminoglutamase (hutG) |
| CC0959 | 4.0 | 4.9 | Histidine ammonia lyase (hutH) |
| CC0960 | 4.8 | 5.8 | Imidazolonepropionase (hutI) |
| Serine biosynthesis | |||
| CC3215 | 2.6 | 2.1 | d-3-phosphoglycerate dehydrogenase (serA) |
| CC3216 | 3.1 | 3.2 | Phosphoserine aminotransferase (serC) |
| LPSa synthesis | |||
| CC0118 | 3.6 | 3.7 | Glucosamine-fructose-6-phosphate aminotransferase |
| CC1985 | 10.9 | 12.2 | UDP-3-O-acyl-N-acetylglucosamine deacetylase (lpxC). Catalyzes the second step in lipopolysaccharide biosynthesis |
| Others | |||
| CC1506 | 2.8 | 2 | Arsenic resistance protein (arsH) |
LPS, lipopolysaccharide.
Several genes which are known to be involved in response to oxidative stress were up-regulated under either chromate or dichromate stress. The superoxide dismutase (Mn, sodA) had 9-fold to14-fold induction, while the other two superoxide dismutases of different cofactors were not up-regulated more than twofold. As with cadmium stress, glutathione S-transferase and thioredoxin were up-regulated. However, under cadmium stress, two different glutathione S-transferases were up-regulated (only two- to threefold) and growth was less severely affected. Under chromium stress, one glutathione S-transferase (CC2311) was up-regulated about sixfold (Table 7), but unlike under cadmium stress, glutathione synthetase (gshB, CC0141) was not up-regulated. As the physiological states of the cells were quite different between chromium and cadmium stresses (substantially slower growth under chromium stress), this suggests that C. crescentus may employ different processes to counteract oxidative stress, depending on the physiological state of the cell.
DNA damage by reactive oxygen species upon chromium exposure is well documented (4, 73). Up-regulation of recA is known to be induced by DNA breakage in E. coli, and previous studies have shown that chromate-induced DNA damage strongly depends on the reactive intermediates. Frequently, chromate causes DNA single-strand breakage and 8-hydroxydeoxyguanosine formation (correlated with hydroxyl radical as the DNA-damaging species) (5, 26). In this study, DNA repair enzymes, such as CC2272 and CC2200, were up-regulated (Table 7). This suggests that chromate-induced damage in C. crescentus cells may indeed be mediated by hydroxyl radicals generated through nonspecific chromate reduction.
Studies of animal and plant cells have shown that chromium can cause membrane damage through direct or oxidative stress-mediated interactions (14, 25, 50), and our transcriptional data indicate that chromium exposure induces a membrane response. We observed induction of two OmpA family proteins [ompA mutants exhibit increased sensitivity to environmental stresses (72)], TonB-dependent outer membrane receptors, and lipopolysaccharide biosynthesis (Table 7). Our data indicate that the TonB receptor family of proteins was involved in the response to several different metals, such as chromium (Table 7), cadmium (Table 3), and uranium (Table 4), and yet, in most cases, different proteins in the family were up-regulated under specific metal stresses. However, it is not clear whether the up-regulated TonB receptor gene interacts with TonB protein, since the expression of the predicted TonB protein (CC2327) was suppressed and not activated under heavy metal stress. It is likely that the receptor merely binds to the substrates (heavy metals) and communicates extracellular environmental information.
We observed up-regulation of several genes and pathways which typically occur during stationary phase or under nutrient-limiting conditions (Table 7), including acquisition of ammonium, phosphate-starvation response, poly β-hydroxybutyrate (PHB) biosynthesis (including genes in the fatty acid oxidation pathway which can provide precursors for synthesis of medium chain PHBs), and energy/carbon utilization (PHB depolymerase and glucan glucohydrolase).
