Abstract
The Bacillus subtilis aprE gene, which encodes the extracellular alkaline protease, is regulated by many positive and negative transcriptional regulators. SenS is one such positive regulator consisting of 65 amino acids. We found that the senS gene on a multicopy plasmid, pSEN24, caused an increase in aprE expression in strains carrying the upstream region of aprE up to −340 with respect to the transcription initiation site but not in a strain carrying the region up to −299, which is within the binding site of the negative regulator ScoC (Hpr). Epistatic analysis showed that the pSEN24 effect was lost in a scoC-deleted mutant. In accordance with these results, the scoC transcription level as assayed by a scoC-lacZ fusion and Northern analysis was greatly reduced in the cells carrying pSEN24. From these results we conclude that multicopy senS enhances aprE expression by suppressing the transcription of scoC.
Bacillus subtilis secretes degradative enzymes after the end of logarithmic growth, apparently for degrading high-molecular-weight materials around the cell to cope with adverse nutritional conditions (17, 23). Among the extracellular proteases produced by this organism, the alkaline and neutral proteases constitute the major part of the protease activities. Since these enzymes are produced on the order of grams per liter, expression of the genes coding for the enzymes (aprE and nprE, respectively) has to be strictly controlled to avoid extravagant use of energy and materials. Thus, the aprE gene is regulated by many positive and negative regulators. The positive regulators include Spo0A, the DegS-DegU two-component system, DegR, DegQ, ProB, TenA, RelA, SalA, and SenS, while the negative regulators include AbrB, ScoC (previously called Hpr), SinR, and Pai (2, 9, 10, 20). Among these factors, AbrB, DegU, ScoC, and SinR are known to regulate aprE expression directly by binding to upstream regions of the aprE coding sequence (1, 4, 11, 22). Therefore, it can be summarized that the major regulatory pathways controlling aprE expression are the routes via AbrB, ScoC, SinR, and DegU. The positive effect of Spo0A is through inhibition of abrB expression (20), while those of DegR, DegQ, ProB, and TenA are through functional DegU (6, 8, 9, 15). It was shown recently that disruption of salA caused a decrease in aprE expression, and this was attributed to the enhanced synthesis of ScoC (10). In addition, the stringent factor RelA was shown to be required for the efficient expression of aprE (2). The mode of action of the pai gene product remains to be studied.
The DNA-binding transcription regulators, such as ScoC, SinR, DegU, and AbrB, have their own specific target sites in the control region of aprE, i.e., for ScoC, these are nucleotides (nt) spanning −324 to −267 and −79 to −14; for SinR, −268 to −220; for DegU, −164 to −113 and/or −70 to −27; and for AbrB, −59 to +25 (3, 4, 19, 22).
Wong et al. discovered a B. subtilis Natto gene, senN, which on a multicopy plasmid enhances the expression of B. subtilis aprE (26). The authors later identified the B. subtilis counterpart of senN named senS that also functions as a positive regulator of aprE expression. SenS is a positively charged, 65-amino-acid protein with a helix-turn-helix motif in the molecule (25). It was demonstrated that SenS exerts its positive effect by acting on the region between nt −177 and −415 upstream of the transcription start site of aprE (5), but further details, including the target of SenS and the relationship with the other transcriptional regulators, have not been investigated. In this study, we show that multicopy senS enhances aprE expression by reducing the expression of scoC.
Stimulation of aprE-lacZ expression by multicopy senS.
