The Hda protein, a recently identified DnaA-related protein from Escherichia coli, is part of the AAA+ ATPase family known to be involved with various aspects of initiation of DNA replication in prokaryotes. We report here that overexpression of this membrane-associated protein inhibits multiplication, affects membrane permeability, and is also an unexpected initiator of the bacterial SOS response, which may represent a major new pathway for inducing DNA damage repair mechanisms
We recently identified a small membrane-associated protein (28.4 kDa) in Escherichia coli that is related to the DnaA host initiation protein and that affected the initiation of the broad-host-range plasmid RK2 (8). By interacting physically with the plasmid-encoded initiation protein (TrfA), it acted as a steric inhibitor of either or both of TrfA's two functions: cooperating with the DnaA protein (which is also required by RK2) to open the replication bubble and guiding the DnaB-DnaC complex into the open site (8, 9, 10). The protein is identical to the Hda protein (“homologous to DnaA” protein) that is responsible for controlling overinitiation in E. coli by accelerating the ability of the β-clamp subunit of DNA polymerase III to convert the active form of DnaA (ATP-bound DnaA) to its inactive form (ADP-bound DnaA) (1, 7, 11). Hda has a high sequence homology to the domain III ATPase region of DnaA (1, 7, 8, 13) and is important as an accessory component for initiation and, subsequently, replication in prokaryotes.
In further assessing the role of Hda protein in RK2 metabolism, we previously constructed a compatible plasmid that placed hda under the control of an inducible promoter and monitored the effects of increasing levels of Hda induction in vivo. Profound inhibitory effects on both maintenance and replication of RK2 were observed (8). Of additional interest, Hda overexpression also inhibited cell multiplication, with only a limited effect on optical density profiles. In this study, we investigated the basis for these inhibitory effects and determined that they involve induction of the SOS response system that may be actuated by perturbation of membrane integrity and/or permeability.
Effects of Hda on growth and viability of E. coli.
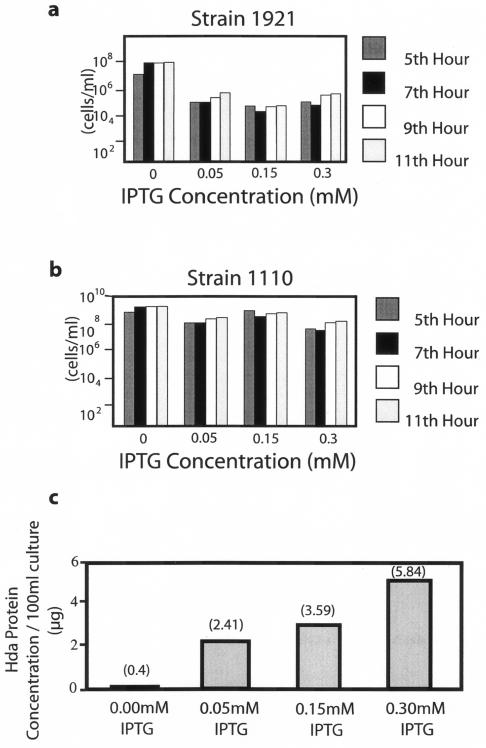
BL21(DE3)/pLysS, which contains an IPTG [isopropyl-(3-D-thiogalactoside)]-inducible T7 RNA polymerase gene (21), was transformed with plasmid pPK101 or the pET17B vector, as described by Hanahan (6), resulting in strain 1921 or 1110, respectively. pPK101 is a pET17B derivative (Novogen) that expresses a functional N-terminal T7 epitope-tagged Hda protein that was constructed previously (8). These two strains were grown to mid-logarithmic phase, induced with different IPTG concentrations, and viable cells were quantified. Whereas IPTG induction had a significant inhibitory effect on the viability of strain 1921 at all concentrations utilized (Fig. 1a), there was only a modest effect on the viability of the control 1110 strain (Fig. 1b). Of note, there was a threshold level of inhibition of cell viability by the Hda protein which could not be increased further despite increasing levels of Hda induction, as shown by Western blot analysis of sonicated cell extracts (Fig. 1c).
FIG. 1.