The cells appeared to exhibit an increased demand for ammonium and glutamate, since ammonium transport, the glutamate synthase, and the histidine degradation pathway (generating ammonium and glutamate) were up-regulated (Table 7). Glutamate is one of the central amino acids that links nitrogen (glutamine synthetase-glutamine 2-oxoglutarate-aminotransferase cycle) and carbon/energy metabolism (trichloracetic acid cycle) through α-keto-glutarate. There appeared to be an increased flow of glutamate to α-keto-glutarate, as indicated by the up-regulation of the serine biosynthesis pathway. In addition, glutamate synthase contains an iron-sulfur cluster and has close structural homology with glutamine phosphoribosylpyrophosphate amidotransferase (71). This latter enzyme from B. subtilis is known to be inactivated by O2 in stationary phase (9). Although up-regulation of ntrBC and PII usually indicates nitrogen limitation response, the key enzyme (glutamine synthetase) was not up-regulated. It is possible that increased ammonium uptake was the first step towards responding to nitrogen demand, and thus it seems plausible that simply obtaining more ammonium from the environment would provide a more energy efficient response than provoking a complete nitrogen starvation response. If this hypothesis is correct, our data suggest that the three PII proteins of C. crescentus may be regulated separately by specific physiological conditions and that ntrBC may be involved in activating the transcription of not only glutamine synthetase but also other operons, such as ammonium transport. Therefore, it may be that nitrogen regulation in C. crescentus is more complicated than in E. coli, at least at the transcriptional level.
Differential gene expression under selenium stress.
The response to sodium selenite was mild compared to other metal stresses. Only 12 transcripts were up-regulated and at most by four- to fivefold (see the supplemental data). All of these were also observed to be up-regulated under chromium or cadmium stresses and included membrane components, glutathione S-transferases, and transport proteins.
Conclusions.
In this study, we investigated whole-genome transcriptional response of Caulobacter crescentus to the stress of several heavy metals, including chromium and uranium, which are significant environmental contaminants and a current focus of bioremediation efforts. In addition to the surprising finding that C. crescentus CB15N is tolerant to uranium, our studies combining physiology observation, transcriptional measurement, and imaging analysis clearly showed that Caulobacter formed a calcium-uranium-phosphate precipitate extracellularly, in contrast to the intracellular sequestration mechanism of other resistant bacteria, such as Arthrobacter spp. This observation was consistent with a limited response to oxidative stress, such as that seen with other metals. The stress response strategy of lowering intracellular metal concentration was also present in cadmium and chromium response. Efflux pumps were up-regulated under cadmium stress. C. crescentus does not seem to have a specific extrusion mechanism for chromium; however, the cells down-regulated a sulfate transporter, which may reduce the uptake of chromate.
In broader terms, cells exposed to cadmium share many up-regulated transcripts with those under chromium stress. Most of those up-regulated genes respond to oxidative stress, such as superoxide dismutase, glutathione S-transferase, thioredoxin, and glutaredoxin. However, on closer inspection, the individual proteins up-regulated, and the fold changes were specific to each metal (for example, different sets of glutathione S-transferase were up-regulated under cadmium and chromium stress), indicating the subtle difference of each metal stress and physiological conditions. We also observed up-regulation of TonB-dependent outer membrane receptors which may serve as sensors for environmental signals. While the detailed mechanisms of their involvement are still not known, our results suggest that they may be an important member of the response network.
We believe that not all of the observed up- or down-regulation was a direct response to the metal toxicity. This was particularly evident in the case of chromium and/or cadmium stress. While the direct involvement of an arsenic reductase operon in cadmium response is unclear, the likely mechanism is that cadmium binds to the repressor of the operon, resulting in its up-regulation. The response of C. crescentus under chromium stress was clearly different from that of other oxidative stress (for example, cadmium), yet it may be complicated by secondary responses. Future transcriptomics studies, with various concentrations of chromium, augmented with proteomic analyses may help elucidate the complex response observed to this heavy metal such as the role of cytochrome oxidases and the apparent nutrient-limitation response.
Our data have also clearly demonstrated the importance of interrogating the whole genome on both strands. We have identified antisense transcripts which are differentially regulated specific to metals, which, as either proteins or RNAs, may play an import part in the response model.
Supplementary Material
Acknowledgments
This study was funded by the Department of Energy Genomes to Life Microbial Cell Program. This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Berkeley National Laboratory, under contract no. DE-AC03-76SF00098. H. McAdams was funded under Department of Energy grant DE-FG03-01ER63219.
We thank Susan Lynch and A. C. Matin at Stanford University for the generous gift of bacterial strains Escherichia coli K12 and Pseudomonas putida KT2440. We thank M. Laub at Harvard University for the generous gift of C. crescentus two-component signal transduction deletion mutants ΔCC1293 and ΔCC1294.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ackerley, D. F., C. F. Gonzalez, M. Keyhan, R. Blake II, and A. Matin. 2004. Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environ. Microbiol. 6:851-860. [DOI] [PubMed] [Google Scholar]
- 2.Ackerley, D. F., C. F. Gonzalez, C. H. Park, R. Blake, I. I., M. Keyhan, and A. Matin. 2004. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl. Environ. Microbiol. 70:873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Affymetrix. 2001. Statistical algorithms reference guide. Technical report. Affymetrix, Santa Clara, Calif.
