Abstract
Using fast Fourier transform (FFT) analysis, we previously observed that cells within Escherichia coli biofilm are organized in nonrandom or periodic spatial patterns (K. Agladze et al., J. Bacteriol. 185:5632-5638, 2003). Here, we developed a gravity displacement assay for examining cell adherence and used it to quantitatively monitor the formation of two distinct forms of cell attachment, temporary and permanent, during early biofilm development. Temporarily attached cells were mainly surface associated by a cell pole; permanent attachments were via the lateral cell surface. While temporary attachment precedes permanent attachment, both forms can coexist in a population. Exposure of attached cells to gravity liberated an unattached population capable of rapidly reassembling a new monolayer, composed of temporarily attached cells, and possessing periodicity. A csrA mutant, which forms biofilm more vigorously than its wild-type parent, exhibited an increased proportion of permanently attached cells and a form of attachment that was not apparent in the parent strain, permanent polar attachment. Nevertheless, it formed periodic attachment patterns. In contrast, biofilm mutants with altered lipopolysaccharide synthesis (waaG) exhibited increased cell-cell interactions, bypassed the polar attachment step, and produced FFT spectra characteristic of aperiodic cell distribution. Mutants lacking the polysaccharide adhesin β-1,6-N-acetyl-d-glucosamine (ΔpgaC) also exhibited aperiodic cell distribution, but without apparent cell-cell interactions, and were defective in forming permanent attachments. Thus, spatial periodicity of biofilm microstructure is genetically determined and evident during the formation of temporary cell surface attachments.
Surface-associated bacterial microbial communities known as biofilms have a profound impact on processes throughout the biosphere (5, 6), suggesting that mechanisms responsible for the design of their structure are likely to be of wide importance. Biofilm development includes stages of cell attachment, growth, differentiation, and dispersal (15, 21). In a previous study, we used fast Fourier transform (FFT) analysis to examine the cellular architecture or microstructure of Escherichia coli biofilm (1). These analyses revealed that a two-dimensional layer of attached cells exhibits distinct spatial periodicity, which persists within the body of the developing three-dimensional biofilm (1). This periodicity is formed independently of surface chemistry and is similar in wild-type strains and in a csrA mutant, which forms elevated amounts of biofilm (10, 11, 26, 27). Possible models for the origination of the spatial patterns were considered, including an activator-inhibitor or Turing model (1). Computer simulations have also suggested that an autoinhibitory process could guide the development of biofilm structure (4).
In E. coli K-12, attachment to abiotic surfaces and cell-cell adhesion depends on surface organelles and the adhesin poly-β-1,6-N-acetyl-d-glucosamine (PGA) (9, 17, 26, 27). Attachment is also affected by physical factors, including shear force and gravity. Although in some cases shear may enhance cell adhesion (25), we observed that flow tended to remove adhering E. coli cells and created anisotropy in the observed attachment patterns (1). Gravitational settling of bacterial cells facilitates attachment and is widely used for the primary inoculation of biofilm support surfaces (2). However, under static conditions or at low flow rates, when the bacterial cells are allowed to settle freely, it can be difficult to distinguish between attached versus settled cells.
Two major classes of bacterial cells are immediately apparent within a biofilm-forming population: unattached planktonic cells and attached cells. Studies dating back to the 1940s reveal that bacterial interaction with a surface often involves an initial reversible or temporary attachment, followed by irreversible or permanent attachment (reviewed in reference 24). Temporary attachments are reversible within a short period of time, within seconds or a few minutes, and are thought to afford the bacterium an opportunity to sample the environment before forming permanent attachments or releasing from the surface (discussed in reference 3). Permanently attached cells remain surface associated for at least tens of minutes and provide the foundation for further biofilm development. Although the molecular mechanisms responsible for these two forms of attachment remain to be defined, they were recently shown to be genetically distinguishable in Pseudomonas species. Mutants lacking lapA or sadB in Pseudomonas fluorescens and P. aeruginosa, respectively, were unable to undergo the transition from reversible to irreversible attachment (3, 8).
The attachment force should be different for temporary versus permanently attached cells. In principle, sheer force can be used to distinguish strength of attachment, e.g., in a flow cell. Although bulk flow rate is easily measured, the actual speed of the flow and thus the shear force may vary significantly within the chamber. Unlike shear force, gravitational field is homogeneous and affects all bacterial cells equally. With this idea, we developed a quantitative assay to distinguish cells according to their strength of attachment by monitoring their displacement by gravitational field.
The gravity displacement assay revealed two classes of surface attachment: temporary and permanent. As shown for Pseudomonas species (3, 8), temporary attachment was mainly by a cell pole, whereas permanent attachment was mainly by the lateral cell surface. A csrA mutant of E. coli (reviewed in reference 18) formed an increased proportion of permanently attached cells and a new subclass of cells that were permanently attached via a cell pole. The major role of CsrA in biofilm formation is posttranscriptional repression of pgaABCD expression, which is needed for PGA synthesis (28). Mutants lacking the glycosyltransferase that synthesizes PGA (ΔpgaC) still formed polar attachments but did not undergo the transition to permanent attachment and formed aperiodic or quasirandom attachment patterns. In contrast, polar attachments were not observed for lipopolysaccharide mutants (waaG), which formed permanently attached cells and cell clusters that exhibited quasirandom FFT patterns. Thus, waaG mutants appear to bypass the temporary attachment step in development.