Viability of E. coli cultures containing different levels of Hda protein. Fresh colonies of strain 1921 (a) and 1110 (b) were inoculated into 20 ml of Luria-Bertani broth (10 g of tryptone, 5 g of yeast extract [Difco], and 10 g of NaCl/liter) containing selective antibiotics (50 to150 μg/ml of penicillin and 30 to 50 μg/ml of chloramphenicol) and shaken at 37°C. At mid-logarithmic-phase growth (−3 h), IPTG was added as indicated, and viable cell counts were performed by serial dilution and plating over the indicated period. The values shown are representative of numerous experiments. For the analysis of Hda levels in strain 1921, induced for 30 to 60 min at mid-log phase by various concentrations of IPTG (c), extracts were prepared from cell precipitates (100-ml cultures) by sonication with the Fisher 50 sonic dismembrator and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting. Relative Hda concentrations were determined by densitometry, using the ZUV transilluminator with Kodak IP image analysis software. Actual protein concentrations were determined by Bio-Rad, using a purified T7-tagged Hda standard (8).
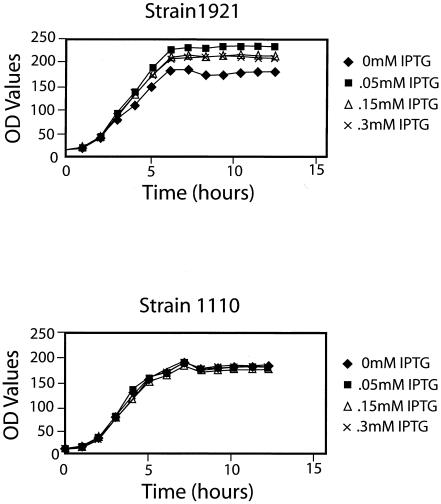
Optical density profiles of strains 1921 and 1110 were also determined during the above experiment. Despite the significant inhibition of viable cell counts for strain 1921 after IPTG induction, there was only a slight effect on its optical density profiles (Fig. 2). This result suggested that many of the nonviable cells produced during growth of this strain were not lysed, but rather, they continued to increase in cell number or length. Further support for this interpretation, at least indirectly, comes from results of Ryan-Arends and Weiss (19), who demonstrated that inhibition of cell division has little if any effect on gene expression.
FIG. 2.
Optical density profiles of E. coli cultures containing different levels of Hda protein. A Klett colorimeter (green filter) was used to collect optical density (OD values) profiles of the indicated cultures grown and induced with IPTG as described in the Fig. 1 legend. The slightly reduced optical density profile of strain 1921 without IPTG was not representative of additional experiments.
Microscopic observations of E. coli after overexpression of Hda.
In order to directly examine cell length, microscopic examination of the cultures was performed. The results show convincingly that after induction of strain 1921 with IPTG, the percentage of cells longer than 25 μm was much greater (up to fourfold greater) than that of strain 1110 at each time period (Table 1). The presence of some longer cells in the control may be due to a variety of factors, including imbalanced transcription caused by activation of the powerful T7 promoter, the presence of two antibiotics that are inactivated only over time, the IPTG inducer itself, and normal population dynamics of the culture. Nevertheless, we conclude that Hda overexpression causes excess cell filamentation. Although a variety of cell lengths under 25 μm were also observed in the culture overexpressing Hda protein, we did not attempt to quantify this data, since it would not provide additional mechanistic insight into this system. The linkage between Hda protein overexpression and cell filamentation is explored in more detail in the section on SOS response.
TABLE 1.
Percentage of filaments in E. coli strains containing different levels of Hda proteina
| Strain | Average % of cells > 25 μm in length at indicated time (h) after induction
|
||
|---|---|---|---|
| 3 | 5 | 8 | |
| 1921 | 29 | 47 | 84 |
| 1110 | 13 | 10 | 27 |
After strains 1921 and 1110 were grown to mid-logarithmic phase, 0.3 mM IPTG was added to the cultures, followed by further incubation as indicated. A loopful of each culture was spread on a glass slide at each time period, heat fixed, and stained with Huckers ammonium oxalate crystal violet. A Zeiss Axioplan light microscope was used to visualize the cells, and 6 to 10 microscopic fields were viewed to record the length of individual cells in micrometers after capturing the image with Kodak Elite II 200 ASA slide film and digitizing them on a Macintosh G4 computer with a Polaroid sprint scan attachment. A final magnification of 1250 X was used.