- 4.Aiyar, J., H. J. Berkovits, R. A. Floyd, and K. E. Wetterhahn. 1990. Reaction of chromium(VI) with hydrogen peroxide in the presence of glutathione: reactive intermediates and resulting DNA damage. Chem. Res. Toxicol. 3:595-603. [DOI] [PubMed] [Google Scholar]
- 5.Aiyer, J., K. M. Borges, R. A. Floyd, and K. E. Wetterhahn. 1989. Role of chromium(V), glutathione thiyl radical and hydroxyl radical intermediates in chromium(VI)-induced DNA damage. Toxicol. Environ. Chem. 22:135-148. [Google Scholar]
- 6.Axelsen, K. B., and M. G. Palmgren. 1998. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46:84-101. [DOI] [PubMed] [Google Scholar]
- 7.Bairoch, A. 1993. A possible mechanism for metal-ion induced DNA-protein dissociation in a family of prokaryotic transcriptional regulators. Nucleic Acids Res. 21:2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benyehuda, G., J. Coombs, P. L. Ward, D. Balkwill, and T. Barkey. 2004. Metal resistance among aerobic chemoheterotrophic bacteria from the deep terrestrial subsurface. Can. J. Microbiol. 49:151-156. [DOI] [PubMed] [Google Scholar]
- 9.Bernlohr, D., and R. L. Switzer. 1981. Reaction of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase with oxygen: chemistry and regulation by ligands. Biochemistry 20:5675-5681. [DOI] [PubMed] [Google Scholar]
- 10.Butcher, B. G., S. M. Deane, and D. E. Rawlings. 2000. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl. Environ. Microbiol. 66:1826-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cánovas, D., I. Cases, and V. de Lorenzo. 2003. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 5:1242-1256. [DOI] [PubMed] [Google Scholar]
- 12.Cervantes, C., J. Campos-Garcia, S. Devars, F. Gutierrez-Corona, H. Loza-Tavera, J. C. Torres-Guzman, and R. Moreno-Sanchez. 2001. Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 25:335-347. [DOI] [PubMed] [Google Scholar]
- 13.Feng, P. C. C. 1991. Soil transformation of acetochlor via glutathione conjugation. Pestic. Biochem. Physiol. 31:84-90. [Google Scholar]
- 14.Fernandes, M. A. S., M. S. Santos, M. C. Alpoim, V. M. C. Madeira, and J. A. F. Vicente. 2002. Chromium(VI) interaction with plant and animal mitochondrial bioenergetics: a comparative study. J. Biochem. Mol. Toxicol. 16:53-63. [DOI] [PubMed] [Google Scholar]
- 15.Fontecave, M., R. Eliasson, and P. Reichard. 1989. Enzymatic regulation of the radical content of the small subunit of Escherichia coli ribonucleotide reductase involving reduction of its redox center. J. Biol. Chem. 264:9164-9170. [PubMed] [Google Scholar]
- 16.Gong, P., O. Y. Ogra, and S. Koizumi. 2000. Inhibitory effects of heavy metals on transcription factor Sp1. Ind. Health 38:224-227. [DOI] [PubMed] [Google Scholar]
- 17.Habig, W. H., M. J. Pabst, and W. B. Jakoby. 1974. Glutathione S-transferases: the first enzymatic step in mercapturic acid formation. J. Biol. Chem. 249:7130-7139. [PubMed] [Google Scholar]
- 18.Hottes, A. K., M. Meewan, D. Yang, N. Arana, P. Romero, H. H. McAdams, and C. Stephens. 2004. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 186:1448-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inagaki, F., K. Takai, H. Hirayama, Y. Yamato, K. Nealson, and K. Horikoshi. 2003. Distribution and phylogenetic diversity of the subsurface microbial community in a Japanese epithermal gold mine. Extremophiles 7:307-317. [DOI] [PubMed] [Google Scholar]
- 20.Ivey, D. M., A. A. Guffanti, Z. Shen, N. Kudyan, and T. A. Krulwich. 1992. The cadC gene product of alkaliphilic Bacillus firmus OF4 partially restores Na+ resistance to an Escherichia coli strain lacking an Na+/H+ antiporter (NhaA). J. Bacteriol. 174:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jerden, J. L., Jr., and A. K. Sinha. 2003. Phosphate based immobilization of uranium in an oxidizing bedrock aquifer. Appl. Geochem. 18:823-843. [Google Scholar]
- 22.Jones, P., A. Kortenkamp, P. O'Brien, G. Wang, and G. Yang. 1991. Evidence for the generation of hydroxyl radicals from a chromium(V) intermediate isolated from the reaction of chromate with glutathione. Arch. Biochem. Biophys. 286:652-655. [DOI] [PubMed] [Google Scholar]
- 23.Kachur, A. V., C. J. Koch, and J. E. Biaglow. 1998. Mechanism of copper-catalyzed oxidation of glutathione. Free Radic. Res. 28:259-269. [DOI] [PubMed] [Google Scholar]
