Abstract
Alginate is an industrially widely used polysaccharide produced by brown seaweeds and as an exopolysaccharide by bacteria belonging to the genera Pseudomonas and Azotobacter. The polymer is composed of the two sugar monomers mannuronic acid and guluronic acid (G), and in all these bacteria the genes encoding 12 of the proteins essential for synthesis of the polymer are clustered in the genome. Interestingly, 1 of the 12 proteins is an alginate lyase (AlgL), which is able to degrade the polymer down to short oligouronides. The reason why this lyase is associated with the biosynthetic complex is not clear, but in this paper we show that the complete lack of AlgL activity in Pseudomonas fluorescens in the presence of high levels of alginate synthesis is toxic to the cells. This toxicity increased with the level of alginate synthesis. Furthermore, alginate synthesis became reduced in the absence of AlgL, and the polymers contained much less G residues than in the wild-type polymer. To explain these results and other data previously reported in the literature, we propose that the main biological function of AlgL is to degrade alginates that fail to become exported out of the cell and thereby become stranded in the periplasmic space. At high levels of alginate synthesis in the absence of AlgL, such stranded polymers may accumulate in the periplasm to such an extent that the integrity of the cell is lost, leading to the observed toxic effects.
Alginates are linear polysaccharides composed of the two sugar monomers, β-d-mannuronic acid (M) and its C5-epimer α-l-guluronic acid (G). In contrast to most other such polymers, the alginate monomers are not distributed in repeating units, and it has been found that both the fractional contents and sequential distributions of M and G vary depending on the source of the polymer (11). Historically, the biochemistry and genetics of bacterial alginate synthesis were mainly studied in the opportunistic human pathogen Pseudomonas aeruginosa, and the reason for this is related to the problems this organism and its alginates create in the serious disease cystic fibrosis (21).
In more recent years, alginate synthesis has been studied experimentally in several other bacteria (15), and the organization of the genes involved in the process are also available from many genome sequencing projects. Based on this information, it appears that the genes in all cases are organized similarly and that the biochemistry of the process is presumably similar in different bacteria. Starting from fructose-6-phosphate, the precursor for polymer formation, GDP-mannuronic acid, is formed through four catalytic steps in the cytoplasm. Polymerization takes place by proteins located in the cytoplasmic membrane, and the polymer is then first exported into the periplasmic space and finally through the outer membrane and into the extracellular environment. As the polymer passes through the periplasmic space it is being acetylated at the O-2 and/or O-3 positions (14, 34, 39), and some of the M residues are being converted to G by a C5-epimerase designated AlgG (13, 18, 29). In Azotobacter vinelandii, further epimerization takes place extracellularly through the action of a family of structurally related and modular type mannuronan C5-epimerases, the AlgE family (8, 10, 34). This type of enzyme does not generally appear to be encoded in the genomes of pseudomonads, but one exception to this rule has recently been found (2). Of particular interest for this paper is that there is also an alginate lyase (AlgL) in the periplasm that potentially can depolymerize the polymers produced (9, 27, 28, 32), but its exact biological function is not clear (see below).
The genes directly involved in alginate synthesis (with the exception of algC) in P. aeruginosa, and possibly in all other pseudomonads, are organized in an operon (5) controlled by one single promoter which is regulated in a complex way. The gene encoding AlgC (16, 41) is located elsewhere in the genome, and its gene product is involved in sugar metabolism, catalyzing the formation of mannose-1-phosphate from mannose-6-phosphate. Due to the operon organization of most of the alginate genes in Pseudomonas, knockout mutants in the operon may potentially exert polar effects on downstream genes, and this has been relevant in some studies of AlgL function (4). It has been observed that an algL null mutant of mucoid P. aeruginosa appeared nonmucoid on agar medium (mucoidy is associated with high levels of alginate production). However, these authors found that mucoidy in the mutant could be restored by separately expressing another gene (algA) (17), which is located downstream of algL in the operon. They therefore concluded that the lack of mucoidy in the algL null mutant was due to a polar effect on algA expression and that AlgL therefore was not absolutely needed for alginate synthesis. They instead speculated that AlgL might function by cleaving the alginate chains during export (controlling chain length), by releasing the alginates from the surface or by producing oligouronides that might function as primers for alginate synthesis, thereby stimulating the quantities of polymer produced. In a later paper by Monday and Schiller (26), results of similar types of studies indicated that AlgL is an essential component of normal alginate synthesis (the mucoid phenotype), and these findings were recently further substantiated by Albrecht and Schiller (1). Somewhat conflicting with these results, a similar A. vinelandii AlgL mutant was recently reported to produce reduced, but considerable, amounts of alginate (37).
In our group, we have been interested in producing high-molecular-weight mannuronan (lacking G residues) for our studies of in vitro epimerization of alginates (38). In these studies, we have used the Pseudomonas fluorescens strain Pf20118, an algG-deficient derivative of the alginate overproducer Pf201 (18, 19). As a part of the experiments, we have also tried to modify AlgL activity, and here we show that this activity is critical for cell survival in the presence of high levels of alginate synthesis. Based on this and other results previously reported in the literature, we propose that the main biological function of AlgL may be to clear the periplasm of a subfraction of alginate molecules that fail to become exported out of the periplasm and into the extracellular environment during normal alginate synthesis.
MATERIALS AND METHODS
Growth of bacteria.
Bacterial strains and plasmids used in this study are listed in Table 1. For routine cloning and strain construction, Escherichia coli strains were grown in L broth (per liter, 10 g tryptone, 5 g of yeast extract, and 5 g of NaCl) or on L agar at 37°C. P. fluorescens was routinely grown in L broth, on L agar, or on Pseudomonas isolation agar (PIA; Difco). Matings between E. coli (strains S17.1 and S17.1λpir) and P. fluorescens were performed at 30°C on L agar, and selection of transconjugants were done on PIA with appropriate antibiotics. E. coli S17.1 and S17.1λpir were used as donors for conjugation of pMG48 and pCNB111luc derivatives, respectively. Production of P. fluorescens alginate and growth experiments were performed in liquid PIA medium (shake flasks) or PM5 medium (fermentors) (18). The PM5 medium was supplemented with 60 g/liter fructose, and the cultures in growth experiments were inoculated with 2% of an overnight stationary-phase culture. Otherwise, the growth conditions were as described previously (18). Antibiotics were present at the following concentrations (μg/ml): ampicillin, 100 to 200; kanamycin, 40; tetracycline, 12.5 (E. coli) and 30 (P. fluorescens). When used, m-toluate was added to a final concentration of 1 mM unless otherwise indicated. In liquid cultures, m-toluate was added 2 h after inoculation.
TABLE 1.
