Abstract
FruA is an essential transcription factor for Myxococcus xanthus development. The expression of tps and dofA genes is fruA dependent. In this study, we show by gel shift and footprint assays with the C-terminal DNA-binding domain of FruA and by a lacZ fusion assay that FruA may directly activate dofA expression during development.
Myxococcus xanthus is a gram-negative soil bacterium that exhibits a communal lifestyle during vegetative growth and fruiting body development (3, 4). When nutrients are limited, M. xanthus cells form multicellular fruiting bodies filled with myxospores, which are dormant and resistant to various environmental stresses. The developmental process is achieved by a series of sophisticated intercellular signaling pathways that regulate the expression of a specific set of genes (11, 14).
FruA is a response regulator (RR) of the two-component system that is essential for aggregation, fruiting body formation, and sporulation during development (5, 13). FruA has been proposed to play a key role in the C-signal transduction system (16). C signal is a cell surface-associated protein encoded by the csgA gene and is essential for aggregation, fruiting body formation, and sporulation during development (6, 14). Analysis of the protein expression patterns in the wild-type, fruA::Tc, and csgA731 strains during development indicated that developmental genes under the control of FruA can be classified into two groups: one that is C signal independent and one that is C signal dependent (7). The production of five proteins was found to be fruA dependent but C signal independent, and one protein was dependent on both fruA and C signal. Among them, protein S (the tps gene product) (9) and DofA, both of which are C signal independent, have been identified by sequence analyses. Protein S is a well-characterized spore coat protein which has structural similarity to βγ-crystallins (10). DofA does not show significant similarity to known proteins (7).
The DNA-binding domain of FruA binds specifically to the dofA promoter region.
To examine whether FruA directly regulates the expression of dofA and tps, a gel shift assay was performed with the DNA fragments containing the dofA and tps promoter regions. The C-terminal DNA-binding domain, from Pro at position 152 to Leu at position 229, of FruA tagged with His8 (FruA-DBD-His8) was used in this study. FruA-DBD-His8 was overproduced with the T7 promoter expression system and was purified with the use of Ni-nitrilotriacetic acid resin (QIAGEN) according to the manufacturer's instructions. Since the region from nucleotides (nt) −128 to −57 with respect to the transcription initiation site is sufficient for the developmental induction of dofA (8), for gel shift assays, DNA fragments containing the dofA promoter region from nt −150 to −32 were amplified by PCR with the chromosomal DNA of M. xanthus DZF1 and oligonucleotide primers −150T and −32B (Fig. 1). Forward primers −150T and −250T contain a HindIII site and reverse primers −32B and +10B contain a BamHI site for subsequent cloning. PCR products were digested with HindIII and BamHI and cloned into pBluescript SK− (Stratagene). DNA fragments for the probe were labeled with [α-32P]dCTP by using the Klenow fragment of DNA polymerase I after digestion of the plasmid with HindIII and BamHI and isolating the fragments by polyacrylamide gel electrophoresis (PAGE). The probe containing the tps promoter region, from nt −250 to −40, required for the developmental expression of tps (2), was also prepared by using primers 5′TCAAGCTTGCCGGTACACCCACGAC3′ and 5′TCGGATCCTACAGTACCGTATCCGTC3′ for PCR amplification as described above.
FIG. 1.
The promoter region of the dofA gene. The transcription initiation site is indicated by a bold letter and +1. The translation initiation codon is indicated by bold letters and Met. The regions from nt −82 to −67 and from nt −57 to −42, protected from DNase I in the presence of FruA-DBD-His8 in the footprint assay, are indicated with bold lines. A highly GC-rich inverted repeat sequence is indicated by arrows. The sequences of the primers used in PCR amplification, −250T, −150T, −32B, and +10B, are underlined.