- 24.Kapranov, P., S. C. Cawley, J. Drenkow, S. Bekiranov, R. L. Strausberg, S. P. A. Fodor, and T. R. Gingeras. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296:916-919. [DOI] [PubMed] [Google Scholar]
- 25.Karbownik, M., J. J. Garcia, A. Lewińsiki, and R. J. Reiter. 2001. Carcinogen-induced, free radical-mediated reduction in microsomal membrane fluidity: reversal by indole-3-propionic acid. J. Bioenerg. Biomembr. 33:73-79. [DOI] [PubMed] [Google Scholar]
- 26.Kawanishi, S., S. Inoue, and S. Sano. 1986. Mechanism of DNA cleavage induced by sodium chromate(VI) in the presence of hydrogen peroxide. J. Biol. Chem. 261:5952-5958. [PubMed] [Google Scholar]
- 27.Klein, C., E. Snow, and K. Frenkel. 1998. Molecular mechanisms in metal carcinogenesis: role of oxidative stress, p. 79-137. In O. I. Aruoma and B. Halliwell (ed.), Molecular biology of free radicals in human diseases. OICA International, London, England.
- 28.Kobayashi, M., Y. Ohara-Nemoto, M. Kaneko, H. Hayakawa, M. Sekiguchi, and K. Yamamoto. 1998. Potential of Escherichia coli GTP cyclohydrolase II for hydrolyzing 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 273:26394-26399. [DOI] [PubMed] [Google Scholar]
- 29.Koh, Y., J. Choih, J. Lee, and J. Roe. 1996. Regulation of ribA gene encoding GTP cyclohydrolase II by the soxRS locus in Escherichia coli. Mol. Gen. Genet. 251:591-598. [DOI] [PubMed] [Google Scholar]
- 30.Kratochvil, D., and B. Volesky. 1998. Advances in the biosorption of heavy metals. Trends Biotechnol. 16:291-300. [Google Scholar]
- 31.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 32.Laub, M. T., S. L. Chen, L. Shapiro, and H. H. McAdams. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 99:4632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leichert, L. I., and U. Jakob. 2004. Protein thiol modification visualized in vivo. PloS. Biology 2:e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowther, W. T., N. Brot, H. Weissbach, and B. W. Mathews. 2000. Structure and mechanism of peptide methione sulfoxide reductase, an “anti-oxidation” enzyme. Biochemistry 39:13307-13312. [DOI] [PubMed] [Google Scholar]
- 35.Macaskie, L. E., K. M. Bonthrone, P. Yong, and D. T. Goddard. 2000. Enzymatically mediated bioprecipitation of uranium by a Citrobacter sp.: a concerted role for exocellular lipopolysaccharide and associated phosphatase in biomineral formation. Microbiology 146:1855-1867. [DOI] [PubMed] [Google Scholar]
- 36.MacRae, J. D., and J. Smit. 1991. Characterization of Caulobacter isolated from wastewater treatment systems. Appl. Environ. Microbiol. 57:751-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Männistö, M. K., M. A. Tiirola, M. S. Salkinoja-Salonen, M. S. Kulomaa, and J. A. Puhakka. 1999. Diversity of chlorophenol-degrading bacteria isolated from contaminated boreal groundwater. Arch. Microbiol. 171:189-197. [DOI] [PubMed] [Google Scholar]
- 38.Marchesi, J. R., A. J. Weightman, B. A. Cragg, R. J. Parkes, and J. C. Fry. 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. 34:221-228. [DOI] [PubMed] [Google Scholar]
- 39.Marrs, K. A. 1996. The functions and regulation of glutathione S-transferase in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47:127-158. [DOI] [PubMed] [Google Scholar]
- 40.McAdams, H. H., and L. Shapiro. 1995. Circuit simulation of genetic networks. Science 269:650-656. [DOI] [PubMed] [Google Scholar]
- 41.McAdams, H. H., and L. Shapiro. 2003. A bacterial cell-cycle regulatory network operating in time and space. Science 301:1874-1877. [DOI] [PubMed] [Google Scholar]
- 42.Moskovitz, J., M. A. Rahman, J. Strassman, S. O. Yancey, S. R. Kushner, N. Brot, and H. Weissbach. 1995. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J. Bacteriol. 177:502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neyt, C., M. Iriarte, V. H. Thi, and G. R. Cornelis. 1997. Virulence and arsenic resistance in Yersiniae. J. Bacteriol. 179:612-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nieboer, E., and A. A. Jusyss. 1988. Biological chemistry of chromium, p. 27-28. In J. O. Nriagu and E. Nieboer (ed.), Chromium in the natural and human environments. John Wiley & Sons, New York, N.Y.