Gravity displacement was reversible; a displaced population was able to rapidly reestablish temporary cell attachments possessing spatial periodicity. These findings suggest that the decision to remain temporarily attached or return to the planktonic phase is a pivotal step in E. coli K-12 biofilm formation, designed to promote the formation of spatial periodicity within the developing microstructure, and that PGA synthesis is integral to this process.
MATERIALS AND METHODS
Growth and media.
Bacteria (Table 1) were routinely grown in Luria-Bertani (LB) medium (13) overnight at 37°C with shaking. Such cultures were used to inoculate (1:100) biofilm cultures, which were incubated at ambient temperature (∼24°C) in colonization factor antigen (CFA) medium (per liter 10 g of Casamino Acids [Difco, Detroit, MI], 1.5 g of yeast extract [Difco], 50 mg of MgSO4, and 5 mg of MnCl2 [pH 7.4]) (12). For microscopy studies, sterile borosilicate coverslips were placed into a petri dish containing 25 ml of a freshly inoculated medium, and colonized slides were withdrawn at defined time intervals. Antibiotics were used as required at the following concentrations: ampicillin, 150 μg/ml; chloramphenicol, 10 μg/ml; kanamycin, 100 μg/ml; and spectinomycin, 100 μg/ml.
TABLE 1.
E. coli K-12 strains used in this study
| E. coli strain | Description or genotype | Source or reference |
|---|---|---|
| MG1655 | Prototrophic | Michael Cashel |
| TR1-5 | MG1655 csrA::kanR | 19 |
| TRXWMGΔC | TR1-5 ΔpgaC nonpolar deletion | 26 |
| DJ24 | TR1-5 ΔmotB uvrC279::Tn10 | 11 |
| DJ25 | DJ24 ΔfimB-H | 11 |
| 12E12-6 | DJ25 waaG::cam (+163)a | This study |
| F470 | Prototypical R1; R-LPS strain | 7 |
| CWG303 | F470 waaG::aacC1 nonpolar insertion | 28 |
| MC4100 | F− Δ(argF-lac)U169 rpsL relA flhD deoC ptsF rbsR | Thomas J. Silhavy |
| TR51 | MC4100 cpxR1::spc | Thomas J. Silhavy |
| WBS262 | MC4100 nlpE::spc | Thomas J. Silhavy |
The transposon insertion site is given relative to the waaG initiation codon.
Slide preparation and handling for microscopy.
After a defined period of time, a coverslip containing attached bacteria was removed from the incubation medium, immersed into a petri dish with sterile CFA medium, and gently rinsed for 30 s to remove unbound cells. A microscopic coverslip-slide was then assembled by placing the coverslip onto a sterile slide, separated by a Parafilm gasket (∼0.18 mm). All borosilicate coverslips and slides that were used to assess cell binding were pretreated with poly-l-lysine, which stabilizes cell-surface interactions but does not alter the cell attachment patterns or biofilm microstructure (1). The space between coverslip with the attached bacteria and the opposite slide contained a drop of sterile CFA medium. This arrangement allows the observation of cells attached to either glass surface, as well as unattached cells within the central space of the slide. Immediately after preparation, the slide was maintained so that the surface of the coverslip with the attached bacteria was kept facing up, i.e., the cells were on the bottom surface of the enclosed space. Within 1 to 2 min after the preparation of the slide, the cell attachment pattern was visually examined and photographed. Afterward, the slide was inverted, so that the glass surface with the attached bacteria was facing down, exposing cells to gravitational detachment. The pictures of cell distribution were taken immediately after inverting the slide and at measured time intervals thereafter. The number of attached cells was electronically counted and plotted versus time. As a rule, the counting was conducted two to four times in different fields of the slide, and the average value was used. This precaution was found to be more than adequate; variability was low for cell densities between 0.5 × 104 and 8 × 104/mm2, which was no more than 3 to 5%. At lower cell densities, statistical disorder increased variability. Error was also increased at higher cell densities because of the small distances between cells and cell overlap, causing the counting of individual cells to be less reliable, e.g., influenced by the settings of the software. A final consideration of this method is that the presence of the cell clusters will lead to underestimates of cell numbers.
Imaging.
The cells were observed under phase contrast with an Olympus BX60 microscope (Thornwood, NY). Images were recorded with a COHU-4910 charge-coupled device camera (COHU, Inc., Florence, KY) connected to a frame-grabber board (GMS500; Scion Corp., Frederick, MD), installed in a personal computer (PC). All image processing used Image Pro-Plus 4.5 software (MediaCybernetics, Silver Spring, MD). The PRIOR ProScan motorized microscopic stage was controlled by a PC and was used for aligning the sample as well as for focusing. The periodicity of the cell attachment pattern in a biofilm monolayer was analyzed by two-dimensional FFT as described previously (1).
Quantitative biofilm assay.
Biofilms were assayed by crystal violet staining, as described previously (11). Overnight cultures were diluted 1:100 into fresh medium and grown at 26°C in 96-well polystyrene microtiter plates. Bacterial growth was determined by measuring the absorbance at 630 nm. At least six replicas were conducted for each sample, and each experiment was performed at least twice. The results were calculated as averages and standard errors of two or more experiments.
