Abstract
Analysis of the genome sequence of Caulobacter crescentus predicts 67 TonB-dependent outer membrane proteins. To demonstrate that among them are proteins that transport nutrients other than chelated Fe3+ and vitamin B12—the substrates hitherto known to be transported by TonB-dependent transporters—the outer membrane protein profile of cells grown on different substrates was determined by two-dimensional electrophoresis. Maltose induced the synthesis of a hitherto unknown 99.5-kDa protein, designated here as MalA, encoded by the cc2287 genomic locus. MalA mediated growth on maltodextrins and transported [14C]maltodextrins from [14C]maltose to [14C]maltopentaose. [14C]maltose transport showed biphasic kinetics, with a fast initial rate and a slower second rate. The initial transport had a Kd of 0.2 μM, while the second transport had a Kd of 5 μM. It is proposed that the fast rate reflects binding to MalA and the second rate reflects transport into the cells. Energy depletion of cells by 100 μM carbonyl cyanide 3-chlorophenylhydrazone abolished maltose binding and transport. Deletion of the malA gene diminished maltose transport to 1% of the wild-type malA strain and impaired transport of the larger maltodextrins. The malA mutant was unable to grow on maltodextrins larger than maltotetraose. Deletion of two C. crescentus genes homologous to the exbB exbD genes of Escherichia coli abolished [14C]maltodextrin binding and transport and growth on maltodextrins larger than maltotetraose. These mutants also showed impaired growth on Fe3+-rhodotorulate as the sole iron source, which provided evidence of energy-coupled transport. Unexpectedly, a deletion mutant of a tonB homolog transported maltose at the wild-type rate and grew on all maltodextrins tested. Since Fe3+-rhodotorulate served as an iron source for the tonB mutant, an additional gene encoding a protein with a TonB function is postulated. Permeation of maltose and maltotriose through the outer membrane of the C. crescentus malA mutant was slower than permeation through the outer membrane of an E. coli lamB mutant, which suggests a low porin activity in C. crescentus. The pores of the C. crescentus porins are slightly larger than those of E. coli K-12, since maltotetraose supported growth of the C. crescentus malA mutant but failed to support growth of the E. coli lamB mutant. The data are consistent with the proposal that binding of maltodextrins to MalA requires energy and MalA actively transports maltodextrins with Kd values 1,000-fold smaller than those for the LamB porin and 100-fold larger than those for the vitamin B12 and ferric siderophore outer membrane transporters. MalA is the first example of an outer membrane protein for which an ExbB/ExbD-dependent transport of a nutrient other than iron and vitamin B12 has been demonstrated.
Caulobacter crescentus has been studied well as a model of bacterial differentiation (30, 39, 55). Since the early studies on the conditions for growth of C. crescentus (summarized in references 44 and 54), only a few publications on intermediary metabolism and none on nutrient transport mechanisms have been published.
Analysis of the genome sequence of C. crescentus predicts 67 TonB-dependent outer membrane transport proteins and no porin of the Escherichia coli OmpC/F type (40; A. Lupas, personal communication). In gram-negative bacteria, TonB-dependent outer membrane proteins transport bacterial and fungal Fe3+ siderophores, heme, Fe3+ provided by transferrin and lactoferrin of the mammalian hosts, and vitamin B12 (7, 10, 14-19, 34, 46, 51, 60). The low level of iron available to bacteria requires extraction from media and very strong binding to outer membrane proteins, from which a vectorial transport into the periplasm occurs. In contrast to diffusion through porins, TonB-dependent transport requires energy, which is provided by the proton motive force of the cytoplasmic membrane (5). The crystal structures of TonB-dependent outer membrane transport proteins reveal β-barrels closed by globular domains that insert from the periplasm into the channel formed by the β-barrel and close them completely (14, 24, 25, 38, 62). Electrochemical energy of the cytoplasmic membrane harvested by the TonB-ExbB-ExbD protein complex (6, 46) and transferred via TonB to the transporters must cause a structural change in the transporters that releases the substrates from their binding sites and opens the channels through which the substrates move into the periplasm.