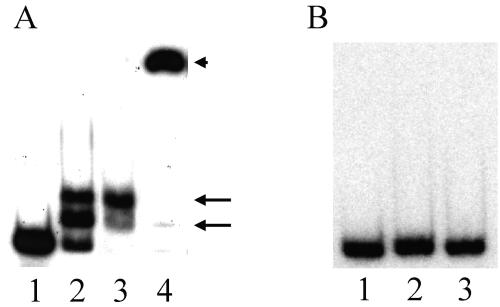
When the gel shift assay was performed as described previously (19), FruA-DBD-His8 was able to bind to the dofA promoter, and two types of complexes were observed (Fig. 2A). The addition of purified anti-FruA antibody to the reaction mixture resulted in the observation of the upshifted band, indicating that the detected complexes indeed consisted of the dofA promoter and FruA-DBD-His8. On the other hand, FruA-DBD-His8 was unable to bind to the tps promoter under the same conditions (Fig. 2B). Although the expression of dofA and tps was not observed in the fruA::Tc strain and both genes were csgA independent (7), FruA appears to directly activate dofA expression and to indirectly regulate tps expression. However, it is possible that the tps promoter tested in this study may have a lower-affinity site for FruA-DBD-His8 or may require another factor for binding of FruA-DBD-His8.
FIG. 2.
Gel shift assay. (A) The dofA promoter. The probe containing the region from nt −150 to −32 was mixed with FruA-DBD-His8, and the binding patterns were analyzed by PAGE. Lane 1, no FruA-DBD-His8; lane 2, 0.5 ng/μl of FruA-DBD-His8; lane 3, 1 ng/μl of FruA-DBD-His8; lane 4, 1 ng/μl of FruA-DBD-His8 and anti-FruA antibody. FruA-DBD-His8/dofA promoter complexes are indicated by arrows. Anti-FruA antibody/FruA-DBD-His8/dofA promoter complexes are indicated by an arrowhead. (B) The tps promoter. The probe contains the region from nt −250 to −41. Lane 1, no FruA-DBD-His8; lane 2, 0.5 ng/μl of FruA-DBD-His8; lane 3, 1 ng/μl of FruA-DBD-His8.
FruA-DBD-His8 binds to two regions in the dofA promoter.
To determine the sequences in the dofA promoter recognized by FruA-DBD-His8, a footprint assay was performed by the DNase I method with a probe containing the promoter region from nt −250 to +10 (Fig. 1). The plasmid containing the region from nt −250 to +10 was constructed with primers −250T and +10B (Fig. 1) as described above. DNA fragments for the probe were prepared by digesting the plasmid with HincII, located in the cloning vector, and BamHI and isolating the fragment by PAGE. The isolated DNA fragments were then labeled with [α-32P]dCTP by using Klenow fragment of DNA polymerase I. Thus, only the top strand was labeled.
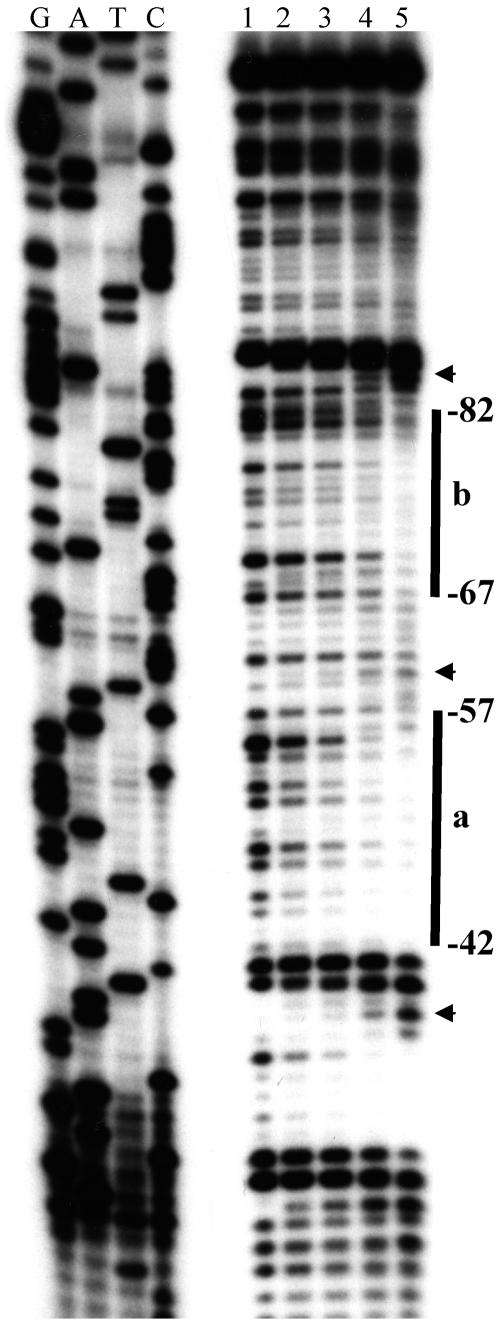
Two regions, from nt −57 to −42 (region a) and from nt −82 to −67 (region b), were found to be protected from DNase I in the presence of FruA-DBD-His8 (Fig. 3). In addition, the sites at −87, −60, and −38 were found to be hypersensitive to DNase I. It appears that region a has a slightly higher affinity to FruA-DBD-His8 than region b does, judging from the results of the footprint assay. Although apparent sequence similarity is not found in these regions, the sequence 5′AGGGC3′ from nt −79 to −75 in region b is found to be the complementary sequence of 5′GCCCT3′ from nt −53 to −49 in region a (Fig. 1). Therefore, it is possible that these sequences are critical for the binding of FruA-DBD-His8 to the dofA promoter. Binding sites located in the reverse orientation are also found in some promoters regulated by the RR. Spo0A, for instance, regulates many genes which have binding sites in both orientations in their promoters (17).