Plasmids and strains used in this work
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17.1 | RP4 2-Tc::Mu-Km::Tn7 pro res mod+ | 33 |
| S17.1(λpir) | λpirn lysogen of strain S17.1 | 7 |
| P. fluorescens | ||
| NCIMB10525 | Nonmucoid P. fluorescens wild type | NCIMB |
| Pf201 | Mucoid P. fluorescens mutant derived from NCIMB 10525 | 19 |
| Pf20118 | Mannuronan-producing derivative of Pf201, algG(R408C) | 18 |
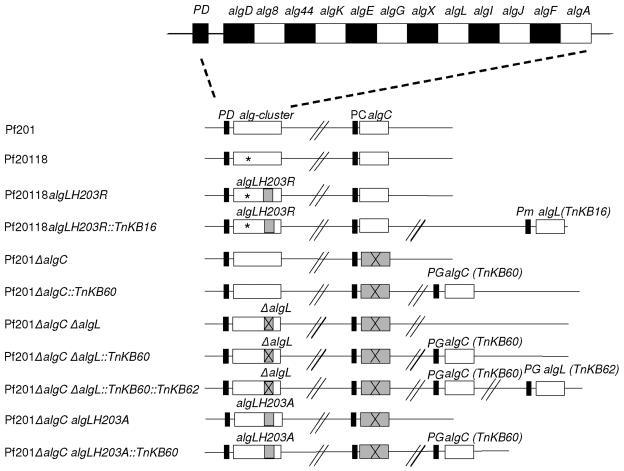
| Pf20118ΔalgL | Derivative of Pf20118 containing ΔalgL from pMG69 | This work |
| Pf20118algLH203R | Derivative of Pf20118 with the algL(H203R) point mutation from pMG70 | This work |
| Pf20118algLH203R::TnKB16 | Derivative of Pf20118algLH203R with transposon TnKB16 from pKB16 | This work |
| Pf201ΔalgC | Pf201 derivative with the ΔalgC deletion from pKB22 | This work |
| Pf201ΔalgC::TnKB60 | Derivative of Pf201ΔalgC with transposon TnKB60 from pKB60 | This work |
| Pf201ΔalgCΔalgL | Pf201ΔalgC with the ΔalgL from pMG69 | This work |
| Pf201ΔalgCΔalgL::TnKB60 | Derivative of Pf201ΔalgCΔalgL with the TnKB60 transposon from pKB60 | This work |
| Pf201ΔalgCΔalgL::TnKB60::TnKB62 | Derivative of Pf201ΔalgCΔalgL::TnKB60 with the TnKB62 transposon from pKB62 | This work |
| Pf201ΔalgCalgLH203A | Pf201ΔalgC with the algL(H203A) point mutation from pMG92 | This work |
| Pf201ΔalgCalgLH203A::TnKB60 | Pf201ΔalgCalgLH203A with the TnKB60 transposon from pKB60 | This work |
| Plasmids | ||
| pLitmus28 | ColE1 replicon; Apr. | New England Biolabs Inc. |
| pUC7Tc | Derivative of pUC7 in which Tcr gene from RK2 is inserted | 3 |
| pLitmus28Tc | Litmus28(BamHI) with an insertion of a 2.3-kb fragment (BamHI) containing tetA and tetR from pUC7Tc; Apr Tcr. | This work |
| pCNB111luc | oriR6K mobRP4, pUT/mini-Tn5 xylS/Pm luc Apr Kmr. | 40 |
| pMG26 | High-copy-number cloning vector encoding alg′ GXLI from P. fluorescens NCIMB 10525; Apr. | 18 |
| pMG48 | RK2-based gene replacement vector; Apr Tcr. | 18 |
| pMG66 | Derivative of pMG26 in which a NruI site was introduced in position 660 of algL. The primer pair algL-NruI-1 and algL-NruI-2 was used for site-specific mutagenesis; Apr. | This work |
| pMG67 | Derivative of pMG26 in which the algL(H203R) mutation was generated by site-specific mutagenesis, using the primer pair algLH203R1 and algLH203R2; Apr. | This work |
| pMG68 | Derivative of pMG66 from which a 0.4-kb NruI fragment was deleted, resulting in an in-frame deletion in algL; Apr. | This work |
| pMG69 | Derivative of NsiI-NotI-restricted pMG48 with insertion of a 2.9-kb fragment from pMG68 (PstI-NotI) encoding ΔalgL; Apr Tcr. | This work |
| pMG70 | Derivative of NsiI-NotI-restricted pMG48 with insertion of a 2.9-kb fragment from pMG67 (PstI-NotI) encoding algL(H203R); Apr, Tcr. | This work |
| pMG91 | Derivative of pMG26 in which the algL(H203A) mutation was generated by site-specific mutagenesis, using the primer pair algLH203A1 and algLH203A2; Apr. | This work |
| pMG92 | Derivative of NsiI-NotI-digested pMG48 with an insertion of a 2.9-kb fragment from pMG91 (PstI-NsiI) encoding algL(H203A); Apr Tcr. | This work |
| pJBphOx-271 | RK2-based expression vector harboring the Pm/xylS-regulated promoter system and the trfA271C copy up mutation; Apr. | 35 |
| pMC2 | Derivative of pKB10 in which algG was replaced with a 1.4-kb PCR-amplified NdeI-NotI fragment containing algC PCR amplified from P. fluorescens NCIMB 10525 total DNA. Primers PfalgCNdeIfor and PfalgCNotlrev were used for amplification; Apr Kmr. | This work |
| pKB10 | Suicide vector encoding a mini-Tn5 transposon with algG under Pm control; Apr Kmr. | 18 |
| pKB16 | Derivative of NdeI-NotI-restricted pCNB111luc in which luc was replaced with a 1.6-kb PCR-amplified fragment (NdeI-NotI) encoding algL. pMG26 was used as PCR template and PfalgLNdeI and M13/pUC reverse primer were used for amplification. Encodes the transposon TnKB16 with algL under the control of Pm; Apr Kmr. | This work |
| pKB20 | Derivative of pJBphOx-271 in which a 1.4-kb EcoRI-XbaI fragment from pMC2 encoding algC was inserted; Apr. | This work |
| pKB21 | Derivative of pKB20 from which a 0.5-kb EcoRI-PmlI fragment was deleted, creating an in-frame deletion in algC; Apr. | This work |
| pKB22 | Derivative of NsiI-NcoI-restricted pMG48 in which a 0.9-kb NsiI-NcoI fragment containing ΔalgC from pKB21 was inserted; Apr Tcr. | This work |
| pKB60 | Derivative of pKD23 in which algL was replaced with a 1.4-kb NdeI-NotI fragment from pMC2 containing algC. Encodes the transposon TnKB60 with algC under the control of PmG5; Apr Kmr. | This work |
| pKB62 | Derivative of pKD23 from which a 1.7-kb NcoI-AvrII fragment encoding the Kmr gene was replaced with a 2.6-kb fragment (AfllII-AvrII) encoding tetA and tetR from Litmus28Tc. Encodes the transposon TnKB62 with algL under the control of PmG5; Apr Tcr. | This work |
| pKD20 | pKB10 (SacI-NotI) with an insertion of a 1-kb PCR fragment (PstI-SacI) encoding Kmr (primers KanSacI and KanPstI) and a 1.8-kb PCR fragment (PstI-NotI) containing the PmG5 promoter and xylS (primers XylSPstI and XylSNotI) both using pHH100-G5 as template; Apr Kmr. | This work |
| pKD23 | Derivative of pKD20(NdeI-NotI) in which a 1.6-kb PCR-amplified fragment (NdeI-NotI) containing algL was inserted. pMG26 was used as PCR template and PfalgLNdeI and M13/pUC reverse primer were used for amplification; Apr Kmr. | This work |
| pHH100-G5 | RK2-based plasmid containing a mutant Pm promoter designated PmG5; Kmr. | 19 |
Standard techniques.