One possible mechanism for the observed decrease in cell viability may involve deleterious effects of Hda overexpression on the integrity of the cell membrane, which is the primary location of this protein (8). Microscopic examination of cells treated with the fluorescent nucleic acid stain SYTOX Green supported this hypothesis. Bacteria are normally impermeable to SYTOX Green (18), but there was a significant percentage of fluorescent nucleoids in strain 1921 after Hda overproduction (Table 2). It seems likely that these stained nucleoids are largely present within the nonviable cell population (Fig. 1), although as we have shown above (Fig. 2 and Table 1), such cells are still increasing in optical density due to cell elongation and are therefore metabolically active. Evaluation of macromolecular (DNA, RNA, and protein) syntheses (as assayed by incorporation of their respective precursors, [6-3H]thymidine, [5,6-3H]uracil, and [35S]methionine) also suggested that membrane integrity might be affected after Hda overexpression, but not in the expected way. Instead, such syntheses were enhanced in strain 1921 after induction at mid-log phase with 0.3 mM IPTG for 30 min (which, as shown in Fig. 1c, was sufficient to increase Hda synthesis significantly) but not in any of the three controls (1921 without induction, 1110 with or without induction). Moreover, the increase in macromolecular syntheses was evident by 5 min after addition of the precursors and was apparent throughout the 30-min time interval in which the analysis was performed (data not shown). These results are consistent with the timing of Hda overexpression as well as an altered membrane permeability, although other causes for the latter result need to be considered (see below).
TABLE 2.
Permeability of E. coli strains expressing different levels of Hda protein as indicated by the fluorescent dye SYTOX Greena
| Strain | Induced with IPTG | % of fluorescent nucleoids |
|---|---|---|
| 1921 | + | 40 |
| − | 0 | |
| 1110 | + | 4 |
| − | 0 |
After strains 1921 and 1110 were grown to mid-logarithmic phase, 0.3 mM IPTG was added to cultures and incubated an additional 3 h. A loopful of each culture was spread on a glass slide, heat fixed, stained with SYTOX Green (using methods recommended by the manufacturer, Molecular Probes), counterstained with methyl green, and visualized by fluorescence microscopy, using a Zeiss LSM 510 microscope with a 488-nm argon laser. Images were captured using Zeiss LSM 510 software and a Zeiss C-Apochromat 63× objective. Between 6 and 10 microscopic fields were examined for the percentage of cells containing fluorescent nucleoids. +, cultures with IPTG; −, cultures without IPTG.
Induction of the SOS response by overexpression of the Hda protein.
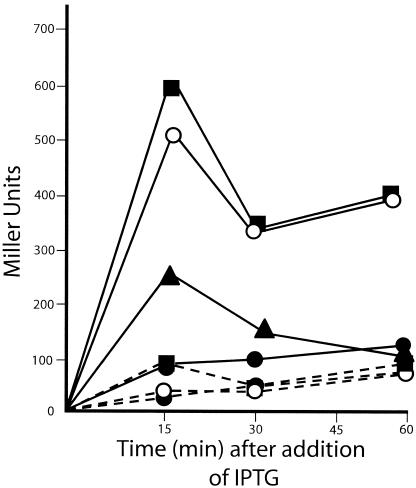
Three observations suggested to us that the SOS DNA repair system was being induced in E. coli after Hda overexpression. They include (i) an inhibition of cell division (Fig. 1), (ii) an increase in the amount of DNA synthesis per cell (based on increased [6-3H]thymidine incorporation per cell) (data not shown and Fig. 2), and (iii) an increase in cell length (Table 1). These features are classic characteristics of the SOS response (14, 15, 23, 24, 25). To directly test for this possibility, we constructed strains that contained a mucB-lacZ fusion. The mucB gene is homologous to the umuC gene product and has been identified as DNA polymerase V, an error-prone replicase that is induced by SOS (3, 23, 24). Strain BL26(DE3), which is isogenic to the BL21 strain except that it has a complete lac deletion (Δ lacU169) (21), was transformed with pSE200 (p15A compatibility group) containing the mucB-lacZ fusion (3) and pPK101 or pET17B to form strain 1922 or 1923, respectively. These two strains were cultured and induced with IPTG, and β-galactosidase activity assays were performed. The results show clearly that Hda overexpression dramatically increased the transcriptional activity of the S0S-responsive mucB promoter (Fig. 3). In fact, the induction was greater than that of the DNA-damaging agent mitomycin-C, used as a control in the experiment. Because expression of mucB occurred soon after IPTG induction, it appears that even modest levels of Hda overexpression rapidly trigger the SOS response. This induction explains our findings concerning inhibition of cell division and ensuing cell filamentation (caused by inhibition of FtsZ), and it also explains the observed increase in DNA synthesis (due to activation of error-prone repair polymerases like polymerase V). However, since increases in RNA and protein syntheses also occur after Hda overproduction, alterations in membrane permeability may be involved in this latter change as well.
FIG. 3.