Biofilm mutants.
The procedures for the isolation and genetic and molecular characterization of biofilm transposon mutants of strain DJ25, which contain random mini-Tn10cam insertions, have been described in detail (26). Briefly, strain DJ25 (MG1655 ΔfimB-H ΔmotB csrA) was infected with λNK1324, containing a mini-Tn10cam transposon, at a multiplicity of infection of 0.2 (12), and chloramphenicol-resistant colonies were isolated and evaluated for growth in planktonic shaking cultures and for biofilm formation by crystal violet staining of adherent cells. The genetic linkage between the biofilm phenotype and the transposon insertion was confirmed by P1vir transduction (13) back into the parent strain, DJ25; the wild-type E. coli K-12 strain MG1655; and/or its csrA mutant (TR1-5). Insertion sites were determined by arbitrarily primed PCR and DNA sequencing as described previously (26). P1vir transduction was also used to move the csrA::kan marker among strains.
RESULTS
Kinetics of cell detachment from a glass surface.
Previous publications reveal that, besides planktonic and sessile bacterial cells, a cell surface interaction referred to as “adsorption” or “reversible attachment” (discussed in reference 2) precedes cell incorporation into a biofilm structure. Our observations further indicated that in static cultures surface-associated cells consist of truly attached cells and loosely sedimented cells (data not shown). The latter cells were removed by gentle rinsing for a few seconds before slides were assembled for microscopic examination (see Materials and Methods).
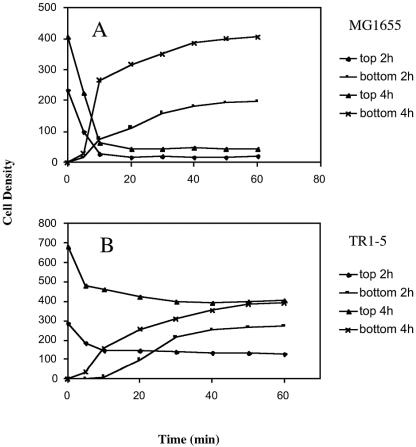
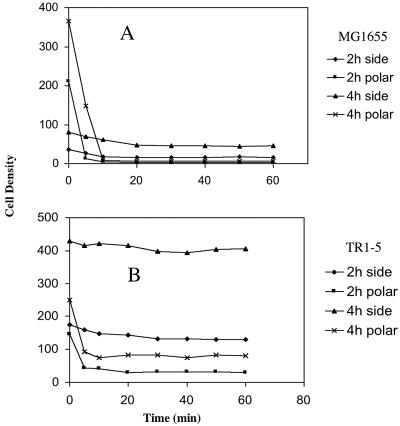
Exposure of surface-attached cells to the gravitational field by inverting the slide caused a portion of the cells to detach. Images taken immediately after inverting the slide and 30 min thereafter are shown in Fig. 1. The kinetics of detachment were monitored by collecting images over time and counting the number of cells remaining on the original attachment surface, now at the top of the assembly, during exposure to gravity. The accumulation of cells on the bottom surface of this slide assembly was simultaneously monitored. The wild-type strain MG1655 (Fig. 2A) and an isogenic high biofilm-producing csrA mutant TR1-5 (Fig. 2B) were both examined in this assay. In addition, cell detachment was examined after the biofilms were grown for 2 or 4 h in petri dishes prior to slide assembly. Within 10 min after the slide was inverted, ca. 90 and 82% of the population of wild-type cells detached from the 2- and 4-h slides, respectively. During that time, the interior space of the slide contained numerous, slowly precipitating cells (data not shown). After 20 min, the number of cells remaining at the upper (original attachment) surface was essentially constant, indicating that these cells were permanently attached. At the bottom surface of the slide assembly, the accumulation of bacteria continued for up to 40 to 50 min. The csrA mutant showed similar behavior, although the proportion of permanently attached cells was much higher, i.e., ca. 50 and 60% of the total for the 2- and 4-h slides, respectively (Fig. 2B). Comparison of total cell numbers for a given strain indicated that only a modest increase in cell number (≤25%) occurred during these experiments.
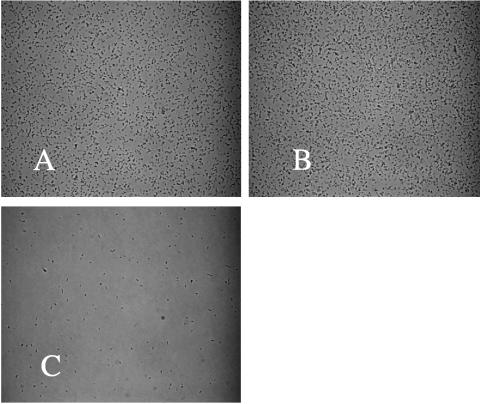
FIG. 1.
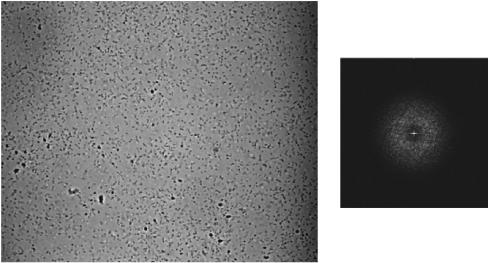
Appearance of bacterial attachments before and after inversion of the microscopic slide assembly. A culture (MC4100) was incubated for 4 h, and images of attached cells were captured before (A), immediately after (B), and 30 min after (C) inversion of the slide and exposure to gravitational displacement. The frame width of each image is 0.3 mm.
