Abstract
The bacterial plant pathogen Pseudomonas syringae pv. tomato DC3000 (DC3000) causes disease in Arabidopsis thaliana and tomato plants, and it elicits the hypersensitive response in nonhost plants such as Nicotiana tabacum and Nicotiana benthamiana. While these events chiefly depend upon the type III protein secretion system and the effector proteins that this system translocates into plant cells, additional factors have been shown to contribute to DC3000 virulence and still many others are likely to exist. Therefore, we explored the contribution of the twin-arginine translocation (Tat) system to the physiology of DC3000. We found that a tatC mutant strain of DC3000 displayed a number of phenotypes, including loss of motility on soft agar plates, deficiency in siderophore synthesis and iron acquisition, sensitivity to copper, loss of extracellular phospholipase activity, and attenuated virulence in host plant leaves. In the latter case, we provide evidence that decreased virulence of tatC mutants likely arises from a synergistic combination of (i) compromised fitness of bacteria in planta; (ii) decreased efficiency of type III translocation; and (iii) cytoplasmically retained virulence factors. Finally, we demonstrate a novel broad-host-range genetic reporter based on the green fluorescent protein for the identification of Tat-targeted secreted virulence factors that should be generally applicable to any gram-negative bacterium. Collectively, our evidence supports the notion that virulence of DC3000 is a multifactorial process and that the Tat system is an important virulence determinant of this phytopathogenic bacterium.
The hallmark of the Pseudomonas syringae infection process is the injection of virulence effector proteins directly into host cells via the type III secretion mechanism (TTSS) (14). For efficient translocation of effector proteins, P. syringae must first establish productive contact with a target plant cell. This entails secretion of exopolysaccharides, cell wall-degrading enzymes, plant hormones, and several low-molecular-weight phytotoxins, such as coronatine and syringomycin (1, 30). Since many of these factors are secreted in a type III-independent manner and since deletion of many of these virulence factors often does not render P. syringae avirulent, it can be assumed that P. syringae virulence is multifactorial. Moreover, given the complexity of interactions that occur at the host-pathogen interface, there are likely many additional virulence determinants in P. syringae yet to be discovered.
One possible contributor to P. syringae pathogenesis is the twin-arginine translocation (Tat) pathway that operates in the inner membrane of many gram-negative bacteria (4, 5, 56). Tat substrates are synthesized with cleavable N-terminal signal peptides that are characterized by a highly conserved twin-arginine motif in the positively charged N-terminal region, a weakly hydrophobic core region, and a positively charged Sec pathway-avoidance signal in the C-terminal region (7, 17). The most remarkable feature of the Tat system is its ability to transport proteins which have already folded in the cytoplasm (5). In fact, emerging evidence indicates that only fully folded proteins are competent for Tat transport (22, 28, 45, 57, 59). Recently, a role for the Tat system in bacterial pathogenesis was demonstrated by Ochsner and coworkers, who reported that the opportunistic human pathogen P. aeruginosa lacking a functional Tat system was attenuated for virulence (51). A similar observation was made for enterohemorrhagic Escherichia coli O:157 (52) and for the phytopathogen Agrobacterium tumefaciens (26). Homologues of the genes encoding members of the TatA and TatC families, which are minimally required for Tat transport, have been found in the genomes of many bacterial pathogens, including A. tumefaciens, E. coli O:157, Helicobacter pylori, Mycobacterium tuberculosis, P. aeruginosa, P. syringae, and Yersinia pestis (67), suggesting a role for this pathway in the virulence of diverse bacterial species.
However, whether the Tat system directly contributes to the secretion of virulence factors in these bacteria is not immediately clear, since most of the known Tat-dependent proteins are periplasmic redox factors that are involved with various respiratory chains that support metabolism and growth of bacteria (4). Consequently, inactivation of the Tat system by genetic deletion results in a pleiotropic phenotype (36, 61) that is likely to compromise a bacterium's fitness in the nutrient-poor intercellular environment of a host organism and attenuate the virulence phenotype. Of further concern is, assuming that Tat virulence factors exist, how would such proteins escape the periplasm and engage host cells? A solution to the latter issue was presented by Voulhoux et al. (64), who demonstrated that the Tat system is essential for secreting two extracellular virulence determinants, both phospholipases, out of P. aeruginosa. This study demonstrated that a protein localized in the periplasm by the Tat system could be translocated to the extracellular environment by the type II (Xcp) secretion machinery. This two-step secretion of a Tat substrate led to the proposition of a mosaic model for the general secretory pathway (GSP) in which proteins are first routed across the inner membrane using either the Sec or Tat translocation pathway followed by translocation across the outer membrane by the type II secretion system.
In the present study, we explored whether Tat transport constitutes a virulence mechanism in P. syringae pv. tomato DC3000 (DC3000). We reasoned that secretion via the Tat system might be important to P. syringae virulence by playing a key role in (i) cellular metabolism and physiology of a bacterium during colonization and growth in planta; (ii) targeting, assembly, and membrane stability of surface structures necessary for specific pathogen-host interactions; and (iii) two-step secretion of proteins that interact directly with plant cells as part of the host-pathogen confrontation. To test this hypothesis, we created and characterized a tatC mutant strain of DC3000. Using this strain in conjunction with a bioinformatic algorithm for identifying putative Tat substrates along with a novel broad-host-range genetic reporter for Tat transport based on the green fluorescent protein (GFP), we identified several P. syringae Tat substrates. Two of these were secreted phospholipases, PSPTO3648 and PSPTOB0005, that each carried Tat-targeting signals and utilized the Tat pathway as the first step in the extracellular secretion process. In addition, we confirmed that these proteins are translocated across the outer membrane by the P. syringae type II system encoded by the gsp operon, indicating that the Tat and type II systems operate in synergy to form a two-step process for the secretion of P. syringae virulence factors.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. All Pseudomonas strains were routinely grown in King's B (KB) medium (39) or modified Luria broth (LM) (31) at 30°C, and all E. coli strains were grown in Luria-Bertani broth (LB) at 37°C. Antibiotics were added to the media at the following concentrations: ampicillin, 100 μg ml−1; rifampin, 50 μg ml−1; spectinomycin, 25 μg ml−1; gentamicin, 10 μg ml−1; and chloramphenicol, 25 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| MC4100 | Escherichia coli F− ΔlacU169 araD139 rpsL150 relA1 ptsF rbs flbB5301 | 10 |
| B1LK0 | MC4100 ΔtatC | 8 |
| DADE | MC4100 ΔtatABCD ΔtatE | 65 |
| DC3000 | Pseudomonas syringae pv. tomato DC3000 wild type, Rifr | 18 |
| PBTAT1 | DC3000 tatC::pKnockout, Spr Strr | This study |
| PBGspD | DC3000 gspD::pKnockout, Spr Strr | This study |
| PBGspE | DC3000 gspE::pKnockout, Spr Strr | This study |
| Plasmid | ||
| pTrc99A | trc promoter, ColE1 ori, Ampr | Amersham Pharmacia |
| pTatCDC3000 | DC3000 tatC gene in pTrc99A | This study |
| pTatABCDC3000 | DC3000 tatABC operon in pTrc99A | This study |
| pKnockout-Ω | Smr Sprmob in T-vector | 66 |
| pJN105 | araC-PBAD-controlled broad-host-range expression vector | 49 |
| pBS1 | Gateway cassette in pJN105 | This study |
| pBS1-TatCDC3000 | DC3000 tatC gene in pBS1 | This study |
| pBS1-PlcA1 | DC3000 plcA1 gene in pBS1 | This study |
| pTGS | Tripartite fusion of ssTorA-GFP-SsrA in pBAD33 | 21 |
| pMMB-207 | tac-controlled wide-host-range plasmid based on pMMB66EH | 47 |
| pMMB-TGS-Cm | ssTorA-GFP-SsrA in pMMB-207 | This study |
| pMMB-TGS-Gm | Cmr cassette of pMMB-TGS replaced by Gmr cassette | This study |
| pMMB-GGS | Replacement of ssTorA with Gateway cassette in pMMB-TGS-Gm | This study |
| pGus-GS | β-Glucuronidase in pMMB-GGS | This study |
| pssPlcA1-GS | DC3000 PlcA1 signal peptide in pMMB-GGS | This study |
| pssCopA-GS | DC3000 CopA signal peptide in pMMB-GGS | This study |
| pssCumA-GS | DC3000 CumA signal peptide in pMMB-GGS | This study |
| pPlcA1-GS | DC3000 PlcA1 in pMMB-GGS | This study |
| pPlcA2-GS | DC3000 PlcA2 in pMMB-GGS | This study |
| pCopA-GS | DC3000 CopA in pMMB-GGS | This study |
| pCumA-GS | DC3000 CumA in pMMB-GGS | This study |
Plasmids pTatCDC3000 and pTatABCDC3000 were constructed by insertion of PCR-amplified DNA encoding the DC3000 tatC or tatABC genes into plasmid pTrc99A between NcoI and HindIII. All pBS1-based expression plasmids were created using Gateway cloning technology (Invitrogen, Carlsbad, CA). Briefly, the gene or genes of interest were amplified by PCR from the DC3000 genome with Deep Vent (New England Biolabs) using primers that introduced a 5′ CACC overhang. These PCR products were combined with pENTR/SD/D-TOPO per the manufacturer's protocol (Invitrogen). All pENTR clones were checked by sequencing, and expression clones were made by combining the pENTR plasmids with the desired expression vector along with Gateway LR clonase II enzyme. The pENTR plasmid was recombined with the destination vector pBS1, a derivative of the araC-PBAD-controlled broad-host-range vector pJN105 (49) that contains a Gateway cassette to allow arabinose-inducible expression of insert genes and generates protein products with a carboxy-terminal hemagglutinin tag. Recombination reactions were carried out as suggested by the manufacturer (Invitrogen). Plasmid pMMB-TGS-Cm was constructed by insertion of a 902-bp EcoRI/HindIII fragment from pTGS (21) into the same sites of pMMB-207 (47). To enable conversion of pMMB-TGS-Cm into a destination vector, the Cmr cassette was replaced by the gentamicin resistance gene, resulting in plasmid pMMB-TGS-Gm. The DNA encoding the E. coli TorA signal peptide (ssTorA) was excised and replaced with a Gateway cassette resulting in plasmid pMMB-GGS, a derivative of the broad-host-range Tat reporter vector pMMB-TGS-Gm that allows insertion of genes upstream of the Tat-specific reporter GFP-SsrA via Gateway technology.