FIG. 3.
Footprint assay. Lane 1, no FruA-DBD-His8; lanes 2 to 5, 0.25, 0.5, 1, and 2 ng/μl of FruA-DBD-His8, respectively. Lanes G, A, T, and C represent sequence ladders generated by a primer, 5′GATCCGCCTGACGGTCTGCGCACCCA3′, which can hybridize to the 3′ end of the top strand of the probe. The sites that are hypersensitive to DNase I, at −87, −60, and −38, are indicated by arrowheads.
To examine the roles of these regions on dofA expression in vivo, various regions of the dofA promoter were translationally fused to the lacZ gene and introduced into the phage Mx8 attachment gene attB in the M. xanthus chromosome (18). Since it has been shown that the dofA promoter region up to nt −128 is sufficient for the developmental regulation of dofA expression (8), the dofA promoter regions up to nt −128, −91, −62, and −41 were examined. The activity of β-galactosidase was measured at 20 h of development. The deletion of the highly GC-rich inverted repeat sequence (Fig. 1) (promoter −91) decreased β-galactosidase activity to 45% of that of the promoter up to nt −128 (promoter −128), and the deletion of region b (promoter −62) and region a (promoter −41) decreased its activity to 13% and 4%, respectively. These results agree well with the results previously described (8). Therefore, regions a and b were important for the developmental regulation of dofA expression. It is worth noting that the spacing between regions a and b was unimportant, since deletion of the region from nt −68 to −60 had no effect (8).
Implications.
Since region a seems to have a slightly higher affinity to FruA-DBD-His8 than region b as described above, FruA may bind in vivo first to region a and then to region b to activate the dofA gene. Furthermore, as the activity of the promoter containing only region a was less that half of that containing both regions a and b, it is likely that regions a and b synergistically activate the dofA gene. Moreover, because the deletion of the highly GC-rich inverted repeat sequence drastically affected the activity of the dofA promoter, some additional factor(s) may be required for the full activation of the dofA gene during development.
It will be important in the future to characterize the nature of the entire FruA protein, namely, the function of the N-terminal receiver domain of FruA, since the C-terminal domain of the RR does not always function in the same fashion as the entire RR. In the case of Spo0A, Spo0A functions as both an activator and a repressor, and it has been proposed that the N-terminal domain of Spo0A inhibits transcription activation unless it is phosphorylated (17). It is possible that FruA also functions as a repressor, since some proteins are not repressed in ΔfruA mutants during development (7).
The fruA gene is essential for development, and the dofA gene is not (8). Therefore, FruA appears to regulate other genes that are essential for development. It has been shown that FrzCD methylation and devRS expression are under the control of both FruA and C signal (5, 15). It is possible that FruA regulates the expression of frzF, which encodes the methyltransferase for FrzCD, although frzF is located in the frz operon containing frzA, frzB, frzCD, frzE, and frzG upstream of frzF (12). On the other hand, the devRS genes are part of the dev operon, which contains three genes upstream of the devRS genes (1). Thus, identification of the promoter regions of frzF and devRS is essential for understanding how FruA regulates FrzCD methylation and devRS expression.