Plasmid isolations, enzymatic manipulations of DNA, agarose gel electrophoresis, and other routine DNA manipulations were performed according to the methods of Sambrook and Russell (31). A QIAquick gel extraction kit and a QIAquick PCR purification kit (QIAGEN) were used for DNA purifications from agarose gels and enzymatic reactions, respectively. Transformations of E. coli were performed according to a Northwest Fisheries Science Center protocol (available at http://micro.nwfsc.noaa.gov/protocols/rbcl.html). PCR for cloning and allele identification was performed using an Expand high-fidelity PCR system (Boehringer Mannheim) or the Pfx-polymerase-Pfx PCR system (Gibco-BRL). For site-specific mutagenesis, a QuickChange site-directed mutagenesis kit (Stratagene) was used. After cloning by PCR and site-specific mutagenesis, the DNA was verified by sequencing using a BigDye Terminator v1.1 cycle kit (Applied Biosystems). The sequences of the oligonucleotides used in this work are available upon request.
Construction of P. fluorescens mutants and insertion of transposons.
Derivatives of pMG48 were used as tools for generating algC and algL mutants by homologous recombination procedures, as previously described (18). To generate an algC deletion mutant, about 40% of algC in plasmid pKB20 was deleted in frame, and the flanking genomic DNA was cloned into suicide vector pMG48, generating pKB22. This internal deletion in algC corresponds to a deletion of amino acid residues 158 to 332 in the protein. pKB22 was transferred by conjugation into strain Pf201, and cells with a chromosomally integrated plasmid were selected on PIA medium containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and tetracycline. Transconjugants were grown in a series of overnight liquid cultures in the absence of tetracycline to allow excision of the vector by a second homologous recombination event, resulting in tetracycline-sensitive cells (18). The algC deletion mutant was verified by PCR analyses, using primers PfalgCNdeIfor and PfalgCNotIrev. For construction of the algL deletion mutants, about 36% of algL in pMG26 were deleted in frame, and the truncated gene was subsequently cloned into pMG48, generating pMG69. This algL deletion corresponds to a deletion of amino acid residues 88 to 221 in the protein. Transconjugants were selected as described above and verified by PCR analyses with primers PfalgL-BspHI-pMG26 and PfalgLRev1 or PfAlgX1 and PfAlgI1. The algL(H203R) and algL(H203A) alleles were introduced to P. fluorescens via pMG48-based suicide vectors pMG70 and pMG92, respectively, as described above. The chromosomal insertions of these mutations were verified by digestion of the PCR-amplified algL allele with appropriate enzymes and by sequencing. For this purpose, restriction enzyme sites were introduced along with the H203 mutations: AgeI [algL(H203R)] and AflIII [algL(H203A)]. The primers PfalgL-BspHI-pMG26 and PfalgLRev1 were used for the PCR amplifications. Transposons were inserted into the P. fluorescens genome by conjugation, using the appropriate pCNB111luc derivatives as donor plasmids. The derivatives used were pKB60 (TnKB60), pKB16 (TnKB16), and pKB62 (TnKB62). algC was expressed from the mutant PmG5 promoter in TnKB60, while algL was expressed either from Pm wild type (TnKB16) or from PmG5 (TnKB62).
Isolation and analysis of alginate.
Bacterial cells in the cultures were removed by centrifugation, and the alginate was deacetylated directly in the supernatant by mild alkaline treatment (12). When necessary, culture samples were diluted 10-fold in 0.2 M NaCl to reduce viscosity. Quantification of alginate was performed by using M-specific lyase (abalone) and G-specific lyase (Klebsiella aerogenes) as described by Østgaard (26a). Isolation and preparation of alginate samples for 1H-nuclear magnetic resonance (NMR) and size exclusion chromatography with on-line multi-angle laser light scattering (SEC-MALLS) analyses was performed as described previously (18). The 1H-NMR spectra were obtained using a Bruker 400-MHz spectrometer. Integration of the spectra and further calculations and assignment of peaks were performed as described by Grasdalen (22). Alginate molecular mass determinations were performed using SEC-MALLS, as described by Christensen et al. (6). Calculations of molecular mass distributions and averages were based on an exponential fit of log M versus elution volume.
Measurements of alginate lyase activity.
Cell samples for measurement of intracellular alginate lyase activity were collected by centrifugation, and the pellets were frozen, thawed, and resuspended in buffer (Tris-HCl [50 mM], NaCl [0.25 M]; pH 7.5) to an optical density at 660 nm (OD660) between 3 and 10. Proteins were extracted from the cells by addition of B-PER II (bacterial protein extraction reagent; PIERCE) using the manufacturer's protocol. The lyase activities in these extracts were determined by measuring the degradation rate of mannuronan by using an Ubbelohde capillary viscometer as described previously (18).
RESULTS AND DISCUSSION
Initial characterization of the effects of algL mutations on alginate biosynthesis in strain Pf20118.
An algL in-frame deletion (ΔalgL) and a point mutation (H203A) were introduced in strain Pf20118, and the resulting mutants were found to be nonmucoid on agar medium, did not produce alginate in liquid cultures, and could not be complemented by wild-type AlgL. A corresponding point mutation has previously been shown to inactivate AlgL from Pseudomonas syringae (28). Interestingly, when this amino acid was instead substituted by the more similar (to histidine) amino acid arginine, it was found that the resulting mutant (Pf20118algLH203R; Fig. 1) was mucoid on agar medium and produced alginate in similar yields as Pf201 and Pf20118 in fermentor-grown cultures. No alginate lyase activity was detectable after expression of algL(H203R) in E. coli or in extracts of Pf20118algLH203R, while it was readily detected in extracts from Pf20118. Wild-type AlgL activity could also easily be measured in E. coli.
FIG. 1.
Genetic maps of Pf201 derivatives used in this study. Mutated genes relevant for this study are shaded in gray, and deletions are marked with black crosses. Double slashes indicate a compressed part of the chromosome. The type of algL mutation is specified above each mutation. The algG(R408C) mutation in Pf20118 is indicated with an asterisk. Vertically aligned boxes correspond to the same genes. Their identity is indicated on top of the column or above each box. Black boxes indicate the promoter of the gene(s) to the right. The type of promoter is specified above each box. PD, the algD promoter; PC, the algC promoter; PG, the PmG5 promoter; Pm, the Pm promoter. alg-cluster, the alginate biosynthesis gene cluster.