Construction of the DC3000 tatC mutant was carried out by PCR amplification of an internal 600-bp sequence of tatC, corresponding to nucleotides 100 to 700 of the coding sequence, from the DC3000 genome and cloning into pKnockout-Ω(66). The resulting plasmid was transferred into DC3000 via conjugation. As pKnockout cannot replicate in DC3000, single-crossover integrants were selected by resistance to spectinomycin. Orientation of integration was determined by PCR. All plasmids were confirmed by sequencing. Strains with insertion deletions of gspD and gspE were constructed in a similar fashion using PCR-amplified DNA encoding nucleotides 227 to 800 and 322 to 843 for gspD and gspE, respectively.
Growth conditions and phenotypic assays.
Growth analysis of DC3000 and the tatC mutant strain PBTAT1 was performed in KB medium, LM medium, low-phosphate medium (58), and an hrp-derepressing minimal medium (34). Siderophore production was monitored by streaking cells on mannitol-glutamate (MG) medium (38) supplemented with 2,2′-dipyridyl (Sigma) at various concentrations. Motility of DC3000 was assayed by inoculating cells with an applicator stick to the center of a semisolid (0.4% [wt/vol] agar) nutrient broth-yeast extract plate (63) supplemented with 50 μg ml−1 of rifampin. Results were obtained after approximately 48 h of incubation at 28°C. To test for copper susceptibility, stationary DC3000 cultures grown in KB medium were washed three times in MG medium and resuspended in a 96-well plate containing MG media and various amounts of CuSO4. Cell growth was recorded by measuring optical density at 600 nm (OD600). Phospholipase expression was induced using low-phosphate medium. The phospholipase overexpression system, methods for concentrating supernatants, and assays using the synthetic substrate nitrophenyl-phosphatidylcholine (pNPPC) have been described previously (16).
Bioinformatic prediction of Tat substrates.
Specifically, a hidden Markov model (HMM) was constructed using a set of signal peptide sequences selected from experimentally confirmed Tat substrates, primarily from E. coli (5) and P. aeruginosa (51). The null model was taken as the empirical distribution of amino acids in positions 2 through 50 from the set of all annotated proteins in all complete bacterial genome sequences available from the National Center of Biotechnology Information. These frequencies, along with the set of confirmed Tat signal peptide sequences, were used to create an HMM for the Tat motif using hmmbuild (http://hmmer.wustl.edu/) (27). After calibration using hmmcalibrate, the resulting Tat substrate model was used to search the annotated proteins from the chromosome of P. syringae pv. tomato DC3000 (GenBank accession numbers NC_004578, NC_004632, and NC_004633) (9) using hmmsearch. Peptide sequences identified by the HMM that did not fall within the first 35 amino acids of the protein were not considered. All predicted substrates were crosschecked using the signal peptide prediction tool SignalP (50).
Flow cytometric analysis.
Overnight cultures of E. coli or DC3000 cells harboring GFP-based plasmids were subcultured into fresh medium with chloramphenicol or gentamicin and induced with 0.2% (wt/vol) arabinose (for pTGS) or 0.05 mM isopropyl-β-d-thiogalactopyranoside (IPTG; for pMMB-TGS-Cm and pMMB-TGS-Gm) in mid-exponential-phase growth. After 6 h, cells were washed once with phosphate-buffered saline, and 5 μl of washed cells was diluted into 1 ml of phosphate-buffered saline prior to analysis using a BD Biosciences FACSCalibur.
Subcellular fractionation.
DC3000 supernatant fractions were prepared as follows: cultures were grown overnight in low-phosphate media and diluted into fresh media at a final OD600 of 0.15. Cells were grown to an OD600 of ∼0.7, aliquots were removed, and the supernatant was separated by centrifugation in a microcentrifuge and further purified by passage through a 0.2-mm syringe filter. Residual media was removed from the cell pellet by aspiration, and the pellet was resuspended in 10 ml of TE (10 mM Tris-Cl [pH 7.5], 2.5 mM Na-EDTA) and lysed by sonication. Intact cells and cellular debris were removed by centrifugation (5 min at 10,000 × g), and the supernatant was retained as the soluble pellet fraction. E. coli periplasmic fractions were prepared by subjecting equivalent amounts of cells to the lysozyme-EDTA-cold osmotic shock procedure (55). Following release of periplasmic proteins, cells were resuspended in 10 ml of TE (10 mM Tris-Cl [pH 7.5], 2.5 mM Na-EDTA) and lysed by sonication, and intact cells and cellular debris were removed by centrifugation (5 min at 10,000 × g). The resulting supernatant was retained as the soluble cytoplasmic fraction. Protein concentrations were determined with a Bio-Rad protein assay reagent kit with bovine serum albumin as the standard. β-Galactosidase activity was used as a cytoplasmic marker of fractionation efficiency (23). Only data from fractionation experiments in which ≥95% of the β-galactosidase activity was in the cytoplasmic fraction are reported.
Western blot analysis.
Western blotting was performed as previously described (21). All lanes of sodium dodecyl sulfate (SDS)-12% polyacrylamide gels were loaded with samples prepared from an equivalent number of cells harvested from each experiment. The following primary antibodies were used: monoclonal mouse anti-GFP (Clontech) diluted 1:2,000 and polyclonal rabbit anti-GroEL (Sigma) diluted 1:20,000. The secondary antibody was 1:2,000 goat anti-mouse or goat anti-rabbit horseradish peroxidase (Promega). Following development of blots probed with GFP antibodies, membranes were stripped in Tris-buffered saline-2% (wt/vol) SDS-0.7 M β-mercaptoethanol. Stripped membranes were reblocked and probed with anti-GroEL antibody.
Plant assays.