If the prediction of 67 TonB-dependent proteins in C. crescentus holds true, substrates other than ferric iron, ferric iron complexes, and vitamin B12 are likely to be transported by TonB-dependent proteins. C. crescentus is mainly found in freshwater lakes, streams, and ponds with low nutrient content. Strong binding of the scarce nutrients other than iron and vitamin B12 to surface-exposed proteins and subsequent active transport across the outer membrane could be a mechanism to acquire sufficient nutrients. To test this hypothesis, we identified one of the 67 outer membrane proteins as being involved in the uptake of maltodextrins and showed that uptake requires the presence of an outer membrane protein and proteins homologous to the E. coli ExbB ExbD proteins. Since ExbB ExbD homologs are also required for growth promotion of C. crescentus by Fe3+-rhodotorulate on an iron-limited medium, it is likely that the requirement of ExbB ExbD reflects the requirement for energy-dependent outer membrane transport.
MATERIALS AND METHODS
Bacteria, plasmids, and media.
The C. crescentus and E. coli strains and the plasmids used in this study are listed in Table 1. C. crescentus cells were grown at 30°C in PYE rich medium or in M2 salt minimal medium (22) supplemented with 0.3% glucose or 0.3% maltose. E. coli cells were grown at 37°C on tryptone-yeast extract medium (33). Antibiotics were used at the following concentrations (in micrograms per milliliter) for C. crescentus: ampicillin, 50 on plates, 7.5 in liquid culture; chloramphenicol, 2; tetracycline, 2 on plates, 1 in liquid culture; gentamicin, 50 on plates, 25 in liquid culture; spectinomycin, 50 on plates, 25 in liquid culture; and streptomycin, 5. Antibiotics were used at the following concentrations (in micrograms per milliliter) for E. coli: ampicillin, 50 in liquid culture; chloramphenicol, 25; gentamicin, 50 in liquid culture; spectinomycin, 50 in liquid culture; streptomycin, 30; kanamycin, 30; and nalidixic acid, 20. C. crescentus JS1003 is resistant to nalidixic acid, ampicillin, and kanamycin. Growth promotion was tested on M2 salt agar plates on which 108 bacteria were seeded in 3 ml M2 salt soft agar. Ten microliters of the nutrients to be tested were placed on sterile filter paper disks with a 6-mm diameter. Bacterial growth was scored around the disks after 16 and 24 h of incubation at 30°C. The maltodextrins usedwere purchased from Sigma-Aldrich Chemie, Taufkirchen, Germany, and their purity was verified by high-pressure liquid chromatography (HPLC) analysis. [14C]maltotriose, [14C]maltotetraose, and [14C]maltopentaose were kindly provided by Winfried Boos, University of Konstanz, Germany. They were prepared by a published procedure (41) and had a specific radioactivity of 300 μCi/μmol.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| C. crescentus | ||
| NA1000 | syn-1000, synchronizable derivative of C. crescentus wild-type strain | 23 |
| JS1003 | NA1000 rsaA::KSac Kmr cassette, Ampr | 21 |
| HB2003 | JS1003 malA::Ω cassette, Kmr Ampr Spcr Strr | This study |
| HB2004 | JS1003 tonB::Ω cassette, Kmr Ampr Spcr Strr | This study |
| HB2006 | HB2004 exbBD1::Gmr cassette, Kmr Ampr Gmr | This study |