FIG. 2.
Bacterial cell density versus time on the top and bottom surfaces of microscopic slide assemblies. Wild-type E. coli strain MG1655 (A) and its isogenic csrA mutant, TR1-5 (B), were allowed to form biofilm for 2 or 4 h on coverslips in CFA medium, as described in Materials and Methods. A slide was assembled and inverted so that the coverslip with the attached bacteria was now facing down. Cell detachment from the coverslip and cell accumulation on the bottom surface of the slide assembly were monitored over time. Density values were calculated as the number of cells per square millimeter divided by 100.
Kinetics of permanent attachment.
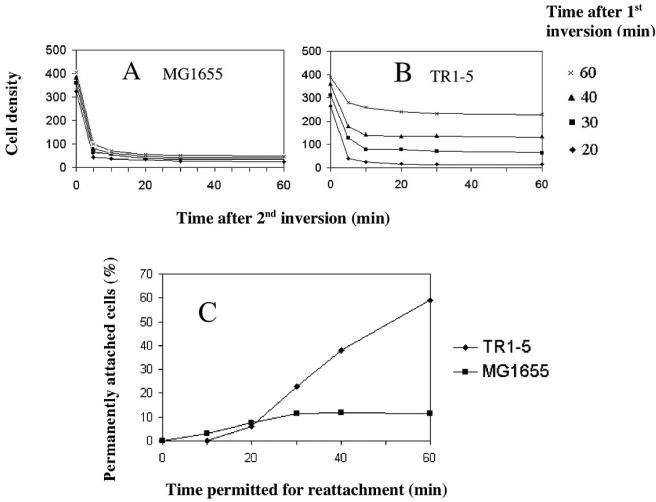
The experiment described above examined the proportion of cells that were temporarily or permanently attached at 2 or 4 h after inoculation of batch cultures. In order to examine systematically the time required for cells to attach permanently to a surface, the attachment process was monitored over time, within the slide assembly. For this experiment, a slide containing the rinsed cells was assembled and then inverted to expose attached cells to gravity, as before. The cells were allowed to detach and precipitate toward the bottom surface for 20, 30, 40, or 60 min, at which times cells on the bottom of the assembly were photographed. Thereafter, the slide was overturned a second time, and the cells that were now at the top of the assembly and exposed to gravity were monitored for detachment up to 1 h. The dynamics of the latter displacement process allowed us to determine the time required for the wild-type and csrA mutant strains to form temporary and permanent attachments (Fig. 3). For both strains, the majority of cells that reached the bottom surface of the slide within the first 20 min were either temporarily attached or had settled but not yet attached. More than 90% of these cells dissociated from the surface within 5 to 10 min. Thereafter, the number of bound cells remained relatively constant, a finding indicative of permanent attachment (Fig. 3A and B). Thus, ∼10% of the cells were permanently attached after 20 min of incubation. Permanent attachment in the wild-type strain increased only modestly beyond 20 min, accounting for ∼12% of the population at 60 min (Fig. 3A and C). In contrast, both the number and the proportion of permanently attached csrA mutant cells continued to increase throughout the experiment, reaching ∼60% of the population by 60 min (Fig. 3B and C).
FIG. 3.
Kinetics of permanent attachment. A series of coverslips were incubated for 4 h with MG1655 (A) or its isogenic csrA mutant, TR1-5 (B), and slides were assembled and inverted, as described in Fig. 2. Individual slides were inverted a second time at intervals of 20 to 60 min thereafter, as indicated, and the densities of bacterial cells attached to the upper surfaces of the slides were monitored over time. This allowed both the number of cells and the proportion of permanently attached cells to be determined at each time interval. (C) The proportion of permanently attached cells from panels A and B were plotted against the time that was permitted for the gravity-displaced cells to reattach. Cell density values were determined as in Fig. 2.
Gravity displacement of cells with polar versus lateral attachments.