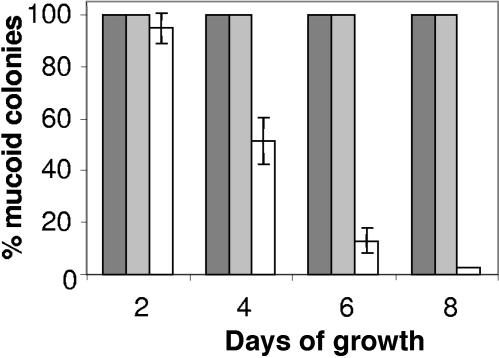
In the studies of strain Pf20118algLH203R, we often observed nonmucoid colonies after plating on agar medium, while this was not seen for the parental strain, Pf20118. To characterize this apparent instability more quantitatively, mucoid Pf20118algLH203R was grown in three parallel liquid cultures, and after 2 days, a small fraction of each culture was used for reinoculation (2%) of a new culture. This culture was again left for further growth for 2 more days, and the procedure was repeated two more times such that the cells were incubated for a total of 8 days. At the end of each 2-day interval, cells were plated on agar medium and inspected for the presence of nonmucoid colonies (Fig. 2). As can be seen, nonmucoid derivatives were already present in the first culture of the mutant (12%), and after 8 days of incubation, the vast majority of the cells were nonmucoid (97%). A Pf20118algLH203R derivative with wild-type algL expressed from a transposon (Pf20118algLH203R::TnKB16; Fig. 1) and strain Pf20118 served as controls in these experiments. In the former strain, wild-type algL was expressed from the Pm promoter, induced by addition of m-toluate (25 μM) to the culture medium. All three strains were therefore grown in the presence of this inducer. No nonmucoid colonies were observed from the control strains, clearly confirming that the observed instability of alginate production was caused by the algL(H203R) mutation.
FIG. 2.
Stability of alginate production in Pf20118algLH203R. Pf20118 (dark gray bars), Pf20118algLH203R (white bars), and Pf20118algLH203R::TnKB16 (light gray bars) were grown in liquid cultures (shake flasks) and reinoculated in fresh medium (2%) every second day. The percentage of mucoid colonies was determined by plating of samples of each culture on agar medium (PIA). The error bars indicate the standard deviations of three parallel cultures.
One possible interpretation of the results described above is that the complete absence of AlgL is toxic to the cells in the presence of alginate synthesis. This would imply that all mutants leading to complete AlgL inactivation could only survive if alginate synthesis is simultaneously turned off by secondary mutations in genes that would turn off synthesis of the polymer. This could involve genes in the alg operon, algC, or regulatory genes, and spontaneous mutations affecting alginate synthesis are well known to occur in the more extensively studied species P. aeruginosa (20, 25). The hypothesis explains why deletion mutants and certain point mutations [like algL(H203A)] are nonmucoid and cannot be complemented by the algL wild-type gene, and the phenotype of strain Pf20118algLH203R might accordingly be the result of a low but undetectable AlgL activity.
Construction of a Pf201 derivative in which alginate synthesis can be turned on and off in the absence of AlgL.
To substantiate the hypothesis described above and to investigate the biological function of AlgL in more detail, we decided to switch to strain Pf201, since it would allow us also to study possible effects of AlgL deficiency on the epimerization process. In order to directly observe the proposed toxicity caused by lack of AlgL, we constructed a strain in which alginate synthesis could be induced in the absence of AlgL. This was done by first constructing an in-frame deletion in algC, generating strain Pf201ΔalgC (Fig. 1). This strain was, as expected, found to be nonmucoid on agar medium and produced virtually no alginate (data not shown). A Pf201ΔalgC derivative with inducible alginate synthesis was then obtained by inserting algC under the control of the PmG5 promoter into the Pf201ΔalgC chromosome, resulting in strain Pf201ΔalgC::TnKB60 (Fig. 1). The PmG5 promoter is a mutant derivative of Pm wild type and was used because it provides lower background expression in the absence of inducer than Pm wild type in P. fluorescens (19). The vast majority of the resulting colonies were found to be mucoid on agar medium in the presence of inducer, even at low concentrations, like 25 μM. In liquid cultures, the production was similar to that of the parental strain (Pf201). In contrast, in the absence of inducer the colonies were nonmucoid, and the cells produced very little alginate in liquid cultures (data not shown).
An in-frame deletion in algL was then constructed in Pf201ΔalgC, as described for Pf20118 above, generating strain Pf201ΔalgCΔalgL (Fig. 1). Wild-type algC was introduced by inserting transposon TnKB60, generating strain Pf201ΔalgCΔalgL::TnKB60 (Fig. 1). The inducer of PmG5 was kept absent at all stages in the construction procedures to avoid potential toxic effects caused by alginate biosynthesis in the absence of algL. To ensure that the machinery for alginate biosynthesis in uninduced Pf201ΔalgCΔalgL::TnKB60 was still intact, we inserted a wild-type copy of algL under the control of PmG5 (transposon TnKB62) into its chromosome, resulting in strain Pf201ΔalgCΔalgL::TnKB60::TnKB62 (Fig. 1). This strain was found to be nonmucoid in the absence of inducer, while its colonies appeared similar to Pf201 (mucoid) in the presence of inducer. Pf201ΔalgCΔalgL::TnKB60 could therefore be used to study the potential toxic effects caused by activating alginate synthesis in the complete absence of AlgL activity.
Induction of alginate synthesis in the absence of AlgL activity leads to cell death.
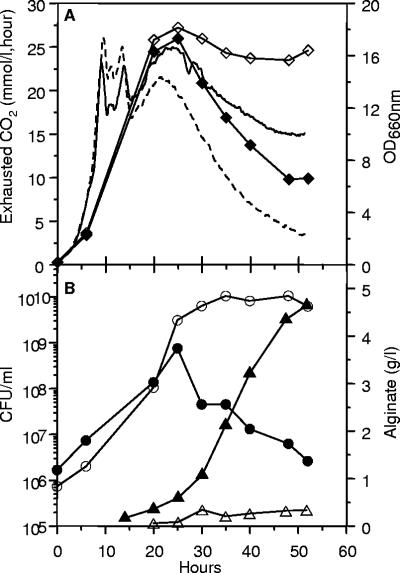
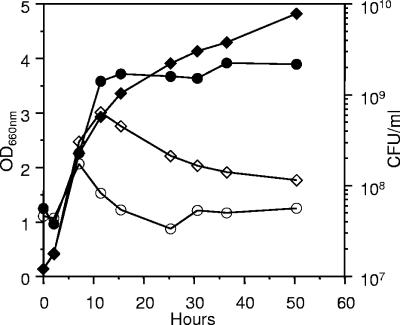
Strain Pf201ΔalgCΔalgL::TnKB60 was grown in liquid culture (fermentor) in the absence and presence of inducer and CO2 evolution, culture turbidity, and viable counts were monitored (Fig. 3). CO2 evolved similarly in both cultures near up to an OD660 of 12 to 16, but upon prolonged incubation CO2 production was reduced, as expected, since metabolic activities must obviously slow down in stationary phase. Interestingly, however, the reduction in gas evolution was much more drastic in the induced culture. This effect was even more pronounced in the OD660 measurements (Fig. 3A), as the density of the uninduced culture remained reasonably constant upon prolonged incubation, while in the induced culture the OD660 value dropped to about half after 50 h.