To study the effect of SenS on aprE expression, we used pSEN24 in which the Shine-Dalgarno sequence and the following senS coding region were placed under the control of the isopropyl-1-thio-β-d-galactopyranoside (IPTG)-inducible Pspac promoter (Table 1). The plasmid was constructed in two steps. A PCR fragment prepared with the primer pair SENSF (5′-AGTTAAGCTTATCGTTTAGATAAGGGCC-3′) and SENSR (5′-AGTTGTCGACAAAAACCCGTTGTAGTCAGC-3′) and B. subtilis CU741 DNA as a template was digested with HindIII and SalI (sites are underlined) and inserted into pDG148 that had been treated with the same restriction enzymes. The ligated sample was transformed into strain CU741 as described previously (13), and the resultant Nmr transformants were screened for the ability to produce larger halos on casein- and gelatin-containing Luria-Bertani plates (24) than those produced by the transformants carrying pDG148. The 5′ end of the senS region on pSEN24 is the 51st nucleotide upstream of the senS coding sequence (25).
TABLE 1.
B. subtilis strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| CU741 | trpC2 leuC7 | S. A. Zhaler |
| OAM145 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) | 10 |
| OAM146 | trpC2 leuC7 amyE::aprE-lacZ(−340 [SG35.21], Cmr) | 10 |
| OAM147 | trpC2 leuC7 amyE::aprE-lacZ(−299, Cmr) | 10 |
| OAM218 | trpC2 leuC7 amyE::aprE-lacZ(−267, Cmr) | 10 |
| TT715 | trpC2 leuC7 aprE-lacZ (Cmr) | 7 |
| MU38 | trpC2 leuC7 amyE::degR-lacZ (Cmr) mecA::KmrdegU::Spr | 12 |
| TU38 | trpC2 leuC7 aprE-lacZ (Cmr) degU::Spr | MU38→TT715 |
| OAM157 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) scoC::Emr (lacZ::Tcr) | 10 |
| OAM221 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) sinR::Pmr | 10 |
| OAM169 | trpC2 leuC7 amyE::aprE-lacZ(−412 [SG35.18], Cmr) spoOA::SprabrB::Kmr | 10 |
| TSU2 | trpC2 leuC7 amyE::scoC-lacZ (Cmr) | pSCO1→CU741 |
| KAW1 | trpC2 leuC7 amyE::scoC-lacZ (Cmr) spoOA::Spr | OAM169→TSU2 |
| Plasmids | ||
| pDG148 | Multicopy B. subtilis and Escherichia coli plasmid; Apr, Kmr | 21 |
| pSEN24 | pDG148 carrying senS | This study |
| pIS284 | E. coli plasmid for insertion of lacZ fusions into the B. subtilis amyE locus | I. Smith |
| pSCO1b | pIS284 carrying scoC-lacZ at the amyE locus | This study |
The numbers in parentheses and the letters in brackets indicate the deletion end points upstream of the transcription start point of aprE and promoter constructs, respectively. Emr, erythromycin resistance; Tcr, tetracycline resistance; Pmr, phleomycin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Spr, spectinomycin resistance; Apr, ampicillin resistance. The concentrations of the antibiotics added to the medium were 1 μg/ml for erythromycin, 5 μg/ml for chloramphenicol and phleomycin, 15 μg/ml for neomycin, 10 μg/ml for tetracycline, and 100 μg/ml for spectinomycin.
Plasmid pSCO1 was constructed as follows. A 561-bp PCR fragment spanning nt 76 in the yhaH gene located immediately upstream of scoC to nt 102 in scoC was prepared with primers YhaHF (5′-AGTTGAATTCGCACCTTCCTCAGGAAAGC-3′) and ScoCR (5′-AGTTGGATCCTTCTCGATCGATTTCC-3′) and CU741 DNA as a template, digested with EcoRI and BamHI (sites are underlined), and inserted into the EcoRI and BamHI sites of pIS284.
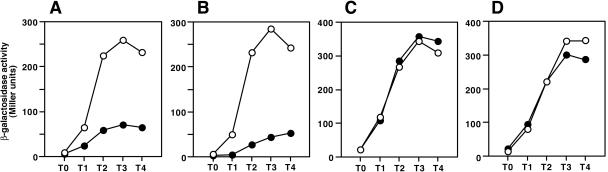
When pSEN24 was introduced into B. subtilis OAM145 carrying aprE-lacZ at the amyE locus and the cells were grown in the presence of 0.2 mM IPTG, the β-galactosidase activity increased 3.7-fold around T3 (3 h after the end of logarithmic growth) compared with that in OAM145 carrying the vector pDG148 (Fig. 1A).