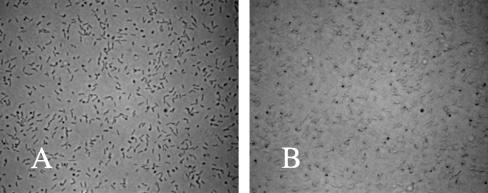
Visual examination of colonized coverslips revealed the presence of polar and laterally attached cells, which were readily distinguished because the former cells were seen to rotate about the attached pole whereas the latter were immobile (data not shown). Furthermore, it was possible to capture still images that could be used to assess cell orientation relative to the substrate and thereby distinguish these two cell types quantitatively. This was accomplished by focusing 1.5 μm below or above the attachment surface, depending upon whether the cells were attached to the top or bottom of the slide assembly, respectively. Cells attached to the upper surface of a slide assembly in which the focal plane coincides with the surface of the glass are shown in Fig. 4A. A micrograph of the same field, taken with the focal plane 1.5 μm below the glass, is shown in Fig. 4B. Polar attached cells are clearly seen in the latter image as the dark round spots (the focal plane cuts through them). Laterally attached cells are no longer in focus and become pale in the latter image. By selecting an appropriate gray level cutoff, it was possible to electronically distinguish images of laterally attached cells from the darker images of polar attached bacteria and thus to automatically count the two cell types. This approach gave satisfactory reproducibility (variability, ≤5%) for up to 3 × 104/mm2 cell density and somewhat lower reproducibility (variability, ≤10%) for up to 6 × 104/mm2. Microscopic observations of cells incubated for 4 h with coverslips in CFA medium showed that a substantial fraction of the attached cells were adhering by a pole: ca. 80% for MG1655 and ca. 40% of the population for its isogenic csrA mutant (Fig. 5). The number of polar and laterally attached cells versus time at the upper surface of the slide is shown for MG1655 (Fig. 5A) and its isogenic csrA mutant TR1-5 (Fig. 5B). From these data, it is apparent that under gravitational field, the number of polar attached cells of both strains decreases rapidly, whereas the number of laterally attached cells remains relatively constant. This indicates that temporarily attached cells are associated with the surface by a cell pole, while permanently attached cells are bound laterally. Interestingly, the csrA mutant exhibited a substantial subpopulation of cells that were permanently attached by a cell pole (Fig. 5B). This behavior appeared to be negligible in the wild-type strain.
FIG. 4.
Images of polar and laterally attached bacteria. Cells of TR1-5 (csrA) were grown and allowed to attach to a coverslip for 4 h. A slide was assembled containing the cells at the upper surface (see Materials and Methods). Images (×640 magnification) of these cells were taken at the surface of the coverslip (A) or at 1.5 μm below the surface (B). The frame width for each image is 0.2 mm.
FIG. 5.
Bacterial cells density versus time for polar and laterally attached bacteria. Analysis of the gravitational detachment of polar and laterally attached cells of MG1655 or its csrA mutant, TR1-5, grown for 2 or 4 h, was performed as described in Fig. 4 legend. Cell density values were determined as in Fig. 2.
These relationships were also supported by direct microscopic observations of single cell interactions with the surface, which revealed that detachment generally involved cells that had been attached by a pole (data not shown). Conversely, when cells of either strain were seen to reattach, they predominantly did so by a cell pole, as observed by their movement around an anchoring point (data not shown).
Correlation of temporary attachment with periodic cell density patterns.
The periodicity of the spatial patterns produced by attached bacteria was examined by FFT analysis of images. The spatial distribution of cells that attach to the bottom surface of the slide after detachment and sedimentation was nonrandom and, after 15 to 20 min of accumulation, the FFT spectrum showed distinct periodicity (Fig. 6), indicated by the symmetrical ring with a central bright spot (discussed in reference 1). Taking into account that 5 min after overturning the slide, almost no cells were observed on the bottom surface, the estimated time for the periodic pattern to form was ca. 10 to 15 min. The images of cells that were collected at the latter times appeared uniformly shorter than those observed in longer incubations because the majority of the cells were in a relatively upright position on the surface. In conjunction with observations on the kinetics of permanent attachment (Fig. 3), this confirms that these cells were associated with the surface by reversible polar attachments. These findings reveal that cell attachments exhibit spatial periodicity during the temporary attachment phase of biofilm formation.
FIG. 6.
Density pattern of cells detached from a coverslip by gravity and allowed to settle at the bottom of the slide assembly. Strain MG1655 was inoculated and allowed to attach to a coverslip for 4 h. A slide was prepared and inverted, and cells were permitted to detach, settle, and reattach for 20 min before they were photographed (×400 magnification). The corresponding two-dimensional FFT spectrum of the image is shown on the right.
Altered attachment behavior of biofilm mutants.
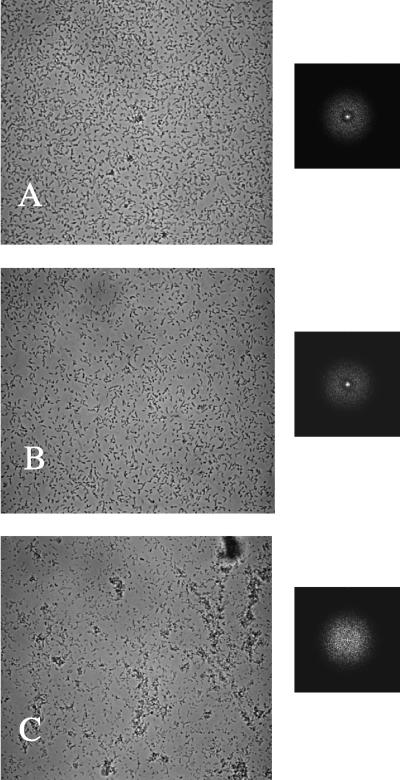
While the csrA mutant forms periodic cell density patterns (1), examination of additional biofilm mutants revealed two classes that exhibited distinct behavior in FFT analysis. In a random transposon mutagenesis screen, we isolated E. coli biofilm mutants that were altered in genes affecting LPS biosynthesis (X. Wang and T. Romeo, unpublished observations), as was previously reported for Salmonella enterica LPS mutants (14). The FFT spectrum of one of these mutants, 12E12-6, its isogenic parent DJ24 (ΔmotB ΔfimB-H csrA), and TR1-5 (csrA) are compared in Fig. 7. Strain 12E12-6 contains a transposon insertion in waaG, which encodes the glucosyltransferase that attaches glucose I to heptose II in the LPS core (28). Although strains TR1-5 and DJ24 both bound to the slide as single cells and exhibited spectra indicative of periodic cell distribution (Fig. 7A and B), 12E12-6 tended to form clusters of cells adhering to the microscope slide (Fig. 7C). Its FFT spectrum showed a uniform circular cloud, indicative of a quasirandom cell distribution (1). A well-studied nonpolar waaG mutant (28) behaved similarly in both csrA mutant and wild-type genetic backgrounds and formed cell clusters and an aperiodic cell distribution pattern as assessed by FFT (data not shown). All of the waaG mutants examined formed few or no polar attachments. A very small proportion of the population of waaG mutants was observed to detach during these experiments, although, because of cluster formation, accurate cell counting was not possible. The latter finding suggests that an increased proportion of the population was permanently attached in these strains.