Arabidopsis thaliana, Lycopersicum esculentum cv. Money Maker, and Nicotiana tobacco cv. Xanthi plants were grown and inoculated with bacteria as described previously (29). For virulence assays in tomato, whole plants were dipped for 30 s in a bacterial suspensions containing 106 CFU ml−1 in 10 mM MgCl2 (pH 5.5) solution containing 0.02% (vol/vol) Silwet L-77 (Lehle Seeds). For assays with Arabidopsis, the bacteria were resuspended in a 10 mM MgCl2 (pH 5.5) solution containing 0.01% (vol/vol) Silwet L-77 at a concentration of 105 CFU ml−1 and were vacuum infiltrated into 3-week-old plants. Plants were monitored daily over a 4- to 14-day period for symptom development and bacterial multiplication. Bacterial levels in planta were determined by cutting tomato leaf disks with a boring tool (inner diameter, 0.7 cm), removing whole Arabidopsis leaves, and placing the plant material in 500 μl of 10 mM MgCl2. The disks were completely homogenized, and the resulting suspension, containing the bacteria, was diluted and plated on KB plates with rifampin (50 μg ml−1) and cycloheximide (2 μg ml−1). To determine whether the P. syringae pv. tomato DC3000 strains elicited hypersensitive response (HR) in tobacco, various dilutions of bacterial suspensions were infiltrated into individual leaf panels using a blunt syringe. The areas of infiltration were marked lightly on the back of leaves. Plant assay experiments were repeated at least three times with similar results.
Cya translocation assays.
Cya assays were performed as previously described (60). Briefly, L. esculentum cv. Money Maker plants were inoculated via a blunt syringe with DC3000 strains containing plasmids expressing Cya fusions. Plant tissue was sampled 8 h later using a boring tool with a 0.7-cm inner diameter, placed in 300 μl of 0.1 M HCl, and ground into a fine suspension. Samples were incubated at −20°C overnight, and cyclic AMP (cAMP) levels were determined using the Correlate-EIA cAMP immunoassay kit according to the manufacturer'sdirections (Assay Designs).
RESULTS
DC3000 encodes a functional Tat system.
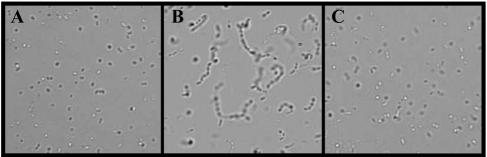
Based on a scan of the recently published DC3000 genome sequence (9), we determined that the DC3000 chromosome possesses a complete tat gene operon comprised of a single copy each of tatA (PSPTO5155), tatB (PSPTO5156), and tatC (PSPTO5157). This was not entirely surprising given that several other pseudomonads were reported to possess Tat export systems (44, 51, 64). The DC3000 tatABC genes shared >83% identity to the same genes from P. aeruginosa (62) and P. putida (48) (data not shown). As a first step towards determining whether the DC3000 tat system was functional, we cloned the DC3000 tatC (tatCDC3000) gene as well as the entire tatABC (tatABCDC3000) operon into plasmid pTrc99A and tested for the ability of these constructs to complement E. coli tat mutants. It is well-known that E. coli strains that are deficient in Tat transport exhibit several phenotypes, including deficiency in cell septation as evidenced by the formation of chains of up to 10 cells and hypersensitivity to hydrophobic drugs and to the anionic detergent SDS (61). Wild-type E. coli strain MC4100 was observed to divide normally in the absence (data not shown) and in the presence of pTatCDC3000 (Fig. 1A). Interestingly, E. coli strain B1LK0 (MC4100 ΔtatC), known to be incapable of Tattransport (8), formed long chains (Fig. 1B) that could beeliminated by expression of DC3000 tatC from plasmid pTatCDC3000 (Fig. 1C), indicating that tatCDC3000 can functionally complement for the loss of tatC in E. coli cells. The introduction of pTatABCDC3000 into the Tat-deficient E. coli strain DADE (MC4100 ΔtatABCD ΔtatE) resulted in a similar restoration of proper cell septation (data not shown).
FIG. 1.
Complementation of E. coli with DC3000 tat genes. Microscopy (1,000× magnification) of (A) wild-type E. coli MC4100 carrying pTatCDC3000; (B) B1LK0 (DtatC mutant derived from MC4100) carrying empty pTrc99A; and (C) B1LK0 carrying pTatCDC3000.
In silico prediction of Tat substrates in DC3000.
We next sought to determine the extent to which the Tat pathway is utilized by DC3000. Genome-wide identification of Tat substrates in a number of bacterial species was reported recently by Dilks and coworkers (25); however, this report does not contain data for DC3000. Therefore, to identify putative Tat substrates in DC3000, we developed a bioinformatic prediction algorithm using a hidden Markov model (HMM) and a Tat signal peptide training set (Fig. 2). Additional information, including the aligned training set and amino acid frequencies, is included in the supplemental data. Using this algorithm, we have identified 59 putative Tat substrates in DC3000, which include a large number of periplasmic binding proteins as well as several hydrolases (2), phosphatases (3), and phospholipases (2) (Table 2). In addition, a number of the predicted substrates appear to be involved in iron acquisition as well as in copper tolerance.
FIG. 2.
Tat secretion signal sequence logo. This is the sequence logo representing the subsequences of known Tat substrates used in the HMM training set to find potential Tat substrates in DC3000. The vertical axis is information content in bits. The height of the amino acids reflects the representation of the residue at that position.
TABLE 2.
Tat substrates of DC3000 identified in silico
| No. | Gene | Predicted gene product |
|---|---|---|
| 1 | PSPTO0249 | Peptide ABC transporter (similar to A. tumafaciens OphA) |
| 2 | PSPTO0315 | Iron ABC transporter, permease protein |
| 3 | PSPTO0491 | Phosphatase (PhoX) |
| 4 | PSPTO0738 | Lipoprotein, putative |
| 5 | PSPTO0759 | Bmp family protein |
| 6 | PSPTO0857 | Conserved hypothetical protein (similar to E. coli DsbG) |
| 7 | PSPTO0985a | Conserved hypothetical protein (similar to sulfite oxidase) |
| 8 | PSPTO1010 | Secreted alkaline phosphatase D (PhoD) |
| 9 | PSPTO1023 | MotA/TolQ/ExbB proton channel family protein |
| 10 | PSPTO1164 | OmpA family protein |
| 11 | PSPTO1212 | Conserved hypothetical protein (putative iron uptake) |
| 12 | PSPTO1309 | HlyD family secretion |
| 13 | PSPTO1340 | Carbonic anhydrase CynT (beta-type carbonic anhydrase) |
| 14 | PSPTO1359 | ABC transporter, permease protein |
| 15 | PSPTO1360 | Bmp family protein (BmpA, basic membrane protein, antigenic) |
| 16 | PSPTO1456 | Multicopper oxidase (similar to E. coli SufI, CumA, CopA) |
| 17 | PSPTO1789 | Dimethyl sulfoxide reductase subunit A (similar to E. coli DmsA) |
| 18 | PSPTO1910 | Conserved hypothetical protein |
| 19 | PSPTO2141 | Cation ABC transporter LraI (manganese binding protein) |
| 21 | PSPTO2155 | Aminotransferase, class V (siderophore biosynthesis) |
| 22 | PSPTO2156 | Part of pvd locus responsible for pyoverdine synthesis |
| 23 | PSPTO2157 | Part of pvd locus responsible for pyoverdine synthesis |
| 24 | PSPTO2160 | Part of pvd locus responsible for pyoverdine synthesis |
| 26 | PSPTO2418 | Branched-chain amino acid transport protein |
| 27 | PSPTO2463 | TonB-dependent siderophore receptor, putative (similar to P. aeruginosa FpvA) |
| 28 | PSPTO2470 | Senescence marker protein-30 family protein (gluconolactonase) |
| 29 | PSPTO2608 | Sco1/SenC family protein |
| 30 | PSPTO2629 | Amino acid ABC transporter HisM (glutamine transport) |
| 31 | PSPTO2654 | 4Fe-4S binding domain protein (xanthine dehydrogenase) |
| 32 | PSPTO2658 | Conserved hypothetical protein (xanthine dehydrogenase) |
| 33 | PSPTO2806 | Dienelactone hydrolase family protein (carboxymethylenebutenolidase) |
| 34 | PSPTO2847 | ThiJ/PfpI family protein |
| 35 | PSPTO3102 | TPR domain protein |
| 36 | PSPTO3242 | TonB-dependent siderophore receptor, putative (similar to P. aeruginosa FpvA) |
| 37 | PSPTO3294 | TonB-dependent siderophore receptor, putative (similar to P. aeruginosa FpvA) |
| 38 | PSPTO3574 | TonB-dependent siderophore receptor, putative (similar to P. aeruginosa FpvA) |
| 39 | PSPTO3598 | Dyp-type peroxidase family protein, iron dependent |
| 40 | PSPTO3624 | Ion transport protein, putative |
| 41 | PSPTO3629 | Thiol:disulfide interchange protein DsbE |
| 42 | PSPTO3631 | Cytochrome c-type biogenesis protein (CcmE) |
| 43 | PSPTO3648 | Acid phosphatase (nonhemolytic phophsolipase) |
| 44 | PSPTO3699 | Methyl-accepting chemotaxis protein |
| 45 | PSPTO3830 | 3-Oxoacyl-(acyl-carrier protein) synthase II (FabF) |
| 46 | PSPTO3914 | Copper resistance protein A (similar to E. coli CopA, PcoA) |
| 47 | PSPTO4078 | Branched-chain amino acid transport protein |
| 48 | PSPTO4351 | Conserved hypothetical protein |
| 49 | PSPTO4352 | Hydrolase, putative (β-lactamase class C, cephalosporin specificity) |
| 50 | PSPTO4369 | Lipoprotein, putative |
| 51 | PSPTO4444a | Toluene tolerance protein, putative |
| 52 | PSPTO4480 | Alkaline phosphatase D, PhoD (similar to B. subtilis PhoD) |
| 53 | PSPTO4858 | Thiol:disulfide interchange protein DsbD |
| 54 | PSPTO4885 | Branched-chain amino acid transport protein |
| 55 | PSPTO5180 | Cystine ABC transporter, per cys binding protein |
| 56 | PSPTO5316 | Sulfonate ABC transport protein |
| 57 | PSPTO5528 | N-acetylmuramoyl-l-alanine amidase (similar to E. coli AmiA) |
| 58 | PSPTO5542 | Periplasmic glucan biosynthesis protein (similar to E. coli MdoG, YdcG) |
| 59 | PSPTOB0005 | Phosphoesterase family protein (nonhemolytic phophsolipase) |
Confirmed by two-dimension gel electrophoresis and tandem mass spectrometry identification to be localized in the supernatant of DC3000 cultures in a Tat-dependent manner (data not shown).