- 45.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. Craig Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 47.Nies, D. H. 2000. Heavy metal-resistant bacteria as extremophiles: molecular physiology and biotechnological use of Ralstonia sp. CH34. Extremophiles 4:77-82. [DOI] [PubMed] [Google Scholar]
- 48.North, N. N., S. L. Dollhopf, L. Petrie, J. D. Istok, D. L. Balkwill, and J. E. Kostka. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh, B. C., B. S. Chang, K. H. Park, N. C. Ha, H. K. Kim, B. H. Oh, and T. K. Oh. 2000. Calcium-dependent catalytic activity of a novel phytase from Bacillus amyloliquefaciens DS11. Biochemistry 40:9669-9676. [DOI] [PubMed] [Google Scholar]
- 50.Pesti, M., Z. Gazdag, and J. Belágyi. 2000. In vivo interaction of trivalent chromium with yeast plasma membrane as revealed by EPR spectroscopy. FEMS Microbiol. Lett. 182:375-380. [DOI] [PubMed] [Google Scholar]
- 51.Poindexter, J. S. 1976. The Caulobacters: ubiquitous unusual bacteria. Microbiol. Rev. 45:123-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puzon, G. J., J. N. Petersen, A. G. Roberts, D. M. Kramer, and L. Xun. 2002. A bacterial flavin reductase system reduces chromate to a soluble chromium(III)-NAD+ complex. Biochem. Biophy. Res. Comm. 294:76-81. [DOI] [PubMed] [Google Scholar]
- 53.Rensing, C., B. Mitra, and B. P. Rosen. 1997. Insertional inactivation of dsbA produces sensitivity to cadmium and zinc in Escherichia coli. J. Bacteriol. 179:2769-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritz, D., and J. Beckwith. 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbial. 55:21-48. [DOI] [PubMed] [Google Scholar]
- 55.Schnell, S., and H. M. Steinman. 1995. Function and stationary-phase induction of periplasmic copper-zinc superoxide dismutase and catalase/peroxidase in Caulobacter crescentus. J. Bacteriol. 177:5924-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimizu, S., K. Shiota, S. Yamamoto, Y. Miyasaka, M. Ishii, T. Watabe, M. Nishida, Y. Mori, T. Yamamoto, and Y. Kiuchi. 2003. Hydrogen peroxide stimulates terahydrobiopterin synthesis through the induction of GTP-cyclohydrolase I and increases nitric oxide synthase activity in vascular endothelial cells. Free Radic. Biol. Med. 34:1343-1352. [DOI] [PubMed] [Google Scholar]
- 57.Silver, S., and L. T. Phung. 1996. Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50:753-789. [DOI] [PubMed] [Google Scholar]
- 58.Singh, J., D. L. Carlisle, D. E. Pritchard, and S. R. Patierno. 1998. Chromium-induced genotoxicity and apoptosis: relationship to chromium carcinogenesis. Oncol. Rep. 5:1307-1318. [DOI] [PubMed] [Google Scholar]
- 59.Skerker, J. M., M. S. Prasol, B. S. Perchuk, E. G. Biondi, and M. T. Laub. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3:e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smit, J., C. S. Sherwood, and R. F. Turner. 2000. Characterization of high density monolayers of the biofilm bacterium Caulobacter crescentus: evaluating prospects for developing immobilized cell bioreacters. Can. J. Microbiol. 46:339-349. [PubMed] [Google Scholar]
- 61.Standeven, A. M., and K. E. Wetterhahn. 1991. Possible role of glutathione in chromium(VI) metabolism and toxicity in rats. Pharmcol. Toxicol. 68:469-476. [DOI] [PubMed] [Google Scholar]
- 62.Standeven, A. M., and K. E. Wetterhahn. 1992. Ascobate is the principal reductant of chromium(VI) in rat lung ultrafiltrates and cytosols, and mediates chromium-DNA binding in vitro. Carcinogenesis 13:1319-1324. [DOI] [PubMed] [Google Scholar]