FIG. 7.
Images of attached cells (×400 magnification) after 4 h of biofilm growth and corresponding FFT spectra. (A) TR1-5 (csrA); (B) DJ24 (csrA ΔfimB-H ΔmotB); (C) 12E12-6 (DJ24 waaG).
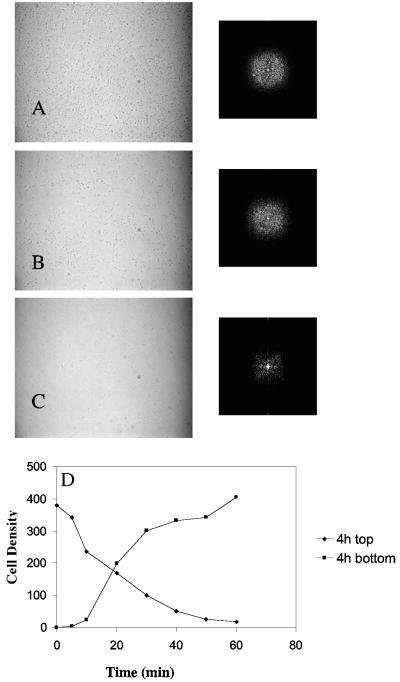
A distinct attachment behavior was observed in mutants altered in the production of the adhesin PGA. These mutants formed single cell attachments to poly-l-lysine-treated glass surfaces (Fig. 8 and data not shown) and to polystyrene (data not shown), although they were defective in adhering to untreated borosilicate glass (26). Analysis of the cell attachment patterns of a nonpolar pgaC mutant, TRXWMGΔC (ΔpgaC csrA), by FFT revealed no detectable periodicity, either initially or upon subjecting the strain to gravitational detachment for various times (Fig. 8). The PgaC gene product is a family-2 glycosyltransferase needed for PGA synthesis (26, 27). The pgaC mutant cells continuously detached from the surface throughout a period of an hour exposure to gravity, until very few cells remained (Fig. 8D). From the experiment depicted in Fig. 8, the initial density of cells was 380 × 102/mm2, while the initial density of polar attached cells was 360 × 102/mm2 (data not shown). Thus, almost 95% of the initial attachments were by a cell pole, a higher proportion than for its isogenic parent, TR1-5 (csrA), or MG1655 (Fig. 5). Therefore, these cells did not establish a permanently attached, adherent population. Upon continued incubation, pga mutants formed closely packed cells with little intervening spacing and yet failed to form the cell-cell interactions required to construct three dimensional biofilm (26; data not shown).
FIG. 8.
Cell attachment patterns and displacement kinetics of a ΔpgaC mutant of TR1-5. The culture was grown for 4 h in CFA medium at ambient (∼24°C) temperature, slides were assembled, and photographs were taken immediately (A), 20 min (B), and 60 min (C) later. FFT spectra are shown to the right of the corresponding images. Cell density versus time under gravitational field was calculated as in Fig. 2 and is shown for the top and bottom surfaces of the slide assembly in panel D.
cpxR and nlpE mutants.
The Cpx two-component signal transduction system and a Cpx-regulated lipoprotein, NlpE, play a role in surface sensing and regulation of gene expression in response to adhesion and have been proposed to affect mechanisms leading to stable surface adhesion (16, 20). Thus, we tested mutations in the genes for the response regulator, cpxR, and the lipoprotein, nlpE, on the attachment process in four strain backgrounds. When present in the original MC4100 strain, MG1655, or in csrA mutants of these two strains, neither mutation had an effect on biofilm formation in the crystal violet microtiter plate assay or spatial periodicity in FFT analysis (data not shown). Thus, under our experimental conditions, this surface sensing system did not appear to influence biofilm development.
DISCUSSION
The use of gravitational field allowed subpopulations of attached bacteria to be quantitatively monitored in a simple format, to our knowledge for the first time. Our primary reason for devising this gravitational displacement assay was to have a tool for studying processes involved in the generation of spatial periodicity of cells within the microstructure of E. coli biofilm. In addition, this approach should be useful for studying other attachment and/or detachment processes on surfaces amenable to microscopic imaging. This assay revealed that surface-attached E. coli cells can be physically separated into reversibly or temporarily attached versus permanently attached forms, which react differently with respect to persistent exposure to gravity (Fig. 2 and 3).