Mutation of DC3000 tatC results in pleiotropic phenotypes.
Previous studies have demonstrated that tat mutants often result in pleiotropic phenotypes. For example, Stanley and coworkers observed that E. coli strains with defective Tat systems exhibit a mutant cell septation phenotype, where chains of up to 10 cells are evident, and a number of other characteristics that are consistent with an outer membrane defect (61). Similarly, Ochsner and coworkers recently showed that a tatC mutant of P. aeruginosa is deficient in a number of virulence-associated phenotypes, including the export of phospholipases, pyoverdine-mediated iron uptake, anaerobic respiration, osmotic stress defense, motility, and biofilm formation (51). Therefore, to test the role of the Tat system with respect to DC3000 physiology and virulence, we explored whether a tatC mutant of DC3000 also exhibited similar pleiotropic phenotypes. This was accomplished by generating an insertion mutation of tatC in DC3000 using suicide plasmid pKnockout-W (66) carrying an internal fragment of tatC. The plasmid was transferred into DC3000 by mating, and single-crossover events were selected by antibiotic resistance. The resultant strain (PBTAT1) was confirmed to have an insertion in tatC. As expected, PBTAT1 exhibited many of the same phenotypes that have previously been reported for tat mutants. For instance, PBTAT1 lacked the fluorescent pigment pyoverdine, which is a siderophore essential for iron acquisition. As shown in Fig. 3A, DC3000 emitted characteristic yellow-green fluorescence when grown on low-iron medium containing 50 mM of the iron-chelating agent 2,2′-dipyridyl, whereas PBTAT1 cells were completely nonfluorescent. Importantly, transformation of pBS1-TatCDC3000, an arabinose-inducible plasmid carrying a wild-type copy of tatCDC3000, into PBTAT1 cells was found to complement for the loss of chromosomal tatC in the presence of 0.2% arabinose (Fig. 3A) but not when arabinose was omitted from the growth plates (data not shown). Furthermore, PBTAT1 cells were completely incapable of growth in the presence of higher concentrations (>100 mM) of the iron chelating agent 2,2′-dipyridyl (Fig. 3B).
FIG. 3.
Multiple phenotypes of DC3000 tatC mutants. (A) Growth of DC3000 and PBTAT1 (tatC mutant derived from DC3000) (upper plate) and DC3000 pBS1-TatCDC3000 and PBTAT1/pBS1-TatCDC3000 (lower plate) on low-iron medium containing 50 μM of 2,2′-dipyridyl. Arabinose at 0.2% was added to the lower plate to induce tatCDC3000 synthesis. DC3000 and PBTAT1 cells were also tested for (B) growth in the presence of 100 μM of the iron chelating agent 2,2′-dipyridyl; (C) formation of swarm halos on semisolid (0.4%, wt/vol) agar plates; and (D) cell division via microscopy (1,000× magnification). (E) Copper susceptibility assays of DC3000 (gray bars) and PBTAT1 (white bars) over a range (0 to 4 mM) of CuSO4. All growth data was normalized to growth of DC3000 in the absence of CuSO4. (F) Subcellular distribution of phospholipase (gray bars) and phosphatase (white bars) activity measured by pNPPC and pNPP assay, respectively. Supernatant and pellet fractions of DC3000 and PBTAT1 cells were prepared as described in Materials and Methods. Data were normalized to the phospholipase or phosphatase activity in supernatant fraction isolated from wild-type DC3000 cells. wt, wild type; sup, supernatant.
PBTAT1 cells were also significantly less motile compared to the wild type, as evidenced by attenuated swarm halos on semisolid (0.4%, wt/vol) agar plates (Fig. 3C). Since no motility-related proteins were found that carried putative Tat signals (Table 2), we suspect that the reduced motility of tatC mutants is due to the characteristic membrane defect commonly associated with Tat-deficient bacteria (61). A similar loss in motility was shown for P. putida, which exhibited destabilized outer membranes due to deletion of tol genes (43). It should be noted that the extent of the outer membrane defect in DC3000 is unclear, as we found that PBTAT1 cells did not exhibit any deficiency in cell septation (Fig. 3D) and were not sensitive to SDS or other hydrophobic drugs (data not shown). The incomplete cell septation phenotype in E. coli results from the cytoplasmic accumulation of two Tat-transported N-acetylmuramoyl-l-alanine amidases, namely, AmiA and AmiC, in tat mutants (6, 37). In DC3000, there are at least four N-acetylmuramoyl-l-alanine amidases (PSPTO0338, PSPTO4634, PSPTO4945, and PSPTO5528), but only one of these, PSPTO5528, carries a putative Tat signal sequence, which likely explains why PBTAT1 cells divide correctly.
The DC3000 tatC mutant is hypersensitive to copper.
In P. syringae, copper resistance is mediated by a number of periplasmic and outer membrane proteins, in particular the CopA, CopB, and CopC proteins, which sequester copper outside of the cytoplasm (11). Interestingly, DC3000 CopA(PSPTO3914), a copper efflux transporter, and CumA (PSPTO1456), a multicopper oxidase CumA, were predicted to be Tat substrates according to our bioinformatic analysis (Table 2). Therefore, we reasoned that PBTAT1 cells would be incapable of transporting CopA and, as a result, might exhibit increased susceptibility to copper. To test this hypothesis, we compared the growth of DC3000 and PBTAT1 cells in media containing increasing concentrations of CuSO4 (Fig. 3E). Both DC3000 and PBTAT1 cells grew equally well in media that contained no CuSO4, and both were completely incapable of growth at higher concentrations of CuSO4 (≥4 mM). Remarkably, wild-type DC3000 cells were fully resistant to CuSO4 concentrations over a range of 0 to 0.25 mM and only exhibited susceptibility when the CuSO4 concentration was supplied at concentrations between 0.5 and 2.0 mM. On the contrary, PBTAT1 cells were extremely sensitive to copper, exhibiting dramatically reduced growth at CuSO4 concentrations as low as 0.03 mM.
Extracellular phospholipase activity is Tat-dependent in DC3000.