FIG. 3.
The responses to algC induction in fermentor-grown Pf201ΔalgCΔalgL::TnKB60. Open symbols, uninduced; closed symbols, induced. OD600, ⧫ and ⋄; alginate production, ▴ and ▵; CFU/ml, • and ○. Dotted line, evolved CO2, induced culture; continuous line, evolved CO2, uninduced culture. The experiment has been carried out two times with essentially the same result, and in each experiment duplicate or more samples were used to determine alginate production and CFU values. The mean values for each point are shown in the figure.
The drop in OD660 might mean that extensive cell lysis was taking place in the presence of induction of alginate synthesis, and this hypothesis was supported by the viable count determinations (Fig. 3B). In the absence of inducer, the CFU leveled off and remained constant after about 30 h, while in the presence of inducer, the viable counts dropped drastically after about 25 h, and after 50 h less than 1% of the cells were able to form colonies on agar medium, even in the absence of inducer.
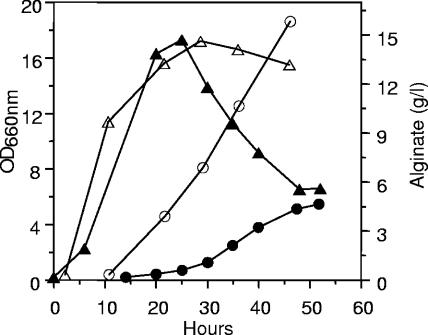
Alginate synthesis was also monitored in these experiments, and as expected, virtually no alginate was made in the absence of inducer (Fig. 3B). In the presence of inducer, alginate was produced, but the amounts of polymer accumulated (about 4.5 g/liter) were lower than normally observed for Pf201. We therefore carried out a separate experiment in which we specifically compared the level of alginate synthesis in Pf201ΔalgCΔalgL::TnKB60 with that of Pf201ΔalgC::TnKB60 (Fig. 4). This experiment clearly showed that the lack of AlgL caused reduced levels of alginate synthesis also before toxicity was observed as reduced growth rate and drop in optical density (Fig. 3A).
FIG. 4.
Growth and alginate production of strains Pf201ΔalgC::TnKB60 (open symbols) and Pf201ΔalgCΔalgL::TnKB60 (filled symbols). OD660, ▴ and ▵; alginate production, • and ○. The strains were grown in fermentors in the presence of m-toluate. Reproducibility was ensured as described in the legend to Fig. 3.
Alginate polymer formation has previously been shown to depend on the presence of the periplasmic AlgG, AlgK, and AlgX proteins, and based on the phenotypes of the corresponding mutants it may be hypothesized that these proteins are parts of a protein complex involved in alginate biosynthesis (18, 24, 30). Since AlgL is periplasmic, it appeared possible that this protein is also a part of such a proposed complex. If this is the case, there might be a difference in the phenotypes of ΔalgL mutants, compared to corresponding point mutants in the same gene. In other words, AlgL might have a structural role in addition to its lyase activity. To investigate this, we constructed a strain with inducible alginate production in an algL(H203A) background. The algL(H203A) mutation was assumed to be catalytically equivalent to the ΔalgL mutation with respect to lyase activity (see above for experiments in Pf20118). The algL(H203A) allele was first introduced into algL wild type in strain Pf201ΔalgC, generating Pf201ΔalgCalgLH203A. To obtain inducible alginate synthesis in this strain, TnKB60 was then inserted, resulting in Pf201ΔalgCalgLH203A::TnKB60 (Fig. 1). Cell growth and viable counts were then analyzed in liquid cultures under uninduced and induced conditions, as for the corresponding ΔalgL mutant. The results of this experiment (Fig. 5) were essentially the same as for strain Pf201ΔalgCΔalgL::TnKB60. Cell growth under uninduced and induced conditions proceeded similarly up to the beginning of the transition to stationary phase, after which the optical density in the induced culture dropped. This effect also corresponded well with a drop in viable counts. No drop in optical density or CFU was observed under uninduced conditions. Based on the assumption that the structure of AlgL(H203A) is similar to wild-type AlgL, these results clearly show that loss of AlgL lyase activity alone is sufficient to cause toxicity during alginate production.
FIG. 5.
The responses of algC induction on growth and cell viability in Pf201ΔalgCalgLH203A::TnKB60. Open symbols, induced; closed symbols, uninduced. OD660, ⧫ and ⋄; CFU/ml, • and ○. The strain was grown in shake flasks, and the experiment was carried out three times with similar results. The graph shows the results obtained in one of the three experiments, and each CFU value represents the mean of three samples.
The toxic effect caused by lack of AlgL during alginate synthesis correlates with the algC induction level.
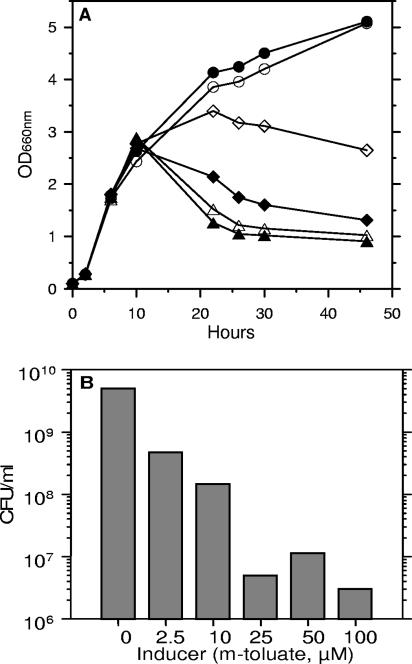
Strain Pf201ΔalgCΔalgL::TnKB60 did not appear to be affected in its growth properties in the absence of AlgL as long as algC expression was not induced. The significance of the amounts of alginate produced could be studied directly since the Pm promoter system controlling algC expression has the important advantage that the expression level of genes cloned downstream of it can be fine-tuned by varying the inducer concentration (40). As can be seen from Fig. 6A, there is no drop in OD660 at late stages of growth in the presence of a very low inducer concentration (2.5 μM). In the presence of 10 μM inducer, the cell density also did not drop much, but it leveled off at a much lower value than in the absence of inducer or at 2.5 μM of it. At higher concentrations (25 to 100 μM) the OD660 values dropped, consistent with the results shown in Fig. 3. Analyses of cell viability (Fig. 6B) confirmed these results, by showing that it dropped as algC was more induced, also demonstrating a significant cell death (about 90%) even at only 2.5 μM inducer. At 100 μM inducer, less than one in a thousand of the cells had survived. This could potentially explain the somewhat conflicting observations reported in the literature (see the introduction), since the strains used presumably produce different amounts of alginate.