FIG. 1.
Effects of deletions of the upstream region of aprE on stimulation of aprE-lacZ expression by multicopy senS on pSEN24. Deletions were up to −412 (A, strain OAM145), −340 (B, OAM146), −299 (C, OAM147), and −267 (D, OAM218). The solid and open circles indicate β-galactosidase activities in the cells carrying pDG148 and pSEN24, respectively. Cells were grown in Schaeffer's medium (18) containing 0.2 mM IPTG, and β-galactosidase activities were determined for the samples taken at the indicated times as described previously (10).
It has been well studied that many positive and negative transcription factors of aprE exert their effects at specific regions upstream of the aprE gene (see above). To locate the cis-acting site of multicopy senS in the control region of aprE, we used aprE-lacZ fusions with various deletions upstream of the transcription initiation site of aprE (3, 10) and examined the effect of multicopy senS on aprE-lacZ expression. The OAM145 strain used above carries a region up to −412 relative to the transcription initiation site of aprE (Table 1). In strain OAM146, in which the upstream region of aprE is deleted up to −340, the stimulating effect of multicopy senS was still exhibited (Fig. 1B), but in OAM147 and OAM218, carrying deletions up to −299 and −267, respectively, the positive effect was lost (Fig. 1C and D). The results show that the SenS target is located in a region upstream of or including −299. Since, among the known aprE regulators, ScoC is the only known regulator whose target DNA region includes −299 (4, 10), the results suggested that SenS might affect scoC expression. It should be noted that the deletions up to −299 and −267 resulted in derepression of aprE (Fig. 1), probably because part or all of the scoC target sequence is deleted in these strains. A similar deletion effect was observed previously (3).
Effect of multicopy senS on aprE-lacZ expression in mutants affecting aprE expression.
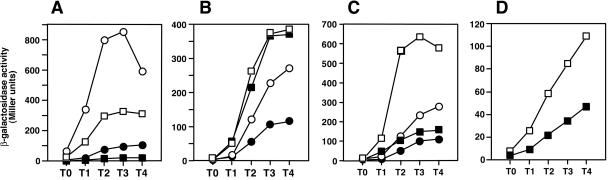
If scoC is the target of SenS, it is expected that the positive effect of multicopy senS would be lost in a scoC-deleted strain. To investigate this possibility and also the relationship between SenS and the transcription regulators that directly affect aprE expression, we studied the multicopy effect of senS by quantifying aprE-lacZ expression in strains with deletions of the degU, scoC, sinR, and spo0A-abrB genes. The results presented in Fig. 2 show that, under the condition where there was 2.5- to 8.5-fold stimulation by pSEN24 in the wild-type strains (compare circles), the enhancing effect was also observed in the degU (Fig. 2A), sinR (Fig. 2C), and spo0A-abrB (Fig. 2D) mutants but not in the scoC mutant (Fig. 2B) (compare squares). These results, together with those described in the previous section, support the notion that multicopy senS stimulates aprE expression through inhibition of scoC expression. The stimulatory effect of pSEN24 in strain TT715 (Fig. 2A) is somewhat stronger than that in strain OAM145 (Fig. 1A and 2B and C). This is probably due to the difference in the location of the aprE-lacZ fusions, i.e., the original aprE locus in TT715 and the amyE locus in OAM145 (Table 1), although the reason remains to be determined.
FIG. 2.