Induction of the SOS Response by Hda overexpression. Strains 1922 (pSE200, pPK101) and 1923 (pSE200, pET17B) were grown until mid-logarithmic phase and induced (or not) with 0.5 mM or 1.0 mM IPTG. β-galactosidase activity was measured in Miller units as described by Maniatis et al. (12). As a positive control of the SOS response, mitomycin-C was added at a final concentration of 100 μg/ml to a culture of strain 1922 2 h prior to assay (14). All points represent an average of three separate experiments. Solid line, strain 1922; dashed line, strain 1923; •, no induction; ○, 0.5 mM IPTG; ▪, 1.0 mM IPTG; ▴, 100 μg/ml mitomycin-C.
The primary signal for production of the SOS response is the presence of single-stranded DNA (20), which is produced by either DNA damage or an inhibition of DNA synthesis at the replication fork (24). From our previous results with plasmid RK2, where a direct inhibition of membrane-associated DNA replication was observed after Hda overproduction (8), it seems highly likely that a similar inhibition also occurred for the bacterial chromosome. In effect, we are proposing that destabilization of membrane integrity by Hda overexpression (which was directly demonstrated by the results with SYTOX Green staining and the presumed increased permeability to nucleic acid and protein precursors) has damaged the ability of cells to carry out normal DNA replication leading to the SOS response. In this context, it is important to recognize that numerous studies, including our own, have shown that, in vivo, DNA replication is membrane associated (for reviews, see references 4, 5, and 22). Thus, both Hda and the DnaA initiator protein are membrane localized (8, 16), anionic phospholipids activate the initiation protein (26), and oriC itself binds to a subfraction of the inner membrane (2). Therefore, it is not surprising that when a protein such as Hda is overexpresssed in its membrane environment, profound physiological changes can result. Exactly what the linkage is between membrane perturbation and a defect in initiation control is speculative. In that regard it is of interest that overexpression of either DnaA or DnaB initiation proteins does not induce the SOS response (14). Hda may function more analogously to the Bacillus subtilis YabA protein, which has a function in initiation control similar to that of Hda (although they are unrelated) and interacts with certain transmembrane receptors that function in initiation at oriC (17). Recently, another analogous result has been reported in which β-lactam antibiotics that inhibit E. coli cell wall synthesis activate the SOS response through a two-component signal transduction system (14, 15). This inhibition transiently halts cell division, enabling the cells to survive lethal exposure to the antibiotics. Although Hda overexpression affects membrane permeability, cell division is also inhibited, and both the membrane and cell wall are integral components of the cell surface. Thus, our results fit into a pattern of SOS induction triggered by changes in the cell surface.
In conclusion, there are many questions that remain to be answered with this novel system, among them the conundrum of cause and effect. Do the various observations regarding Hda induction, inhibition of cell division, increases in cell length, apparent alteration of membrane permeability, and SOS induction make sense in terms of their timing? Although a logical case for this conclusion has been made here (e.g., induction of SOS is an early response to Hda overexpression and would trigger many of the observed changes in cell division, length of the cells, and DNA synthesis), it is unclear whether further kinetic analysis will enlighten the problem, since kinetics cannot in itself prove cause and effect. Another question concerns whether other membrane perturbants unrelated to Hda could induce the effects we have observed. Again, there is no simple answer to this question because, even if a number of such components tested did not elicit the same response, such results would be equivocal, since the membrane damage promoted by Hda overproduction may be rather specific. Clearly, a variety of approaches will be necessary to properly dissect these phenomena. Nevertheless, a novel pathway has been uncovered for inducing DNA damage repair mechanisms via the SOS system that implicates one important component of the cell surface, namely, the Hda protein within the cell membrane. Furthermore, the inducer comes from a class of proteins that plays accessory (nonessential) roles in prokaryotic DNA replication involved in initiation of DNA synthesis and, indirectly, elongation (1, 7, 8, 11, 17). Additional studies to elucidate the linkages and presumed coordination between these critical cellular processes are now possible.
Acknowledgments
We are grateful to Steven Sandler of the University of Massachusetts, Amherst for the gift of pSE200, Jeff Gilarde for his help with fluorescence microscopy and measurements of cell size, and Keith Weaver of the University of South Dakota for helpful discussions.
This work was supported in part by NIH grant GM0636134.