FIG. 6.
The effect of the level of the inducer concentration on growth and cell viability of Pf201Δ algCΔalgL::TnKB60. (A) Growth (OD660). Uninduced, ○; 2.5 μM, •; 10 μM, ⋄; 25 μM, ⧫; 50 μM, ▵; 100 μM, ▴. (B) CFU/ml after 30 h of growth. Cells were grown in shake flasks. The experiment was carried out two times with similar results, and one of these experiments is shown in the graph. CFU values were determined as described in the legend to Fig. 5.
The structures of the alginates produced in the absence of AlgL are different from those produced by the Pf201 wild type.
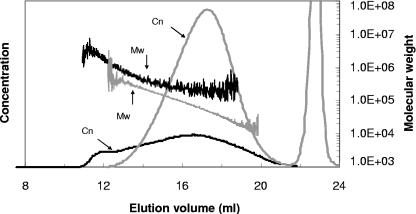
Since AlgL is capable of degrading alginates, one might expect that the molecules produced in its absence would have a very high molecular mass. Although bacterial exopolysaccharides are generally heterogeneous in their degree of polymerization, one can determine average molecular masses and size distributions. As shown in Fig. 7, the weight-average molecular weight (Mw) of the alginates produced by Pf201 is about 75,000, with a polydispersity (Mw/Mn) of 1.7. The amount of alginate with a molecular mass above 200 kDa was negligible. In contrast, strain Pf201ΔalgCΔalgL::TnKB60 produced an alginate with a much broader distribution, containing a significant amount of material (>40%) with a molecular mass above 200 kDa and at the same time low-molecular-mass polymers extending to about 10 kDa (extrapolated from the molecular mass calibration curve). The Mw was estimated to 330,000, which is significantly higher than for Pf201. Moreover, consistent with the broader molecular mass distribution, the polydispersity had increased to 3.2. The population of high-molecular-mass polymers was expected since AlgL activity no longer reduces the molecular mass. However, the production of low-molecular-mass polymers was not predicted but may be the result of interrupted polymerization.
FIG. 7.
Molecular mass distributions of alginates from Pf201 (gray) and Pf201ΔalgCΔalgL::TnKB60 (black) by SEC-MALLS. Cn, concentration signals (refractive index detector response [arbitrary units]); Mw, molecular mass calibration curves (calculated on-line from light scattering). The molecular mass corresponding to a point on the concentration curve is given by the intersection of a vertical projection from the concentration signals to the molecular mass calibration curves (molecular mass axis). The noise occurring at high elution volumes is due to weak scattering signals. The calibration curves (molecular mass curves) are shifted along the volume axis to different degrees due to dependence of the elution profile on the sample concentration. The peak at 23.5 ml represents the salt peak. The cells used for production of the alginates were grown in shake flasks.
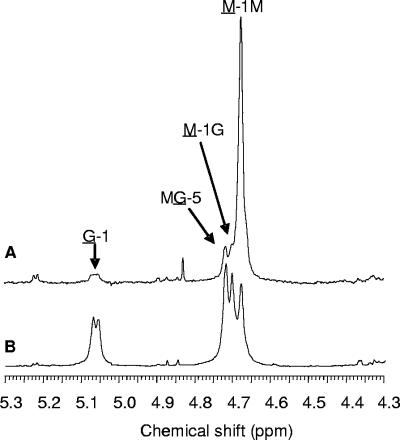
If the low-molecular-mass polymer populations originated from deleterious effects on the cytoplasmic membrane alginate polymerase, it seemed possible that other associated proteins could also be affected by the AlgL deficiency. AlgG is localized in the periplasm and, if the function of this protein is affected, one would predict that the monomer composition of the alginates produced in the absence of AlgL would be different from those of the wild type. The sugar composition of alginates can readily be analyzed by NMR spectroscopy, and Fig. 8 shows such spectra (1H-NMR) for alginates produced in the presence and absence of AlgL. Interestingly, the G content of alginate from induced Pf201ΔalgCΔalgL::TnKB60 grown in fermentor was only 3.5%, while Pf201 alginate normally contains about 30% G. As a control experiment, alginate produced by induced cells of Pf201ΔalgC::TnKB60 was found to contain wild-type levels of G (32%), excluding the possibility that the low G content was caused simply by effects of inducing alginate biosynthesis via algC. The G content in the alginate produced by strain Pf201ΔalgCalgLH203A::TnKB60 (induced conditions, shake flask cultures) was also found to be strongly reduced, showing that, as for the toxicity, the lack of lyase activity alone is sufficient to cause the disturbing effect on the epimerization process.
FIG. 8.
1H-NMR analysis of alginates produced in the presence and in the absence of AlgL. Alginate from Pf201ΔalgC::TnKB60 (B) contains Pf201 levels of G (32%), while alginate from Pf201ΔalgCΔalgL::TnKB60 (A) contains only 3.5% G. The residues from which each signal originates are underlined. The cells used for production of the alginates were grown in fermentors.
Since toxicity previously was shown to correlate with alginate production, we also investigated whether a similar effect could be observed for epimerization. Pf201ΔalgCΔalgL::TnKB60 was induced at three different levels (shake flasks), and the G contents of the resulting alginates were determined by 1H-NMR, as above. Induction at 0.0025 mM, 0.01 mM, and 1 mM resulted in alginate G contents of 11%, 8%, and 5%, respectively. These experiments therefore further substantiated the hypothesis that alginate production in the absence of AlgL activity is deleterious to periplasmic functions.
AlgL may be a scavenger of alginate strayed in the periplasmic space.