P. aeruginosa expresses extracellular phospholipase C (PLC) that is required for the full expression of pathogenicity in both plants and animals (51-53). Two of these, PlcH and PlcN, carry canonical Tat signal peptides and are secreted through the inner membrane via the Tat pathway prior to their secretion outside the cell by the type II secretion system (64). We identified two homologues of P. aeruginosa PlcH, namely PSPTO3648 (23% identical, 46% similar) and PSPTOB0005 (22% identical, 48% similar), in DC3000 which both carry Tat-targeting signals (Table 2) and share 94% identity at the amino acid level with each other. While a role for secreted phospholipases in DC3000 virulence has not been demonstrated, recent studies suggest that these proteins may have a role in pathogenesis: genome-wide expression profiling indicates that PSPTO3648 and PSPTOB0005 are coordinately expressed with hrpL-dependent genes (Greg Martin, Department of Plant Pathology, Cornell University, personal communication), which has subsequently been confirmed using quantitative PCR experiments (Alan Collmer, Department of Plant Pathology, Cornell University, personal communication).
In the present study, we sought to test the hypothesis that these two phospholipases are secreted through Tat in DC3000. First, we assayed supernatant fractions prepared from DC3000 and PBTAT1 cells for PLC activity. When grown in phosphate-rich media, both strains produced undetectable levels of PLC activity (data not shown). However, growth in low-phosphate medium led to a significant accumulation of extracellular PLC activity in DC3000 (Fig. 3F). Under the same growth conditions, PBTAT1 cells failed to secrete detectable levels of PLC activity (Fig. 3F). Expression of DC3000 tatCDC3000 in trans restored extracellular PLC activity to PBTAT1 cells (data not shown). In addition, we observed little to no PLC activity in the cell pellet fraction of DC3000, indicating that PLC secretion is relatively efficient. Unexpectedly, we did not observe any accumulation of PLC in the soluble pellet fraction of PBTAT1 cells, even though no PLC was secreted from this strain. A similar observation was made by Voulhoux et al., who went on to show that PLC activity in P. aeruginosa tatC mutants is predominantly associated with the cell membrane and not the soluble pellet (64). Finally, it is also noteworthy that no extracellular phosphatase activity was detected in supernatant samples prepared from PBTAT1 cells (Fig. 3F).
Inactivation of the Tat system attenuates DC3000 disease symptoms in Arabidopsis thaliana.
Since the Tat system has been linked to virulence in gram-negative bacteria (26, 51, 52) and since phenotypes associated with pathogenesis (e.g., motility, phospholipase activity) were ablated in tatC mutants, we next tested the ability of PBTAT1 cells to cause disease in host plants. Arabidopsis thaliana were inoculated with bacteria and monitored for pathogen-induced chlorotic cell death. As expected, plants infiltrated with wild-type DC3000 exhibited significant disease symptoms when observed 4 and 5 days postinoculation (Fig. 4A, shown for 4 days postinfection). A. thaliana organisms that were challenged with an equivalent number of PBTAT1 cells showed reproducibly less chlorotic cell death. Nearly identical results were obtained when host tomato plants were dipped into bacterial suspensions (data not shown). Assays of bacterial populations in leaves 4 days postinoculation revealed significant differences in the populations achieved by wild-type DC3000 versus the tatC mutant (Fig. 4B). Importantly, the frequency of PBTAT1 cell doublings was nearly identical to that of DC3000 in a variety of media, including LM, KB, low-phosphate minimal medium, and hrp-inducing medium (data not shown), suggesting that reduced survival of PBTAT1 in planta is not due to a gross defect in growth. Collectively, these data indicate that tatC is necessary for DC3000 to elicit chlorotic cell death in A. thaliana leaves and that this change in virulence phenotype is related in part to bacterial fitness in planta.
FIG. 4.
Infiltration of plants with tatC mutants of DC3000. (A) A. thaliana was infected by vacuum-infiltrating bacterial suspensions containing 105 CFU ml−1. Suspensions contained no bacteria (mock), wild-type DC3000, or PBTAT1. Images were captured 4 days postinoculation. (B) CFU of DC3000 and PBTAT1 per milligram of leaf cells determined by cutting leaf disks with a boring tool (inner diameter, 0.7 cm) and plating completely homogenized material, containing the bacteria, on KB plates with rifampin (50 μg ml−1) and cycloheximide (2 μg ml−1). (C) Elicitation of HR by DC3000 and PBTAT1 cells assayed by syringe infiltration of bacterial suspensions of various concentrations into N. tobacco cv. Xanthi. (D) Cya translocation assays performed by inoculating L. esculentum (tomato) plants via a blunt syringe with wild-type or PBTAT1 strains containing plasmids expressing HopPtoN-Cya. Plant tissue was sampled 8 h later using a boring tool (inner diameter, 0.7 cm), and cAMP levels were determined as described in Materials and Methods. All plant assay experiments were repeated at least three times with similar results. t, time; hpi, hours postinfection.
Loss of tatC affects bacterial elicitation of HR in nonhost tobacco leaves.
P. syringae pathovars or nonpathogenic bacteria such as E. coli K12 expressing functional type III secretion systems (TTSS) typically elicit HR in nonhost tobacco (N. tabacum) leaves (2, 33); thus, HR is a useful diagnostic assay for functional TTSS. Accordingly, we inoculated tobacco leaves via a blunt syringe with DC3000 or PBTAT1 cells. Using a dilution series of inocula, 24 h after inoculation we observed that, relative to DC3000, PBTAT1 exhibited a slight but reproducible decrease in confluent tissue collapse at the threshold level of inoculum needed for elicitation of the HR (Fig. 4C). We reasoned that this difference in HR might be due to a defect in type III secretion in tatC cells, perhaps caused by a compromised outer membrane (61) or a defect in the assembly of the type III secretion machinery. To test whether type III secretion of effector proteins into host plants was affected by loss of tatC, we utilized a calmodulin-dependent adenylate cyclase (Cya) reporter system for Hrp-mediated translocation of P. syringae TTSS effectors into plant cells (60). As shown in Fig. 4D, DC3000 efficiently translocated a type III effector protein fused to Cya (HopPtoN-Cya) into tomato (L. esculentumcv. Money Maker) leaf cells, as indicated by Cya-dependent cAMP production. Tomato leaf cells infiltrated with PBTAT1 produced less cAMP, suggesting that inactivation of the Tat system reduces the efficiency of type III translocation by >1.5-fold.
Construction of a broad-host genetic reporter of Tat transport.
In order to determine the contribution of Tat-secreted proteins to virulence in the absence of the pleiotropic effects caused by deletion of tatC, we developed a reporter system for identifying Tat-secreted virulence factors. A number of powerful genetic reporters have been developed for assaying Tat function (21, 32, 35), including an engineered reporter construct for Tat-specific transport based on the green fluorescent protein (GFP) (20, 21). In the case of the latter, a fusion was made between a Tat signal peptide (ssTorA), GFP, and a C-terminal SsrA degradation peptide. The premise behind this reporter construct is as follows: an unstable version of GFP (GFP-SsrA) is directed to the periplasm via the Tat pathway while any nontransported GFP retained in the cytoplasm is efficiently targeted for degradation by ClpXP by virtue of the carboxy-terminal SsrA degradation signal. As a result, cells with a functional Tat system and expressing TGS accumulate GFP in the periplasm and appear as green “halos” under a fluorescent microscope (24 and data not shown).
In the present study, we sought to generate a broad-host version of this reporter that would reliably report Tat function in P. syringae (or other gram-negative bacteria). This was accomplished using the broad-host Tat reporter plasmid pMMB-TGS-Gm. To test whether this plasmid retained the ability to correctly report functional Tat export, we transformed wild-type E. coli as well as Tat-deficient E. coli strains with pMMB-TGS-Gm and measured their fluorescence output using flow cytometric analysis. We found that wild-type E. coli strain MC4100 carrying pMMB-TGS-Gm was highly fluorescent (Fig. 5A), while both B1LK0 and DADE cells carrying the same plasmid were found to be completely nonfluorescent (Fig. 5A for B1LK0; DADE results not shown). Finally, to confirm that the subcellular distribution of GFP corresponded to the fluorescent phenotype observed via flow cytometry, we performed subcellular fractionation of E. coli cells followed by immunoblotting with antiserum against GFP and found that, as expected, GFP was localized entirely to the periplasm in MC4100 but was virtually absent in both the cytoplasmic and periplasmic fractions of B1LK0 cells (data not shown).
FIG. 5.