| HB2007 | JS1003 exbBD1::Gmr cassette, Kmr Ampr Gmr | This study |
| E. coli | ||
| BL21(DE) omp8 | hsdSB(rB− mB−) gal ompT dcm (DE3) ΔlamB ompF::Tn5 ΔompA ΔompC Neor | 48 |
| S17-1 λ pir | hsdR hsdM+ RP4-2-Tc::Mu Km::Tn7 (Tpr Smr) Tra+ λ pir | 1 |
| KB419 | lamB | 9 |
| H2300 | AB2847 ΔtonB trp | K. Hantke |
| Plasmids | ||
| pBAD/Myc-HisB | araBAD promoter, Ampr | Invitrogen |
| pET-27b(+) | T7 lac promoter, Kmr | Novagen |
| pBHR1 | Broad-host-range vector, Camr Kmr | MoBiTec |
| pHP45Ω | Ω cassette (Smr/Spcr), Ampr | 47 |
| pBSL202 | R6K oriV, RP4 oriT, Ampr Gmr | 1 |
| pBSL204 | R6K oriV, RP4 oriT, Ampr Tetr | 1 |
| pMR20 | RK2-based Tetr broad-host-range vector | 31 |
| pNPTS138 | Kmr derivative of pLITMUS with sacB and oriT | 31 |
| pNPTS138Tet | pNPTS138, Tetr Kmr | This study |
| pMR2287 | pMR20 malA, Tetr | This study |
| pBAD2287His | pBAD/Myc-HisB, MalAHis6, Ampr | This study |
| pHB138 | pNPTS138Tet ΔmalA cc2286Ωcc2288, Kmr Tetr Spcr Strr | This study |
| pHB139 | pNPTS138Tet ΔtonB cc2326Ωcc2328, Kmr Tetr Spcr Strr | This study |
| pHB140 | pNPTS138Tet, ΔexbB exbD, cc2334Gmr cc2337 Kmr Tetr | This study |
| pHB141 | pNPTS138Tet, ΔtolR tolQ, cc3231Gmr cc3234 Kmr Tetr | This study |
| pHB142 | pNPTS138Tet, ΔtolR tolQ, cc3231Ωcc3234, Kmr Tetr Spcr Strr | This study |
| pETcctonB | pET-27(b)+ tonB Kmr | This study |
| pMRexb1 | pMR20 exbBD, Tetr | This study |
| pETextonB | pETcctonB exbBD, Kmr | This study |
Recombinant DNA techniques.
Isolation of plasmids, use of restriction enzymes, ligation, agarose gel electrophoresis, transformation, hybridization, and Southern blotting were performed by standard methods (50). PCR of E. coli DNA was performed as follows: 3 min at 94°C; 35 cycles, with 1 cycle consisting of 1 min at 94°C, 2 min of annealing at 54°C, and 3 min of elongation at 72°C; and then 10 min at 72°C. The GC-rich DNA was amplified as described above for E. coli, but the annealing temperature started at 64°C and was lowered at each cycle by 0.5°C. Information on the primers used for PCR will be provided upon request.
Cloning of genes and mutated genes.
The genes homologous to E. coli malA, tonB, exbB exbD, and tolQ tolR were cloned by PCR with appropriately designed primers from the C. crescentus chromosome. Plasmid pMR2287 contains a 3-kb BglII-KpnI malA fragment in pMR20. pBAD2287His encoding MalA labeled at the C terminus with His6 (MalAHis6) was constructed by cloning the 2.8-kb BglII-KpnI fragment into pBADMyc/HisB. pHB138 contains an Ω insertion in cc2287 (ΔmalA) cloned with SpeI in pNPTS138Tet. pHB139 contains an Ω insertion in cc2327 (ΔtonB) cloned with SpeI in pNPTS138Tet. pHB140 contains a gentamicin resistance cassette insertion in cc2335-2336 (ΔexbBD) cloned with SpeI in pNPTS138Tet. pHB141 contains an Ω insertion in cc3232-3233 (ΔtolRQ) cloned with SpeI in pNPTS138Tet. pHB142 contains a gentamicin resistance cassette insertion in cc3232-3233 (ΔtolRQ) cloned with SpeI in pNPTS138Tet. pETcctonB contains a 743-bp NdeI-EcoRI tonB fragment cloned in pET27b(+). pETextonB (tonB exbB exbD) contains a 1.3-kb EcoRI-SalI fragment in pETcctonB. pMRexb1 contains a 1.5-kb PstI-BamHI fragment cloned into pMR20.