Epistatic analysis of multicopy senS in mutants that affect aprE-lacZ expression. Cells were grown in Schaeffer's medium containing 0.2 mM IPTG. (A) •, TT715 degU+ (pDG148); ○, TT715 degU+ (pSEN24); ▪, TU38 degU (pDG148); □, TU38 degU (pSEN24). (B) •, OAM145 scoC+ (pDG148); ○, OAM145 scoC+ (pSEN24); ▪, OAM157 scoC (pDG148); □, OAM157 scoC (pSEN24). (C) •, OAM145 sinR+ (pDG148); ○, OAM145 sinR+ (pSEN24); ▪, OAM221 sinR (pDG148); □, OAM221 sinR (pSEN24). (D) ▪, OAM169 spo0A abrB (pDG148); □, OAM169 spo0A abrB (pSEN24).
Inhibition of scoC expression by multicopy senS.
We next investigated whether the enhancing effect of SenS on aprE expression is a result of inhibition of scoC transcription.
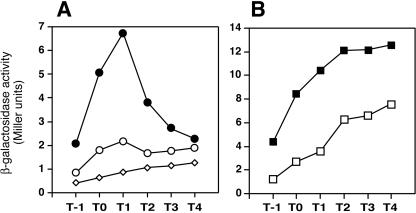
First, we constructed strain TSU2 carrying a scoC-lacZ fusion at the amyE locus and quantified the β-galactosidase levels in the cells harboring either pSEN24 or the pDG148 vector. As shown in Fig. 3A, the β-galactosidase activities in the pSEN24-carrying cells (open circles) were much lower than those in the cells carrying pDG148 (filled circles): the background β-galactosidase levels in strain CU741 lacking a lacZ fusion are also shown (diamonds).
FIG. 3.
Effect of multicopy senS on scoC-lacZ expression in wild-type and spo0A strains. Cells were grown in Schaeffer's medium containing 0.2 mM IPTG. The solid and open symbols indicate the values observed in strains carrying pDG148 and pSEN24, respectively. (A) TSU2 spo0A+. (B) KAW1 spo0A. The diamonds in panel A indicate the values of the blank test with strain CU741.
It was shown previously that scoC expression reaches its highest level at the early stationary phase and that inactivation of spo0A results in overexpression of scoC, because abrB, a positive regulator of scoC expression, is negatively regulated by Spo0A (16). To test whether the AbrB-stimulated scoC expression is also reduced by multicopy senS, we introduced pSEN24 and pDG148 into strain KAW1 carrying a scoC-lacZ fusion in a spo0A background. It was shown that scoC-lacZ expression continued to increase after T1, which is in accordance with the previous result (16), and that this enhanced transcription was also inhibited by multicopy senS on pSEN24 (Fig. 3B).
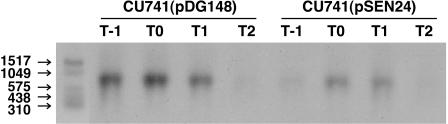
We further analyzed the effect of SenS on scoC expression by Northern analysis. RNA was prepared from strain CU741 carrying either pSEN24 or pDG148, and scoC mRNA was detected with a digoxigenin-labeled PCR fragment derived from a scoC coding region. As shown in Fig. 4, the intensity was lower in the RNA samples from the pSEN24-carrying cells than those from the cells harboring pDG148. The results substantiate the scoC-lacZ fusion experiments described above. We note that the size of the scoC mRNA was around 750 nucleotides (Fig. 4), which is slightly larger than the scoC coding sequence (609 bp) (16). Since the size difference (around 140 bp) is less than the intergenic region (180 bp) between scoC and its upstream yhaH, it is likely that the transcriptional start site of scoC is within this region and scoC is transcribed in a single mRNA.
FIG. 4.