REFERENCES
- 1.Camara, J. E., K. Skarstad, and E. Crooke. 2003. Controlled initiation of chromosomal replication in Escherichia coli requires functional Hda protein J. Bacteriol. 185:3244-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakraborti, A., S. Gunji, N. Shakibai, J. Cubeddu, and L. Rothfield. 1992. Characterization of the Escherichia coli domain responsible for binding oriC DNA. J. Bacteriol. 174:7202-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elledge, S. J., and G. C. Walker. 1983. The muc genes of pKM101 are induced by DNA damage. J. Bacteriol. 135:1306-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Firshein, W., and P. D. Kim. 1997. Plasmid replication and partition in Escherichia coli: is the cell membrane the key? Mol. Microbiol. 23:1-10. [DOI] [PubMed] [Google Scholar]
- 5.Firshein, W. 2002. Prokaryotic DNA replication, p. 3-26. In U. N. Streips and R. E. Yasbin (ed.), Modern microbial genetics, 2nd ed. Wiley-Liss, Inc., New York, N.Y.
- 6.Hanahan, D. 1983. Studies on transformation of Escherichia coli by plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 7.Kato, J. I., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, P. D., T. Banack, D. M. Lerman, J. C. Tracy, J. Eltz Camara, E. Crooke, D. Oliver, and W. Firshein. 2003. Identification of a novel membrane-associated gene product that suppresses toxicity of a TrfA peptide from plasmid RK2 and its relationship to the DnaA host initiation protein. J. Bacteriol. 185:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konieczny, I., K. S. Doran, D. R. Helinski, and A. Blasina. 1997. Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J. Biol Chem. 272:20173-20178. [DOI] [PubMed] [Google Scholar]
- 10.Konieczny, I., and D. R. Helinski. 1997. Helicase delivery and activation of DnaA and TrfA proteins during the initiation of replication of the broad host range plasmid RK2. J. Biol. Chem. 272:33312-33318. [DOI] [PubMed] [Google Scholar]
- 11.Kurz, M., B. Dalrymple, G. Wijffels, and K. Kongsuwan. 2004. Interaction of the sliding clamp β-subunit and Hda, a DnaA-related protein. J. Bacteriol. 186:3508-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maniatis, T., E. F. Fritich, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Messer, W., and C. Wiegel. 1996. Initiation of chromosomal replication, p. 1579-1601. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 14.Miller, C., H. Ingmer, L. E. Thomsen, K. Skarstad, and S. N. Cohen. 2003. DpiA binding to the replication origin of Escherichia coli plasmids and chromosomes destabilizes plasmid inheritance and induces the bacterial SOS response. J. Bacteriol. 185:6025-6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, C., L. E. Thomsen, C. Gaggero, R. Mosseri, H. Ingmer, and S. N. Cohen. 2004. SOS response induction by β-lactams and bacterial defense against antibiotic lethality. Science 305:1629-1631. [DOI] [PubMed] [Google Scholar]
- 16.Newman, G., and E. Crooke. 2000. DnaA, the initiator of Escherichia coli chromosomal replication, is located at the cell membrane. J. Bacteriol. 182:2604-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noirot-Gross, M., E. Dervyn, L. J. Wu, P. Mervelet, J. Errington, and S. D. Ehrlich. 2002. An expanded view of DNA replication. Proc. Natl. Acad. Sci. USA 99:8342-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth, B. L., M. Poot, S. T. Yue, and P. J. Millard. 1997. Bacterial viability and antibiotic susceptibility testing with SYTOX Green nucleic acid stain. Appl. Environ. Microbiol. 63:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan-Arends, S. J., and D. S. Weiss. 2004. Inhibiting cell division in Escherichia coli has little if any effect on gene expression. J. Bacteriol. 186:880-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sassanfar, M., and J. W. Roberts. 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 212:79-96. [DOI] [PubMed] [Google Scholar]
- 21.Studier, F. W., and B. A. Moffat. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 89:113-150. [DOI] [PubMed] [Google Scholar]
- 22.Sueoka, N. 1998. Cell membrane and chromosomal replication in Bacillus subtilis. Prog. Nucleic Acid Res. Mol. Biol. 59:35-53. [DOI] [PubMed] [Google Scholar]
- 23.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umu DC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-4997. [DOI] [PubMed] [Google Scholar]
- 24.Walker, G. C. 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev. 48:60-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasbin, R. E. 2002. DNA repair mechanisms and mutagenesis, p. 27-46. In U. N. Streips and R. E. Yasbin (ed.), Modern microbial genetics, 2nd ed. Wiley-Liss, Inc., New York, N.Y.
- 26.Yung, B. Y. M., and A. Kornberg. 1988. Membrane attachment activates the DnaA protein, the initiation protein of chromosomal replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 85:7202-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]