Construction of C. crescentus mutants.
The malA, tonB, and exbB exbD target genes were mutated by insertion of an Ω or gentamicin resistance cassette (1, 22, 47). The flanking genes of the target gene were amplified by PCR, ligated, and cloned into the pDrive vector. The 2-kb Ω Spcr-Strr fragment of pHP45Ω and the 0.6-kb MluI Gmr fragment of pBSL202 were cloned between the two flanking genes, and the DNA was amplified by PCR and cloned into pNPTS138Tet, which was derived from pNPTS138 by insertion of the 1.4-kb MluI Tetr fragment of pBSL204. The resulting plasmid was introduced into E. coli S17-1 λ pir by transformation and transferred by conjugation on PYE nutrient plates into C. crescentus JS1003. Growth of the donor strain was inhibited by nalidixic acid, and integration of the plasmid into the recipient genome was selected with streptomycin-spectinomycin or gentamicin. A second recombination deleted the target gene along with sacB and the Tetr gene by growing cells on 3% sucrose. The deletion of the target gene was verified by Southern blotting and DNA sequencing.
Isolation of the outer membrane fraction of C. crescentus.
One liter of PYE medium or 2 liters of M2 medium supplemented with 0.3% glucose or 0.3% maltose was inoculated with 20 ml of an overnight culture of C. crescentus and shaken at 30°C for 2 and 3 days, respectively. Cells were harvested by centrifugation for 20 min at 10,000 × g, washed with 50 mM Tris-HCl, pH 8, and suspended in 15 ml of 60 mM Tris-HCl, pH 8, 0.2 mM EDTA, 0.6 ml of Complete protease inhibitor mixture (Boehringer, Mannheim, Germany), 0.15 ml of 1 mM phenylmethylsulfonyl fluoride, and 0.3 ml of DNase (1 mg per ml). Cells were disrupted in a French pressure cell three times at 16,000 lb/in2, and cell debris was pelleted by centrifugation twice at 10,000 × g for 20 min. Fifteen milliliters of extraction buffer (50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 2% Triton X-100) was added. The sediment obtained after 60 min of centrifugation at 38,000 × g contained the outer membrane fraction, which was washed twice with 10 ml water and finally suspended in 0.3 ml water.
Two-dimensional (2-D) gel electrophoresis.
To the outer membrane fraction, 0.6 ml of 66% acetone in water was added, and the mixture was incubated overnight at −20°C to remove lipids. The pellet was washed with 0.1 ml 66% acetone, sedimented by centrifugation, and suspended in buffer containing 9 M urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 1% dithiothreitol, and 2.5% pharmalyte (Amersham Biosciences, Freiburg, Germany). Solubilization of the proteins was aided by sonication. Undissolved proteins were removed by centrifugation. The entire protein solution was subjected to isoelectric focusing on Immobiline DryStrips pH 4-7 (Amersham Biosciences). The proteins were separated in the first dimension by isoelectric focusing for 48 h in a Multiphor II (Amersham Biosciences). For protein separation in the second dimension, the strips were equilibrated in 50 mM Tris-HCl, pH 8.8, 6 M urea, 4% sodium dodecyl sulfate (SDS), and 2% dithiothreitol and then in 50 mM Tris-HCl, pH 8.8, 6 M urea, 4% SDS, and 4.8% iodoacetamide. The SDS-polyacrylamide gels were run for 18 h at 250 mA in an IsoDalt electrophoresis system (Amersham Biosciences). The gels were stained with Coomassie blue if they were to be subsequently analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) or with silver nitrate.
Mass spectrometry for protein identification.