Broad-host genetic reporter of Tat transport based on GFP-SsrA. Flow cytometric analysis of (A) wild-type E. coli MC4100 and B1LK0 and (B) DC3000 and PBTAT1 cells all expressing pMMB-TGS. Histograms depict number of cells (events) versus their fluorescence intensity (FL1-H). Standard error of three replicate experiments was <10%. M, mean fluorescence.
Next, we tested whether pMMB-TGS-Gm could be used to report Tat transport directly in DC3000. We reasoned this would be feasible because (i) unstable GFP-SsrA variants had previously been tested in P. putida and were found to be efficiently degraded (3), and (ii) the E. coli TorA signal sequence was recognized by tatABCDC3000 (see above) and thus should direct transport of TGS into the periplasm of DC3000. Accordingly, wild-type DC3000 was transformed with pMMB-TGS-Gm and assayed for transport of TGS to the periplasm. As shown in Fig. 5B, DC3000 expressing TGS was found to emit a low but reproducible level of cell fluorescence. This level was lower than that observed for E. coli MC4100 expressing TGS from the same vector but was still more than 10-fold more fluorescent than PBTAT1 cells expressing TGS (Fig. 5B). These results confirmed that our broad-host vector was an effective genetic reporter of Tat transport in E. coli and DC3000.
Using this reporter assay, we confirmed Tat-specific transport for the putative virulence factors PSPTO3648 and PSPTOB0005 (hereafter PlcA1 and PlcA2, respectively), which were previously found to be transcriptionally regulated with hrp-regulated genes. For these experiments, we used E. coli as the host organism since our previous data indicated that the E. coli and P. syringae Tat systems are functionally interchangeable. Fusion of the PlcA1 signal peptide (Fig. 6A, ssPlcA1) to GFP-SsrA resulted in efficient translocation of ssPlcA1-GFP-SsrA to the periplasm of wild-type E. coli cells (Fig. 6B). In contrast, B1LK0 cells expressing ssPlcA1-GFP-SsrA were completely nonfluorescent. We did not test ssPlcA2, since this signal peptide was nearly identical to ssPlcA1, varying only by 5 amino acids (Fig. 6A). However, we did confirm two additional Tat signal peptides using this assay, namely ssCopA and ssCumA.
FIG. 6.
Confirmation of DC3000 Tat substrates using GFP-SsrA reporter. (A) Predicted signal peptides for PlcA1 (PSPTO3648) and PlcA2 (PSPTOB0005). Gray shaded area denotes amino acid dissimilarities. Underlined, boldface text denotes a Tat consensus motif. Space between the last three amino acids (A/HE or S/HE) denotes peptidase cleavage site as predicted by SignalP, version 3.0 (http://www.cbs.dtu.dk/services/SignalP/). Mean fluorescence measured by flow cytometric analysis of MC4100 (gray bars) or B1LK0 (white bars) expressing (B) putative Tat signal peptides cloned in pMMB-GGS or (C) full-length Tat substrates cloned in pMMB-GGS. All mean fluorescence (M) data are the averages of three replicate experiments. Western blot analysis of cytoplasmic (cyt) and periplasmic (per) fractions generated from MC4100 cells expressing (D) ssPlcA1-GFP-SsrA or (E) full-length PlcA1 fused to GFP-SsrA. Blots were first probed with anti-GFP serum and then stripped and reprobed with anti-GroEL serum to ensure the quality of the fractionation procedure.
Since features of the mature protein are likely to affect its targeting and transport through the Tat system, we next constructed full-length substrate fusions to GFP-SsrA. Fusions of PlcA1, PlcA2, and CopA all were transported to the periplasm in a Tat-dependent manner as determined by whole-cell fluorescence (Fig. 6C). As a control, the cytoplasmic protein Gus was fused to GFP-SsrA, and as expected, both wild-type and ΔtatC cells expressing this construct were nonfluorescent (data not shown). Interestingly, even though ssCumA could deliver GFP-SsrA to the periplasm, full-length CumA-GFP-SsrA was not transported. The reason for this is not clear, although cells expressing CumA-GFP-SsrA grew very slowly relative to other GFP-SsrA fusion constructs, suggesting that the CumA fusion might be toxic. Finally, to confirm that whole-cell fluorescence emitted by our assay is indicative of periplasmic localization, we performed subcellular fractions of ssPlcA1-GFP-SsrA and full-length PlcA1 fused to GFP-SsrA. Western blot analysis confirmed that both of these reporter constructs were correctly localized in the periplasmic space (Fig. 6D and E).
Extracellular secretion of Tat substrates by the DC3000 Gsp system.
To function as virulence factors, Tat substrates such as PlcA1 and PlcA2 must be subsequently delivered out of the periplasm so that they may interact with the host. The type II secretion system is a likely candidate for this process. Thus, we tested whether Tat substrates could be delivered to the extracellular space by the type II secretion machinery. First, we generated insertion mutations of gspD and gspE in DC3000 using pKnockout-W. The gspD and gspE genes were selected based on the observation that homologues of these genes in Erwinia chrysanthemi are essential for type II secretion (42). The resulting strains, PBGspD and PBGspE, were confirmed to have insertions in gspD and gspE, respectively. We assayed supernatant fractions prepared from PBGspD and PBGspE cells for PLC activity and observed no detectable PLC activity (data not shown). Since PLC expression levels might be downregulated in a gsp or tatC strain background, we transformed PBGspD, PBGspE, PBTAT1, and DC3000 with an arabinose-inducible PlcA1 expression vector. Under low-phosphate growth conditions and in the presence of 0.2% arabinose, only DC3000 cells exhibited extracellular PLC activity while PBGspD, PBGspE, and PBTAT cells all failed to secrete detectable levels of PLC activity (Fig. 7A). The level of PLC activity in DC3000 overexpressing PlcA1 was significantly lower than that observed in wild-type DC3000, suggesting that overexpressing PlcA1 leads to saturation of the Tat and/or type II translocation machinery. This type of saturation has been previously observed for Tat substrates in E. coli (12, 20).
FIG. 7.
Extracellular secretion of Tat substrates by the Gsp system. (A) Subcellular distribution of phospholipase activity measured by the pNPPC assay. Expression of PlcA1 from plasmid pBS1-PlcA1 is denoted as PlcA+. Supernatant and pellet fractions were prepared as described in Materials and Methods. Data were normalized to the phospholipase activity in supernatant fraction isolated from DC3000. (B) A. thaliana was infected by vacuum infiltrating bacterial suspensions containing 105 CFU ml−1. Suspensions contained no bacteria (mock), wild-type DC3000, PBGspD, or PBGspE. Images were captured 4 days postinoculation. (C) Elicitation of HR by DC3000 and PBGspD cells assayed by syringe infiltration of bacterial suspensions of various concentrations into N. tobacco cv. Xanthi. All plant assay experiments were repeated at least three times with similar results.
Finally, we tested the ability of PBGspD and PBGspE cells to cause disease in host plants by inoculating A. thaliana with bacteria and monitoring pathogen-induced chlorotic cell death. Remarkably, plants infiltrated with PBGspD and PBGspE cells showed significantly less chlorotic cell death relative to wild-type DC3000 observed 4 and 5 days postinoculation (Fig. 7B, shown for 4 days postinfection). Moreover, PBGspD and PBGspE elicited HR at a level that was indistinguishable from that of wild-type DC3000 (Fig. 7C) and did not exhibit any growth defects in a variety of liquid growth media (data not shown), indicating that the reduced virulence phenotype was likely due to secreted type II substrates.
DISCUSSION
In the present study we explored the contribution of the Tat pathway to the physiology of P. syringae cv. tomato DC3000. We found that inactivation of the Tat system in DC3000 results in multiple complex phenotypes, including loss of motility on soft agar plates, deficiency in siderophore synthesis and iron acquisition, sensitivity to copper, loss of extracellular phospholipase activity, and attenuated virulence in host plant leaves. Further, we provide evidence that decreased virulence of tatC mutants likely arises from at least three major factors: (i) compromised fitness of bacteria in planta; (ii) decreased efficiency of type III translocation; and (iii) cytoplasmically retained virulence factors.