An important feature of temporary attachment is that it is mediated primarily by the cell pole (Fig. 5). Preferential localization of structures at the bacterial cell pole, i.e., polarity, is well known (22, 23). Thus, polar attachment might reflect polar localization of an adhesin, although direct evidence for this is lacking. The different strengths of temporary versus permanent attachments may result from the smaller area of contact of polar versus lateral attachments with the substrate. The finding that a csrA mutant is able to permanently attach by a cell pole is most likely due to its overproduction of the cell-bound adhesin, PGA (26). Although temporary attachment is reversible, it is nevertheless able to resist modest shear force, as evidenced by the polar attached cells that persist on coverslips after gentle rinsing (Fig. 4).
Although the present study provides a quantitative evaluation of cell populations undergoing reversible attachment and detachment, it cannot determine whether (i) the temporary attachment force is insufficient to resist gravity over time or (ii) temporarily attached cells occasionally detach and are removed by gravity or (iii) both of these processes contribute to the removal of cells. Because cells of early biofilms that are not exposed to gravity occasionally detach and reattach, at least some contribution must be from possibility ii. Detachment in the absence of gravitational force may involve Brownian motion, as well as forces contributed by the bacterial cell, e.g., motility or charge repulsion. Nevertheless, we found that flagellum-dependent motility, type I fimbriae, and cpx signal transduction were not needed for reversible attachment or generation of spatial periodicity (Fig. 7 and data not shown). It is possible that the experimental design of the gravitational displacement studies, i.e., examination of cells within a microscope slide assembly, might have had an influence on the attachment or detachment behavior during the relatively brief (≤1-h) time course of our experiments. However, cells that were allowed to detach under gravity and reform attachments within the slide assembly exhibited the characteristic periodic distribution patterns observed in batch culture (compare Fig. 6 and 7). Furthermore, E. coli attachment or detachment behavior appeared to be similar to that described for Pseudomonas species (3, 8).
The pgaC mutant of E. coli behaves similarly to lapA and sadB mutants of P. fluorescens and P. aeruginosa, respectively (3, 8), in that it fails to make the transition from temporary to permanent attachment. This finding strongly suggests that transition to permanent attachment is mediated by synthesis of the adhesin PGA in E. coli. The lapA and sadB genes encode a large protein that is loosely associated with the outer membrane protein and a cytoplasmic protein of unknown function, respectively. The underlying mechanism of this transition in the two Pseudomonas species and the precise roles played by LapA and SadB are still uncertain. Whether the biofilm matrices of the latter species exhibit spatial periodicity also remains to be determined.
We suggested previously that spatial periodicity might require the sensing of a chemical gradient produced by attached cells, which causes altered detachment frequency (1). Here, we found that a subpopulation of cells, detached by gravity, is able to reconstruct temporary attachments (Fig. 3) that exhibit spatial periodicity within <15 min of reaching the surface (Fig. 6). Thus, the process that generates periodicity in the attachment pattern must be ongoing within this time frame. Other researchers have concluded that temporary attachment affords cells an opportunity to sample the environment before choosing to take up residence (discussed in reference 3). We propose that environmental sampling may serve to determine proximity to other attached cells. Thus, permanent attachment might result from association with a surface long enough for the cell to sense that it is positioned appropriately within the developing biofilm matrix and produce an adhesin. We caution that neither the putative signals that might be sampled nor the relevant attachment and detachment mechanisms have been defined for any developing biofilm, let alone the intervening signal transduction pathway. These three kinds of information are needed for the sampling hypothesis to be validated.
Examination of biofilm mutants by FFT analysis revealed cell distribution patterns distinct from those of parental E. coli strains, providing the first direct evidence that these patterns are genetically based. Mutations in waaG, which result in a shortened LPS molecule (28), increased cell-cell interaction, decreased the formation of polar attachments, caused the loss of spatial periodicity, and stabilized the attachment of cells to the abiotic surface. Although it is possible that LPS might either physically or through a regulatory mechanism have an integral influence on the pattern-generating mechanism, a trivial explanation for this attachment behavior may be that increased cell-cell adhesion by this mutant and the presence of cell clusters overwhelms more-subtle interactions of the pattern-forming process.
Perhaps the more interesting attachment phenotype was exhibited by pga mutants, including a nonpolar deletion of pgaC, which lacks PGA (26, 27). Although PGA is not essential for polar attachment to a poly-l-lysine-treated glass surface, the ΔpgaC mutant nevertheless failed to form periodic cell density patterns and was defective in permanent attachment (Fig. 8). Furthermore, a csrA mutation, which derepresses PGA synthesis (27), causes increased lateral attachment, as well as permanent attachment via the cell pole (Fig. 2, 3, and 5). These findings confirm and extend our understanding of the crucial role played by PGA in E. coli attachment behavior.