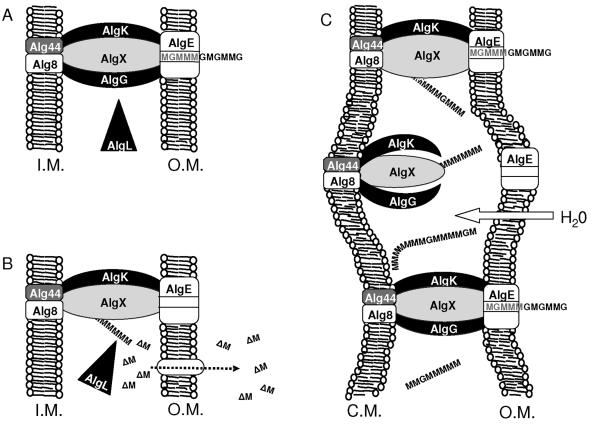
The results reported here clearly demonstrate that the lack of AlgL activity during alginate synthesis has a profound toxic effect on the cells and the alginate biosynthesis machinery. To explain the many phenotypes observed, we propose a relatively simple model which appears to be consistent with all the results reported in the literature and presented here. In this model, a major role of AlgL is to degrade alginates that are not exported to the extracellular environment but rather become stranded in the periplasm. We have previously proposed a multicomponent alginate biosynthesis and export scaffold spanning the inner membrane, the periplasmic space, and the outer membrane (18) in which AlgG, AlgK, and AlgX may be components. The loss of alginate polymer formation and secretion of unsaturated oligouronides in algK, algX, and algG mutants (18, 24, 30) indicate that the corresponding proteins probably normally protect the growing polymer chain from AlgL-mediated degradation, possibly as illustrated in Fig. 9A. There are presumably numerous such protein complexes in each cell at high levels of alginate synthesis, and it therefore seems likely that some of these complexes may not form properly. This might still allow the polymerization reaction in the inner membrane to occur, but it could lead to extrusion of the newly formed polymer into the periplasm. Such stranded molecules might then be degraded to short oligouronides by the periplasmic AlgL and passively leak out through pores in the outer membrane (Fig. 9B). This hypothesis explains the presence of low levels of oligouronides found in the culture medium of the alginate overproducer Pf201 (18). When components of the predicted scaffold are not present due to deletions in one of the corresponding genes, a considerable portion of all polymer molecules would become stranded in the periplasm. If AlgL is always present in surplus, this would still not create an immediate problem to the cells but rather lead to increased production of oligouronides, as observed for algK, algG, and algX mutants. The hypothesis that AlgL is always present in surplus is supported by the observations of the phenotype of the H203R mutation and by the fact that if alginate synthesis is increased by stimulation of alg operon expression, more AlgL would also be made since its gene is encoded in the operon. If, on the other hand, AlgL activity is missing, the polymer molecules would become permanently stranded in the periplasm. This can be predicted to lead to influx of water and swelling of the periplasmic space, possibly leading to the observed cell toxicity effects. The model therefore also explains why the toxicity becomes worse at higher levels of alginate production. The swelling of the periplasmic space might lead to disturbances in the functioning of the scaffold (Fig. 9C), causing reduced epimerization and interrupted polymerization relative to in a wild-type cell.
FIG. 9.
Model for the function of AlgL, AlgG, AlgK, and AlgX. I.M., inner membrane; O.M., outer membrane. (A) Alginate is polymerized in the inner membrane and AlgK, AlgG, and AlgX form an alginate transport scaffold that surrounds the growing polymer and thereby prevents alginate accumulations in the periplasmic space and protects the growing alginate chain from AlgL-mediated polymer degradation. (B) When one of the alginate scaffold components is absent (in this case AlgG), the alginate polymer leaks into the periplasmic space and becomes depolymerized to dimers (ΔM) by AlgL (scavenging). These dimers leak out of the periplasmic space via channels in the outer membrane. (C) Absence of AlgL leads to accumulation of alginate in the periplasmic space. Influx of water to the periplasm maintains osmotic pressure but causes swelling of the periplasmic compartment. This expansion of the periplasm is toxic to the cells and disturbs periplasmic functions (e.g., polymerization and epimerization of alginate). This is indicated with the changed structure of the AlgK-AlgG-AlgX scaffold.
The main advantage of the model proposed above is that one does not need to postulate that AlgL is a protein with multiple functions and that its lyase activity alone is sufficient to explain all the relevant observations reported here and elsewhere in the literature. The observation that alginate synthesis is low in the absence of AlgL also before toxicity is observed (Fig. 4) may be seen as an indication pointing to more than one function of AlgL. Although such a hypothesis cannot be excluded, we believe that an additional function is not needed to explain this particular observation. As long as the cells are growing, more periplasm is continuously being made, while after cell growth has stopped, the synthesis of new periplasm also comes to an end. Since alginate is synthesized at both stages of growth, one would therefore predict that the periplasmic accumulation of polymer becomes more extensive at later stages of growth. During exponential growth in the absence of AlgL, alginate synthesis may be disturbed due to a moderate level of periplasmic swelling, while no direct toxicity is generated. At late stages of growth, on the other hand, both toxicity and inefficient alginate synthesis are observed due to the more extensive accumulation of polymer in the periplasm.
We believe that the model proposed above also resolves some of the apparent inconsistencies reported in the literature, mainly because it explains why phenotypes of algL-deficient mutants are dependent on the amounts of polymer produced. If our hypothesis is correct, it means that AlgL represents a maintenance system that has evolved to take care of errors happening during biosynthesis. Such systems are already well known for other types of macromolecules (DNA, RNA, and proteins), while to our knowledge it has not been previously reported for polysaccharides.
Acknowledgments
This work was funded by a grant from the Norwegian Research Council and by FMC Biopolymers AS.
We are very grateful to Ann-Sissel Ulset (NTNU) for performing the alginate molecular mass determinations (SEC-MALLS) and Wenche Iren Strand (NTNU) for performing the 1H-NMR analyses.
REFERENCES
- 1.Albrecht, M. T., and N. L. Schiller. 2005. Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 187:3869-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjerkan, T. M., C. L. Bender, H. Ertesvåg, F. Drabløs, M. K. Fakhr, L. A. Preston, G. Skjåk-Bræk, and S. Valla. 2004. The Pseudomonas syringae genome encodes a combined mannuronan C-5-epimerase and O-acetyl hydrolase, which strongly enhances the predicted gel-forming properties of alginates. J. Biol. Chem. 279:28920-28929. [DOI] [PubMed] [Google Scholar]
- 3.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. J. Bacteriol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, A., M. Ghosh, T. B. May, D. Shinabarger, R. Keogh, and A. M. Chakrabarty. 1993. Sequence of the algL gene of Pseudomonas aeruginosa and purification of its alginate lyase product. Gene 131:1-8. [DOI] [PubMed] [Google Scholar]
- 5.Chitnis, C. E., and D. E. Ohman. 1993. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8:583-593. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, B. E., A. S. Ulset, M. U. Beer, B. E. Knuckles, D. L. Williams, M. L. Fishman, H. K. Chau, and P. J. Wood. 2001. Macromolecular characterisation of three barley beta-glucan standards by size-exclusion chromatography combined with light scattering and viscometry: an inter-laboratory study. Carbohydr. Polymers 45:11-22. [Google Scholar]
- 7.de Lorenzo, V., I. Cases, M. Herrero, and K. N. Timmis. 1993. Early and late responses of TOL promoters to pathway inducers: identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J. Bacteriol. 175:6902-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ertesvåg, H., B. Doseth, B. Larsen, G. Skjåk-Bræk, and S. Valla. 1994. Cloning and expression of an Azotobacter vinelandii mannuronan C-5-epimerase gene. J. Bacteriol. 176:2846-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ertesvåg, H., F. Erlien, G. Skjåk-Bræk, B. H. Rehm, and S. Valla. 1998. Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J. Bacteriol. 180:3779-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ertesvåg, H., H. K. Høidal, I. K. Hals, A. Rian, B. Doseth, and S. Valla. 1995. A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol. Microbiol. 16:719-731. [DOI] [PubMed] [Google Scholar]
- 11.Ertesvåg, H., and S. Valla. 1998. Biosynthesis and applications of alginates. Polymer Degrad. Stabil. 59:85-91. [Google Scholar]
- 12.Ertesvåg, H., and G. Skjåk-Bræk. 1999. Modification of alginate using mannuronan C-5-epimerases, p. 71-78. C. Bucke (ed.), Methods in biotechnology 10: carbohydrate bio/technology protocols. Humana Press, Inc., Totowa, N.J.