We are now attempting to identify a consensus sequence of FruA binding sites by using randomized oligonucleotides as probes. Since sequencing of the M. xanthus genome has been completed (http://www.ncbi.nlm.nih.gov), it may be possible to identify target genes of FruA by searching the genome for sequences homologous to FruA-binding sites. Identification of genes regulated by FruA is important for elucidation of the molecular mechanisms of the signal transduction pathways during development.
Acknowledgments
We thank L. Vales for a critical reading of the manuscript, C. Xu for DNA manipulation, and H. Nariya for helpful discussions.
This work was supported by a grant from the Foundation of University of Medicine and Dentistry of New Jersey.
REFERENCES
- 1.Boysen, A., E. Ellehauge, B. Julien, and L. Søgaard-Andersen. 2002. The DevT protein stimulates synthesis of FruA, a signal transduction protein required for fruiting body morphogenesis in Myxococcus xanthus. J. Bacteriol. 184:1540-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downard, J. S., S.-H. Kim, and K.-S. Kil. 1988. Localization of the cis-acting regulatory DNA sequences of the Myxococcus xanthus tps and ops genes. J. Bacteriol. 170:4931-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dworkin, M. 1996. Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 60:70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dworkin, M., and D. Kaiser. 1993. Myxobacteria II. American Society for Microbiology, Washington, D.C.
- 5.Ellehauge, E., M. Nørregaard-Madsen, and L. Søgaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol. Microbiol. 30:807-817. [DOI] [PubMed] [Google Scholar]
- 6.Hagen, T. J., and L. J. Shimkets. 1990. Nucleotide sequence and transcriptional products of the csg locus of Myxococcus xanthus. J. Bacteriol. 172:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horiuchi, T., M. Taoka, T. Isobe, T. Komano, and S. Inouye. 2002. Role of fruA and csgA in gene expression during development of Myxococcus xanthus: analysis by two-dimensional gel electrophoresis. J. Biol. Chem. 277:26753-26760. [DOI] [PubMed] [Google Scholar]
- 8.Horiuchi, T., T. Akiyama, S. Inouye, and T. Komano. 2002. Analysis of dofA, a fruA-dependent developmental gene, and its homologue, dofB, in Myxococcus xanthus. J. Bacteriol. 184:6803-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inouye, M., S. Inouye, and D. R. Zusman. 1979. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inouye, S., T. Franceschini, and M. Inouye. 1983. Structural similarities between the development-specific protein S from a gram-negative bacterium, Myxococcus xanthus, and calmodulin. Proc. Natl. Acad. Sci. USA 80:6829-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser, D. 2004. Signaling in myxobacteria. Annu. Rev. Microbiol. 58:75-98. [DOI] [PubMed] [Google Scholar]
- 12.McCleary, W. R., M. J. McBride, and D. R. Zusman. 1990. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J. Bacteriol. 172:4877-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1996. FruA, a putative transcriptional factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22:757-767. [DOI] [PubMed] [Google Scholar]
- 14.Shimkets, L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 15.Søgaard-Andersen, L., and D. Kaiser. 1996. C factor, a cell-surface-associated intercellular signaling protein, stimulates the Frz signal transduction system in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 93:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Søgaard-Andersen, L., M. Overgaard, S. Lobedanz, E. Ellehauge, L. Jelsbak, and A. A. Rasmussen. 2003. Coupling gene expression and multicellular morphogenesis during fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 48:1-8. [DOI] [PubMed] [Google Scholar]
- 17.Spiegelman, G. B., T. H. Bird, and V. Voon. 1995. Transcription regulation by the Bacillus subtilis response regulator Spo0A, p. 159-179. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 18.Stellwag, E., J. M. Fink, and J. Zissler. 1985. Physical characterization of the genome of the Myxococcus xanthus bacteriophage MX-8. Mol. Gen. Genet. 199:123-132. [DOI] [PubMed] [Google Scholar]
- 19.Ueki, T., and S. Inouye. 2003. Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100:8782-8787. [DOI] [PMC free article] [PubMed] [Google Scholar]