Northern analysis of scoC mRNA in cells carrying the pDG148 vector and pSEN24. The leftmost lane shows the RNA markers, whose sizes are indicated by the numbers to the left of the panel. Strain CU741 carrying pDG148 or pSEN24 was grown in Schaeffer's medium containing 0.2 mM IPTG and harvested every hour from T − 1 to T2. RNA was prepared as described previously (27). The RNA samples (20 μg) were electrophoresed in a 1.2% agarose gel, and after the transfer of RNA to the membrane was verified by UV illumination, scoC mRNA was detected as described previously (10). A digoxigenin-labeled PCR fragment containing a coding region of scoC was prepared using a PCR DIG probe synthesis kit (Roche Diagnostics) with primer pair ScoC-1 (5′-ATCGAGTGGAACCGCCCTATGA-3′) and ScoC604R (5′-TTACAGGTTCGAGCTCTTCA-3′) and CU741 DNA as a template. The PCR was carried out by following the procedure provided by the supplier. The scoC-specific probe DNA was purified by agarose gel electrophoresis, and the specificity was confirmed by Southern analysis using DNAs from strains CU741 and OAM157. The size of scoC mRNA was determined by comparing with the size markers contained in RNA Molecular Weight Marker III obtained from Roche Diagnostics.
Binding of His-tagged SenS to the control region of aprE.
It was predicted that SenS contains a DNA-binding, helix-turn-helix motif (24). We prepared a His-tagged SenS protein and subjected it to gel shift analysis with a DNA region spanning the C terminus of the upstream gene to an N-terminal region of scoC. Despite several attempts, however, no shifted band was detected.
The results described here show that multicopy senS stimulates aprE expression through inhibition of the expression of the negative regulator scoC. SenS is the first example that a positive regulator of aprE expression exerts its effect without the participation of DegU. However, since we failed to detect the binding of His-tagged SenS to the upstream region of scoC, it is not known at present whether the SenS effect on scoC expression is direct or via a second factor. An attempt was made to test the second possibility by microarray analysis, but no candidate was found (data not shown).
We have previously shown that scoC expression is regulated by SalA (9). It was shown, however, that the multicopy effect of senS on scoC-lacZ expression was still observed in a salA-deficient strain (data not shown), suggesting that SalA and SenS work in different pathways.
Although multicopy senS caused a decrease in scoC expression, resulting in overexpression of aprE, disruption of the chromosomal senS gene by insertion of the tetracycline resistance or neomycin resistance gene did not affect aprE-lacZ expression (data not shown), apparently indicating that the senS gene in a single-copy state is not expressed to a level that influences the expression of aprE. Although the SenS level may be low in the cells grown in the laboratory condition, i.e., growth in Shaeffer's sporulation medium, it is possible that the senS gene may play a role in some growth condition in the natural habitats of B. subtilis, the soil and the rhizosphere.
We have shown previously that a disruption of chromosomal senS by the spectinomycin resistance gene resulted in a decrease in aprE-lacZ expression (14). This negative effect on aprE expression, however, was observed only when senS was disrupted by the spectinomycin resistance gene in a specific orientation and was attributed to a secondary effect of spectinomycin resistance (data not shown).
REFERENCES
- 1.Gaur, N. K., J. Oppenheim, and I. Smith. 1991. The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J. Bacteriol. 173:678-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hata, M., M. Ogura, and T. Tanaka. 2001. Involvement of stringent factor RelA in expression of the alkaline protease gene aprE in Bacillus subtilis. J. Bacteriol. 183:4648-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henner, D. J., E. Ferrari, M. Perego, and J. A. Hoch. 1988. Location of the targets of the hpr-97, sacU32(Hy), and sacQ36(Hy) mutations in upstream regions of the subtilisin promoter. J. Bacteriol. 170:296-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kallio, P. T., J. E. Fagelson, J. A. Hoch, and M. A. Strauch. 1991. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J. Biol. Chem. 266:13411-13417. [PubMed] [Google Scholar]