An inspection of the putative Tat substrates identified using bioinformatics (Table 2) is helpful in explaining how some of these phenotypes are likely to arise. For instance, at least 10 of the predicted Tat substrates are involved in iron metabolism, most notably the pvd locus responsible for biosynthesis of pyoverdine (Pvd), the TonB-dependent iron transporters resembling P. aeruginosa FvpA, and the ferroxidase CopA. Under iron-limiting conditions, pseudomonads produce iron-scavenging siderophores such as pyoverdine. Pyoverdine is secreted into the extracellular environment where it chelates iron, and the resulting ferri-pyoverdine complexes are transported back into the bacteria by a cell surface receptor protein (e.g., FpvA). In P. aeruginosa, pyoverdine has been linked to pathogenesis in two distinct ways: first, in animal models, infection requires the production of pyoverdine, which enables the bacteria to acquire iron from host proteins (46); second, pyoverdine has been shown to act as a signaling molecule inducing the production of secreted virulence factors (41). Similar to our observations, Ochsner and coworkers did not detect pyoverdine in the supernatants of P. aeruginosa tatC mutants (51). However, a similar connection between pyoverdine and P. syringae pathogenesis has not yet been demonstrated. Nonetheless, it is possible that the observed decrease in virulence exhibited by tatC mutants of DC3000 was due in part to a lack of extracellular pyoverdine or other siderophores, preventing bacteria from acquiring iron in planta or from signaling the induction of virulence factors.
An unexpected discovery was the prediction that the thiol:disulfide interchange proteins DsbD, DsbE, and DsbG, which transit the Sec pathway in E. coli, each contain an Arg-Arg pair in their signal peptides, although DsbD and DsbE lack a canonical Tat motif (S-R-R-X-F-L-K). Another disulfide bond protein, the powerful periplasmic disulfide catalyst DsbA, is required for full virulence of both P. syringae DC3000 and P. aeruginosa on A. thaliana (40, 53) and for efficient type III secretion in DC3000 (40). While DC3000 DsbA does not carry a Tat-specific signal peptide, we have observed that localization of this protein to the periplasm is significantly impaired in tatC mutants relative to wild-type DC3000 based on proteomic analysis (two-dimensional protein electrophoresis; P. A. Bronstein, L. Choe, A. Collmer, K. H. Lee, and M. P. DeLisa, unpublished observations). Taken together, it appears that deletion of the Tat machinery in DC3000 disrupts the disulfide bond machinery, which in turn may contribute to a reduction in virulence of the tatC mutant.
In addition to the above phenotypes, we also identified two putative virulence factors, namely, PlcA1 (PSPTO3648) and PlcA2 (PSPTOB0005), which carry Tat-specific targeting signals. The recent observation that both of these phospholipase C homologs were coregulated with approximately 120 other hrpL-dependent genes (Greg Martin, personal communication) suggests that they may be virulence factors. This is intriguing in light of the fact that even though plcA1 possesses a conserved cis element (called the “hrp box”) in its promoter, neither plcA1 nor plcA2 has been identified using genome-wide approaches for hrpL-regulated effector proteins (13, 15, 68). Evidence that the phospholipase Cs from pseudomonads are significant virulence factors in pathogenesis is well documented. For instance, a plcH/plcN double mutant of P. aeruginosa exhibits attenuated virulence in the lethal paralysis model of Caenorhabditis elegans (19), and a P. aeruginosa plcH mutant has reduced mortality in a mouse burn model and decreased virulence in A. thaliana (53, 54). It should be noted, however, that the phospholipase superfamily of enzymes (to which the P. syringae and P. aeruginosa members belong) is comprised of phospholipase Cs as well as phosphatases. Thus, it is possible that an enzyme with phosphodiesterase activity, which is not necessarily a phospholipase C, could have hydrolyzed NPPC in our assays. Consequently, it is currently unclear whether the NPPC activity measured in DC3000 but not PBTAT1 supernatants is due to a phospholipase C or a phosphatase.
Furthermore, while it remains to be tested whether DC3000 PlcA1 and PlcA2 contribute directly to pathogenesis, our results show that these enzymes are secreted to the extracellular environment, using the Tat system to transit the inner membrane in a folded conformation and the Gsp (type II) to traverse the outer membrane. A similar two-step translocation of phospholipase PlcH and PlcN was observed to occur in P. aeruginosa (64). More importantly, we show for the first time that inactivation of either the Tat or type II secretion systems resulted in a significant attenuation of virulence, suggesting that two-step secretion might provide a complementary virulence mechanism to the predominant TTSS of P. syringae. Collectively, this evidence supports the notion that virulence of DC3000 is multifactorial, relying upon a network of secretion systems that is now extended to include the Tat and Gsp machineries.
Finally, the approach used in this study to demonstrate that PlcA1 and PlcA2 are secreted by a Tat/Gsp-dependent mechanism is illustrative of a powerful screening platform for the identification of Tat-targeted secreted virulence factors that should be generally applicable to any gram-negative bacterium. This screen employs standard tools to perform a genome-wide bioinformatic search to generate a list of putative Tat substrates. These substrates can then be rapidly cloned into the pMMB-GGS broad-host reporter plasmid as either signal peptide or full-length fusions to GFP-SsrA, provided the host organism possesses the SsrA degradation mechanism. The latter is likely as the SsrA RNA molecule is highly conserved in eubacteria. We are currently testing whether this broad-host reporter is useful in other strains of gram-negative bacteria. The demonstration that full-length proteins can be Tat transported as fusions to GFP-SsrA was not previously shown and highlights the versatility of this genetic reporter. A further advantage of this newly developed broad-host vector is its “tune-ability” relative to the original pTGS construct reported previously (21). Whereas the fluorescence emission from cells expressing pTGS was relatively insensitive to a wide range of inducer (arabinose) concentrations, the fluorescence level emanating from wild-type cells carrying pMMB-TGS was easily titrated to very high levels by varying the inducer (IPTG) concentration without concomitant fluorescence emission from tatC mutant cells (data not shown). Once confirmation of Tat transport is complete, the use of gsp mutants can then be used to confirm extracellular localization. To facilitate these latter studies, we are currently exploring whether our full-length Tat/Gsp-specific substrates fused to GFP-SsrA are localized into the extracellular environment. This entire process is entirely iterative, such that experimental confirmation of Tat substrates will allow the Tat prediction algorithm to be extended beyond canonical Tat motifs, thereby improving the predictive capability of the method. We expect that Tat substrate identification combined with known information regarding the three-dimensional structure of these substrates will help to define the true capacity of the Tat transporter.
Acknowledgments
We thank Alan Collmer and Tracy Palmer for helpful discussions of the manuscript and Gina Gremona for her initial construction of plasmid pMMB-TGS. We also thank Kelvin Lee and Leila Choe for their proteomic analysis of DC3000 cultures. We appreciate the kind gift of plasmid pBS1 from Bryan Swingle.
This work was supported by the New York State Office of Science, Technology, and Academic Research (NYSTAR) in the form of a James D. Watson Young Investigator Award to M.P.D.