- 13.Franklin, M. J., C. E. Chitnis, P. Gacesa, A. Sonesson, D. C. White, and D. E. Ohman. 1994. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J. Bacteriol. 176:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin, M. J., and D. E. Ohman. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gacesa, P. 1998. Bacterial alginate biosynthesis—recent progress and future prospects. Microbiology 144:1133-1143. [DOI] [PubMed] [Google Scholar]
- 16.Gaona, G., C. Nuñez, J. B. Goldberg, A. S. Linford, R. Najera, M. Castañeda, J. Guzman, G. Espin, and G. Soberon-Chavez. 2004. Characterization of the Azotobacter vinelandii algC gene involved in alginate and lipopolysaccharide production. FEMS Microbiol. Lett. 238:199-206. [DOI] [PubMed] [Google Scholar]
- 17.Gill, J. F., V. Deretic, and A. M. Chakrabarty. 1986. Overproduction and assay of Pseudomonas aeruginosa phosphomannose isomerase. J. Bacteriol. 167:611-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gimmestad, M., H. Sletta, H. Ertesvåg, K. Bakkevig, S. Jain, S. J. Suh, G. Skjåk-Bræk, T. E. Ellingsen, D. E. Ohman, and S. Valla. 2003. The Pseudomonas fluorescens AlgG protein, but not its mannuronan C-5-epimerase activity, is needed for alginate polymer formation. J. Bacteriol. 185:3515-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gimmestad, M., H. Sletta, P. Karunakaran, K. Bakkevig, H. Ertesvåg, T. Ellingsen, G. Skjåk-Bræk, and S. Valla. February. 2004. Patent WO 2004/011628 A1. New mutant strains of Pseudomonas fluorescens and variants thereof, methods for their production, and uses thereof in alginate production.
- 20.Govan, J. R., J. A. M. Fyfe, and C. McMillan. 1979. The instability of mucoid Pseudomonas aeruginosa: fluctuation test and improved stability of the mucoid form in shaken culture. J. Gen. Microbiol. 110:229-232. [DOI] [PubMed] [Google Scholar]
- 21.Govan, J. R., and J. W. Nelson. 1992. Microbiology of lung infection in cystic fibrosis. Br. Med. Bull. 48:912-930. [DOI] [PubMed] [Google Scholar]
- 22.Grasdalen, H. 1983. High-field, H-1-NMR spectroscopy of alginate: sequential structure and linkage conformations. Carbohydr. Res. 118:255-260. [Google Scholar]
- 23.Jain, S., M. J. Franklin, H. Ertesvåg, S. Valla, and D. E. Ohman. 2003. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol. Microbiol. 47:1123-1133. [DOI] [PubMed] [Google Scholar]
- 24.Jain, S., and D. E. Ohman. 1998. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J. Bacteriol. 180:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. W. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 90:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monday, S. R., and N. L. Schiller. 1996. Alginate synthesis in Pseudomonas aeruginosa: the role of AlgL (alginate lyase) and AlgX. J. Bacteriol. 178:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Østgaard, K. 1992. Enzymatic microassay for the determination and characterization of alginates. Carbohydr. Polymers 19:51-59. [Google Scholar]
- 27.Pecina, A., A. Pascual, and A. Paneque. 1999. Cloning and expression of the algL gene, encoding the Azotobacter chroococcum alginate lyase: purification and characterization of the enzyme. J. Bacteriol. 181:1409-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preston, L. A., C. L. Bender, and N. L. Schiller. 2001. Analysis and expression of algL, which encodes alginate lyase in Pseudomonas syringae pv. syringae. DNA Sequence 12:455-461. [DOI] [PubMed] [Google Scholar]
- 29.Rehm, B. H., H. Ertesvåg, and S. Valla. 1996. A new Azotobacter vinelandii mannuronan C-5-epimerase gene (algG) is part of an alg gene cluster physically organized in a manner similar to that in Pseudomonas aeruginosa. J. Bacteriol. 178:5884-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robles-Price, A., T. Y. Wong, H. Sletta, S. Valla, and N. L. Schiller. 2004. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 186:7369-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schiller, N. L., S. R. Monday, C. M. Boyd, N. T. Keen, and D. E. Ohman. 1993. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J. Bacteriol. 175:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo engineering: transposon mutagenesis in gram negative bacteria. Biotechnology (New York) 1:784-791. [Google Scholar]
- 34.Skjåk-Bræk, G., H. Grasdalen, and B. Larsen. 1986. Monomer sequence and acetylation pattern in some bacterial alginates. Carbohydr. Res. 154:239-250. [DOI] [PubMed] [Google Scholar]
- 35.Sletta, H., A. Nedal, T. E. Aune, H. Hellebust, S. Hakvåg, R. Aune, T. E. Ellingsen, S. Valla, and T. Brautaset. 2004. Broad-host-range plasmid pJB658 can be used for industrial-level production of a secreted host-toxic single-chain antibody fragment in Escherichia coli. Appl. Environ. Microbiol. 70:7033-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svanem, B. I., G. Skjåk-Bræk, H. Ertesvåg, and S. Valla. 1999. Cloning and expression of three new Azotobacter vinelandii genes closely related to a previously described gene family encoding mannuronan C-5-epimerases. J. Bacteriol. 181:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trujillo-Roldan, M. A., S. Moreno, D. Segura, E. Galindo, and G. Espin. 2003. Alginate production by an Azotobacter vinelandii mutant unable to produce alginate lyase. Appl. Microbiol. Biotechnol. 60:733-737. [DOI] [PubMed] [Google Scholar]
- 38.Valla, S., J. Li, H. Ertesvåg, T. Barbeyron, and U. Lindahl. 2001. Hexuronyl C5-epimerases in alginate and glycosaminoglycan biosynthesis. Biochimie 83:819-830. [DOI] [PubMed] [Google Scholar]
- 39.Vazquez, A., S. Moreno, J. Guzman, A. Alvarado, and G. Espin. 1999. Transcriptional organization of the Azotobacter vinelandii algGXLVIFA genes: characterization of algF mutants. Gene 232:217-222. [DOI] [PubMed] [Google Scholar]
- 40.Winther-Larsen, H. C., K. D. Josefsen, T. Brautaset, and S. Valla. 2000. Parameters affecting gene expression from the Pm promoter in Gram-negative bacteria. Metab. Eng. 2:79-91. [DOI] [PubMed] [Google Scholar]
- 41.Zielinski, N. A., A. M. Chakrabarty, and A. Berry. 1991. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 266:9754-9763. [PubMed] [Google Scholar]