- 63.Stohs, S. J., and D. Bagchi. 1995. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18:321-336. [DOI] [PubMed] [Google Scholar]
- 64.Strandberg, G. W., S. E. Shumate II, and J. R. Parrott, Jr. 1981. Microbial cells as biosorbents for heavy metals: Accumulation of uranium by Saccharomyces cerevisiae and Pseudomonas aeruginosa. Appl. Environ Microbiol. 41:237-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki, T., N. Miyata, H. Horitsu, K. Kawai, K. Takamizawa, Y. Tai, and M. Okazaki. 1992. NAD(P)H-dependent chromium(VI) reductase of Pseudomonas ambigua G-1: a Cr(V) intermediate is formed during the reduction of Cr(VI) to Cr(III). J. Bacteriol. 174:5340-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki, Y., and J. F. Banfield. 2004. Resistance to and accumulation of uranium by bacteria from a uranium-contaminated site. Geomicrobiol. J. 21:113-121. [Google Scholar]
- 67.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2003. Microbial populations stimulated for hexavalent uranium reduction in uranium mine sediment. Appl. Environ. Microbiol. 69:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki, Y., T. Sato, H. Isobe, T. Kogure, and T. Murakami. 2005. Dehydration processes of the meta-autunite group minerals, meta-autunite, metasaléeite and metatorbernite. Am. Miner. 70:1308-1314. [Google Scholar]
- 69.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valls, M., and V. de Lorenzo. 2002. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol. Rev. 26:327-338. [DOI] [PubMed] [Google Scholar]
- 71.van den Heuvel, R. H. H., B. Curti, M. A. Vanoni, and A. Mattevi. 2004. Glutamate synthase: a fascinating pathway from L-glutamine to L-glutamate. Cell. Mol. Life Sci. 61:669-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, Y. 2002. The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 292:396-401. [DOI] [PubMed] [Google Scholar]
- 73.Wetterhahn, K. E., J. W. Hamilton, J. Aiyar, K. M. Borges, and R. Floyd. 1989. Mechanisms of chromium(VI) carcinogenesis. Reactive intermediates and effect on gene expression. Biol. Trace Elem. Res. 21:405-411. [DOI] [PubMed] [Google Scholar]
- 74.Wu, J., and P. Rosen. 1993. Metalloregulated expression of the ars operon. J. Biol. Chem. 268:52-58. [PubMed] [Google Scholar]
- 75.Yamada, K., J. Lim, J. M. Dale, H. Chen, P. Shinn, C. J. Palm, A. M. Southwick, H. C. Wu, C. Kim, M. Nguyen, P. Pham, R. Cheuk, G. Karlin-Newmann, S. X. Liu, B. Lam, H. Sakano, T. Wu, G. Yu, M. Miranda, H. L. Quach, M. Tripp, C. H. Chang, J. M. Lee, M. Toriumi, M. M. H. Chan, C. C. Tang, C. S. Onodera, J. M. Deng, K. Akiyama, Y. Ansari, T. Arakawa, J. Banh, F. Banno, L. Bowser, S. Brooks, P. Carninci, Q. Chao, N. Choy, A. Enju, A. D. Goldsmith, M. Gurjal, N. F. Hansen, Y. Hayashizaki, C. Johnson-Hopson, V. W. Hsuan, K. Iida, M. Karnes, S. Khan, E. Koesema, J. Ishida, P. X. Jiang, T. Jones, J. Kawai, A. Kamiya, C. Meyers, M. Nakajima, M. Narusaka, M. Seki, T. Sakurai, M. Satou, R. Tamse, M. Vaysberg, E. K. Wallender, C. Wong, Y. Yamamura, S. Yuan, K. Shinozaki, R. W. Davis, A. Theologis, J. R. Ecker. 2003. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302:842-846. [DOI] [PubMed] [Google Scholar]
- 76.Yoon, K. P., and S. Silver. 1991. A second gene in the Staphylococcus aureas cadA cadmium resistance determinant of plasmid pI258. J. Bacteriol. 173:7636-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.