- 5.McCready, P. M., and R. H. Doi. 1992. Bacillus subtilis SenS exerts its activity through a site in the 5′ flanking region of the aprE promoter. J. Gen. Microbiol. 138:2069-2074. [DOI] [PubMed] [Google Scholar]
- 6.Msadek, T., F. Kunst, A. Klier, and G. Rapoport. 1991. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J. Bacteriol. 173:2366-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukai, K., M. Kawata, and T. Tanaka. 1990. Isolation and phosphorylation of the Bacillus subtilis degS and degU gene products. J. Biol. Chem. 265:20000-20006. [PubMed] [Google Scholar]
- 8.Mukai, K., M. Kawata-Mukai, and T. Tanaka. 1992. Stabilization of phosphorylated Bacillus subtilis DegU by DegR. J. Bacteriol. 174:7954-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogura, M., M. Kawata-Mukai, M. Itaya, K. Takio, and T. Tanaka. 1994. Multiple copies of the proB gene enhance degS-dependent extracellular protease production in Bacillus subtilis. J. Bacteriol. 176:5673-5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogura, M., A. Matsuzawa, H. Yoshikawa, and T. Tanaka. 2004. Bacillus subtilis SalA (YbaL) negatively regulates expression of scoC, which encodes the repressor for the alkaline exoprotease gene, aprE. J. Bacteriol. 186:3056-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogura, M., K. Shimane, K. Asai, N. Ogasawara, and T. Tanaka. 2003. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol. Microbiol. 49:1685-1697. [DOI] [PubMed] [Google Scholar]
- 12.Ogura, M., and T. Tanaka. 1996. Bacillus subtilis DegU acts as a positive regulator for comK expression. FEBS Lett. 397:173-176. [DOI] [PubMed] [Google Scholar]
- 13.Ogura, M., and T. Tanaka. 1997. Bacillus subtilis ComK negatively regulates degR gene expression. Mol. Gen. Genet. 254:157-165. [DOI] [PubMed] [Google Scholar]
- 14.Ogura, M., and T. Tanaka. 1997. Expression of alkaline protease gene in Bacillus subtilis mutants that lack positive regulatory genes degR, degQ, senS, tenA, and proB. Biosci. Biotechnol. Biochem. 61:372-374. [Google Scholar]
- 15.Pang, A. S., S. Nathoo, and S. L. Wong. 1991. Cloning and characterization of a pair of novel genes that regulate production of extracellular enzymes in Bacillus subtilis. J. Bacteriol. 173:46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perego, M., and J. A. Hoch. 1988. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J. Bacteriol. 170:2560-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Priest, F. G. 1977. Extracellular enzyme synthesis in the genus Bacillus. Bacteriol. Rev. 41:711-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaeffer, P. J., J. Millet, and J. Aubert. 1965. Catabolite repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimane, K., and M. Ogura. 2004. Mutational analysis of the helix-turn-helix region of Bacillus subtilis response regulator DegU, and identification of cis-acting sequences for DegU in the aprE and comK promoters. J. Biochem. (Tokyo) 136:387-397. [DOI] [PubMed] [Google Scholar]
- 20.Smith, I. 1993. Regulatory proteins that control late-growth development, p. 785-800. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 21.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 527:697-704. [DOI] [PubMed] [Google Scholar]
- 22.Strauch, M. A. 1995. In vitro binding affinity of the Bacillus subtilis AbrB protein to six different DNA target regions. J. Bacteriol. 177:4532-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strauch, M. A., and J. A. Hoch. 1993. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol. Microbiol. 7:337-342. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka, T., and M. Kawata. 1988. Cloning and characterization of Bacillus subtilis iep, which has positive and negative effects on production of extracellular proteases. J. Bacteriol. 170:3593-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, L.-F., and R. H. Doi. 1990. Complex character of senS, a novel gene regulating expression of extracellular-protein genes of Bacillus subtilis. J. Bacteriol. 172:1939-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong, S. L., L. F. Wang, and R. H. Doi. 1988. Cloning and nucleotide sequence of senN, a novel ‘Bacillus natto’ (B. subtilis) gene that regulates expression of extracellular protein genes. J. Gen. Microbiol. 134:3269-3276. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida, K.-I., K. Kobayashi, Y. Miwa, C.-M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]