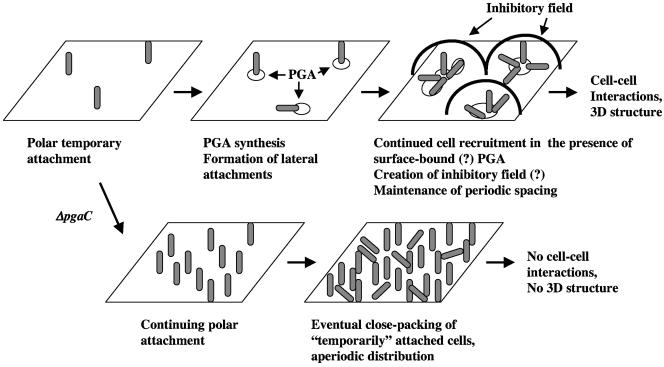
Taken together, our observations support the idea that PGA promotes stabilization of interactions necessary for the cell to undergo the transition from temporary to permanent attachment. However, the transition to permanent attachment alone does not appear to explain the origination of periodicity, since cells that are temporarily attached (Fig. 6) still exhibit periodicity. We suggest that regulated secretion of PGA and/or asymmetric distribution of this polymer on the abiotic surface during the temporary attachment step might account for its role in pattern formation. The later hypothesis is depicted within a working model for the origination of biofilm microstructure in Fig. 9. In this model, periodicity is generated by the interplay of the attachment activator, PGA, and a putative soluble inhibitor of attachment. PGA production permits the conversion from temporary to permanent attachment and is suggested to lead to recruitment of additional cells to the surface at close proximity to existing attachments. A critical density of closely attached cells permits the putative inhibitory signal to accumulate and generates an inhibitory field, which is sensed by newly attached cells. Within this inhibitory field, the cell attachment does not become permanent, perhaps due to inhibition of PGA synthesis in newly attached cell, unless the cell has encountered surface-associated PGA, which is only present very close to cells that have already attached. The inability to synthesize PGA eventually results in a monolayer of closely packed cells with an aperiodic distribution pattern. Because three-dimensional biofilms exhibit the characteristic periodic cell distribution patterns, the process that generates spatial periodicity appears to persist as the biofilm grows (1). We hope that the findings and hypotheses of the present study will foster an increased appreciation of the sophistication and complexity of the biofilm development process.
FIG. 9.
Working model for the initiation of E. coli K-12 biofilm microstructure. This model is described in the Discussion. Hypothetical steps of the model are indicated by “(?)”.
Acknowledgments
We thank T. J. Silhavy, B. Clarke, and C. Whitfield for strains; Rebecca Desplas for arbitrarily primed PCR analyses; and Carlos Goller and June Scott for reviewing the manuscript.
These studies were funded in part by the National Institutes of Health (GM066794). Kane Biotech, Inc., may develop applications related to the findings herein. T.R. serves as Chief Scientific Advisor for, owns equity in, and may receive royalties from this company. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict-of-interest policies.
REFERENCES
- 1.Agladze, K., D. Jackson, and T. Romeo. 2003. Periodicity of cell attachment patterns during Escherichia coli biofilm development. J. Bacteriol. 185:5632-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annachhatre, A. P., and S. M. Bhamidimarri. 1992. Microbial attachment and growth in fixed-film reactors: process startup considerations. Biotechnol. Adv. 10:69-91. [DOI] [PubMed] [Google Scholar]
- 3.Caiazza, N. C., and G. A. O'Toole. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J. Bacteriol. 186:4476-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, I., E. S. Gilbert, N. Eliashberg, and J. D. Kiesling. 2004. A three-dimensional, stochastic simulation of biofilm growth and transport-related factors that affect structure. Microbiology 149:2859-2871. [DOI] [PubMed] [Google Scholar]
- 5.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Heinrichs, D. E., J. A. Yethon, P. A. Amor, and C. Whitfield. 1998. The assembly system for the outer core portion of R1- and R4-type lipopolysaccharide of Escherichia coli: the R1 core-specific β-glucosyltransferase provides a novel attachment site for O-polysaccharides. J. Biol. Chem. 273:29497-29505. [DOI] [PubMed] [Google Scholar]
- 8.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905-918. [DOI] [PubMed] [Google Scholar]
- 9.Itoh, Y., X. Wang, B. J. Hinnebusch, J. F. Preston, III, and T. Romeo. 2005. Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson, D. W., J. W. Simecka, and T. Romeo. 2002. Catabolite repression of Escherichia coli biofilm formation. J. Bacteriol. 184:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 13.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 14.Mireles, J. R., II, A. Toguchi, and R. M. Harshey. 2001. Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol. 183:5848-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 16.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis, and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 18.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 19.Romeo, T., M. Gong, M. Y. Liu, and A.-M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175:4744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz, N., and T. J. Silhavy. 2005. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 8:122-126. [DOI] [PubMed] [Google Scholar]
- 21.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapiro, L., H. H. McAdams, and R. Losick. 2002. Generating and exploiting polarity in bacteria. Science 298:1942-1946. [DOI] [PubMed] [Google Scholar]
- 24.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 25.Thomas, W. E., E. Trintchina, M. Forero, V. Vogel, V., and E. V. Sokurenko. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell 109:913-923. [DOI] [PubMed] [Google Scholar]
- 26.Wang, X., A. K. Dubey, K. Suzuki, C. S. Baker, P. Babitzke, and T. Romeo. 2005. CsrA posttranscriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56:1648-1663. [DOI] [PubMed] [Google Scholar]
- 27.Wang, X., J. F. Preston, III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yethon, J. A., E. Vinogradov, M. B. Perry, and C. Whitfield. 2000. Mutation of the lipopolysaccharide core glycosyltransferase encoded by waaG destabilizes the outer membrane of Escherichia coli by interfering with core phosphorylation. J. Bacteriol. 182:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]