REFERENCES
- 1.Alfano, J. R., and A. Collmer. 1996. Bacterial pathogens in plants: life up against the wall. Plant Cell. 8:1683-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385-414. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjorn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berks, B. C., T. Palmer, and F. Sargent. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47:187-254. [DOI] [PubMed] [Google Scholar]
- 5.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 6.Bernhardt, T. G., and P. A. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogsch, E., S. Brink, and C. Robinson. 1997. Pathway specificity for a delta pH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J. 16:3851-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogsch, E. G., F. Sargent, N. R. Stanley, B. C. Berks, C. Robinson, and T. Palmer. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273:18003-18006. [DOI] [PubMed] [Google Scholar]
- 9.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cha, J. S., and D. A. Cooksey. 1991. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. USA 88:8915-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chanal, A., C. L. Santini, and L. F. Wu. 2003. Specific inhibition of the translocation of a subset of Escherichia coli TAT substrates by the TorA signal peptide. J. Mol. Biol. 327:563-570. [DOI] [PubMed] [Google Scholar]
- 13.Chang, J. H., J. M. Urbach, T. F. Law, L. W. Arnold, A. Hu, S. Gombar, S. R. Grant, F. M. Ausubel, and J. L. Dangl. 2005. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 102:2549-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collmer, A., J. L. Badel, A. O. Charkowski, W. L. Deng, D. E. Fouts, A. R. Ramos, A. H. Rehm, D. M. Anderson, O. Schneewind, K. van Dijk, and J. R. Alfano. 2000. Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc. Natl. Acad. Sci. USA 97:8770-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collmer, A., M. Lindeberg, T. Petnicki-Ocwieja, D. J. Schneider, and J. R. Alfano. 2002. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 10:462-469. [DOI] [PubMed] [Google Scholar]
- 16.Cota-Gomez, A., A. I. Vasil, J. Kadurugamuwa, T. J. Beveridge, H. P. Schweizer, and M. L. Vasil. 1997. PlcR1 and PlcR2 are putative calcium-binding proteins required for secretion of the hemolytic phospholipase C of Pseudomonas aeruginosa. Infect. Immun. 65:2904-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cristobal, S., J. W. de Gier, H. Nielsen, and G. von Heijne. 1999. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 18:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuppels, D. A. 1986. Generation and characterization of Tn5 insertions in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darby, C., C. L. Cosma, J. H. Thomas, and C. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLisa, M. P., P. Lee, T. Palmer, and G. Georgiou. 2004. Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway. J. Bacteriol. 186:366-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLisa, M. P., P. Samuelson, T. Palmer, and G. Georgiou. 2002. Genetic analysis of the twin arginine translocator secretion pathway in bacteria. J. Biol. Chem. 277:29825-29831. [DOI] [PubMed] [Google Scholar]
- 22.DeLisa, M. P., D. Tullman, and G. Georgiou. 2003. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc. Natl. Acad. Sci. USA 100:6115-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derman, A. I., W. A. Prinz, D. Belin, and J. Beckwith. 1993. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science 262:1744-1747. [DOI] [PubMed] [Google Scholar]
- 24.Dery, K. J., B. Soballe, M. S. Witherspoon, D. Bui, R. Koch, D. J. Sherratt, and M. E. Tolmasky. 2003. The aminoglycoside 6′-N-acetyltransferase type Ib encoded by Tn1331 is evenly distributed within the cell's cytoplasm. Antimicrob. Agents Chemother. 47:2897-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dilks, K., R. W. Rose, E. Hartmann, and M. Pohlschroder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding, Z., and P. J. Christie. 2003. Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J. Bacteriol. 185:760-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 28.Fisher, A. C., and M. P. DeLisa. Submitted for publication.
- 29.Gopalan, S., D. W. Bauer, J. R. Alfano, A. O. Loniello, S. Y. He, and A. Collmer. 1996. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 8:1095-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross, D. C., and Y. S. Cody. 1985. Mechanisms of plant pathogenesis by Pseudomonas species. Can. J. Microbiol. 31:403-410. [Google Scholar]
- 31.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning, a practical approach. IRL Press, Oxford, United Kingdom.
- 32.Hicks, M. G., P. A. Lee, G. Georgiou, B. C. Berks, and T. Palmer. 2005. Positive selection for loss-of-function tat mutations identifies critical residues required for TatA activity. J. Bacteriol. 187:2920-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang, H. C., R. Schuurink, T. P. Denny, M. M. Atkinson, C. J. Baker, I. Yucel, S. W. Hutcheson, and A. Collmer. 1988. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J. Bacteriol. 170:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 35.Ize, B., F. Gerard, M. Zhang, A. Chanal, R. Voulhoux, T. Palmer, A. Filloux, and L. F. Wu. 2002. In vivo dissection of the Tat translocation pathway in Escherichia coli. J. Mol. Biol. 317:327-335. [DOI] [PubMed] [Google Scholar]
- 36.Ize, B., I. Porcelli, S. Lucchini, J. C. Hinton, B. C. Berks, and T. Palmer. 2004. Novel phenotypes of Escherichia coli tat mutants revealed by global gene expression and phenotypic analysis. J. Biol. Chem. 279:47543-47554. [DOI] [PubMed] [Google Scholar]
- 37.Ize, B., N. R. Stanley, G. Buchanan, and T. Palmer. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48:1183-1193. [DOI] [PubMed] [Google Scholar]
- 38.Keane, P. J., A. Kerr, and P. B. New. 1970. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust. J. Biol. Sci. 23:585-595. [Google Scholar]
- 39.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanine and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 40.Kloek, A. P., D. M. Brooks, and B. N. Kunkel. 2000. A dsbA mutant of Pseudomonas syringae exhibits reduced virulence and partial impairment of type III secretion. Mol. Plant Pathol. 1:139-150. [DOI] [PubMed] [Google Scholar]
- 41.Lamont, I. L., P. A. Beare, U. Ochsner, A. I. Vasil, and M. L. Vasil. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 99:7072-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindeberg, M., and A. Collmer. 1992. Analysis of eight out genes in a cluster required for pectic enzyme secretion by Erwinia chrysanthemi: sequence comparison with secretion genes from other gram-negative bacteria. J. Bacteriol. 174:7385-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Llamas, M. A., J. L. Ramos, and J. J. Rodriguez-Herva. 2000. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J. Bacteriol. 182:4764-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma, Q., Y. Zhai, J. C. Schneider, T. M. Ramseier, and M. H. Saier, Jr. 2003. Protein secretion systems of Pseudomonas aeruginosa and P. fluorescens. Biochim. Biophys. Acta 1611:223-233. [DOI] [PubMed] [Google Scholar]
- 45.Masip, L., J. L. Pan, S. Haldar, J. E. Penner-Hahn, M. P. DeLisa, G. Georgiou, J. C. Bardwell, and J. F. Collet. 2004. An engineered pathway for the formation of protein disulfide bonds. Science 303:1185-1189. [DOI] [PubMed] [Google Scholar]
- 46.Meyer, J. M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 48.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. Chris Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 49.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197-203. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 51.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pradel, N., C. Ye, V. Livrelli, J. Xu, B. Joly, and L. F. Wu. 2003. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71:4908-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahme, L. G., F. M. Ausubel, H. Cao, E. Drenkard, B. C. Goumnerov, G. W. Lau, S. Mahajan-Miklos, J. Plotnikova, M. W. Tan, J. Tsongalis, C. L. Walendziewicz, and R. G. Tompkins. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97:8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 55.Randall, L. L., and S. J. Hardy. 1986. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell 46:921-928. [DOI] [PubMed] [Google Scholar]
- 56.Robinson, C., and A. Bolhuis. 2001. Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell. Biol. 2:350-356. [DOI] [PubMed] [Google Scholar]
- 57.Rodrigue, A., A. Chanal, K. Beck, M. Muller, and L. F. Wu. 1999. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J. Biol. Chem. 274:13223-13228. [DOI] [PubMed] [Google Scholar]
- 58.Roeder, D. L., and A. Collmer. 1985. Marker-exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi. J. Bacteriol. 164:51-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanders, C., N. Wethkamp, and H. Lill. 2001. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol. Microbiol. 41:241-246. [DOI] [PubMed] [Google Scholar]
- 60.Schechter, L. M., K. A. Roberts, Y. Jamir, J. R. Alfano, and A. Collmer. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanley, N. R., K. Findlay, B. C. Berks, and T. Palmer. 2001. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J. Bacteriol. 183:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 63.Vidaver, A. K. 1967. Synthetic and complex media for the rapid detection of phytopathogenic pseudomonads: effect of the carbon source. Appl. Microbiol. 15:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voulhoux, R., G. Ball, B. Ize, M. L. Vasil, A. Lazdunski, L. F. Wu, and A. Filloux. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20:6735-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wexler, M., F. Sargent, R. L. Jack, N. R. Stanley, E. G. Bogsch, C. Robinson, B. C. Berks, and T. Palmer. 2000. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J. Biol. Chem. 275:16717-16722. [DOI] [PubMed] [Google Scholar]
- 66.Windgassen, M., A. Urban, and K. E. Jaeger. 2000. Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 193:201-205. [DOI] [PubMed] [Google Scholar]
- 67.Yen, M. R., Y. H. Tseng, E. H. Nguyen, L. F. Wu, and M. H. Saier, Jr. 2002. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 177:441-450. [DOI] [PubMed] [Google Scholar]
- 68.Zwiesler-Vollick, J., A. E. Plovanich-Jones, K. Nomura, S. Bandyopadhyay, V. Joardar, B. N. Kunkel, and S. Y. He. 2002. Identification of novel hrp-regulated genes through functional genomic analysis of the Pseudomonas syringae pv. tomato DC3000 genome. Mol. Microbiol. 45:1207-1218. [DOI] [PubMed] [Google Scholar]