In-gel digestion and protein identification were performed as previously described (55, 56) with minor modifications. The protein spots were excised from the gel, washed with Millipore-purified water and with 50% acetonitrile-water. After the protein spots were dried, trypsin (sequencing grade; Promega, Mannheim, Germany) was added to each sample. Tryptic protein fragments were extracted from the gel matrix with 5% formic acid and with 50% acetonitrile-5% formic acid. The extracts were pooled and concentrated in a speed vac concentrator. After purification with ZipTips (C18-ZipTip; Millipore, Bedford, MA), aliquots were deposited on a spot of α-cyano-4-hydroxycinnamic acid-nitrocellulose and analyzed with a Reflex III MALDI-TOF mass spectrometer (Bruker Daltonic, Bremen, Germany). All measurements were performed in the positive-ion reflection mode at an accelerating voltage of 23 kV and delayed-pulsed ion extraction. Sequence verification of tryptic fragments was performed by nanoelectrospray tandem mass spectrometry on a hybrid quadrupole orthogonal acceleration time-of-flight mass spectrometer (QSTAR Pulsar i; Applied Biosystems/MDS Sciex, Foster City, CA) equipped with a nanoflow electrospray ionization source. Purified aliquots were loaded in a nanoelectrospray needle (BioMedical Instruments, Zoellnitz, Germany) and tandem mass spectra were obtained by collision-induced decay of selected precursor ions. The instrument was calibrated externally.
Database searches (NCBInr, nonredundant protein database) were performed using the MASCOT software from Matrix Science (42) with methionine oxidations as variable modifications (probability value P < 0.05).
Purification of MalAHis6 by affinity chromatography on a Ni2+-NTA agarose column.
E. coli BL21 omp8 was transformed with pBAD2287His. The culture was grown at 37°C to an optical density at 578 nm of 0.5, and then arabinose (0.002%) was added to induce transcription. After 1.5 h of further cultivation, the cells were harvested by centrifugation, and the outer membrane fraction was isolated and incubated overnight at 12°C in 1% SDS. The solubilized proteins were identified by SDS-polyacrylamide gel electrophoresis (13% acrylamide). The protein mixture was adjusted to 50 mM NaH2PO4, pH 8, 0.3 M NaCl, 10 mM imidazole, and 0.2% SDS by passage through a PD-10 column (Amersham Biosciences) equilibrated with this buffer. The sample was then applied to a Ni2+-nitrilotriacetic acid (Ni2+-NTA) agarose spin column (QIAGEN, Hilden, Germany), and the column was washed with the same buffer but with increasing concentrations of imidazole (10, 20, and 250 mM). MalAHis6 was eluted with 250 mM imidazole.
[14C]maltose transport.
Cells were grown in M2 medium supplemented with 0.3% maltose as a carbon source, harvested by centrifugation, and washed once in M2 medium. The pellet was suspended in 1.3 ml of M2 medium containing 0.3% glucose, and the optical density at 578 nm was adjusted to 0.5. [14C]maltose (specific activity of 25.1 GBq/mmol) was added to a final concentration of 0.2 μM. For the inhibition experiments, maltose and larger maltodextrins, with final concentrations from 0.1 to 20 μM, were mixed with [14C]maltose. Samples of 0.2 ml were withdrawn after 0.2, 1, 2, 3, 4, and 5 min, collected on cellulose nitrate filters, washed twice with 5 ml M2 salt medium, and dried. The radioactivity was determined in a liquid scintillation counter.
In addition, a rapid dilution method was used to measure concentration-dependent initial maltose transport (19, 32). Cells were incubated for 15 s with 0.01, 0.05, 0.1, 0.2, 0.5, and 2 μM [14C]maltose, and 0.2-ml samples were withdrawn and diluted into 5 ml M2 buffer that contained 0.1 mM maltose. Cells were collected by filtration, washed twice with 5 ml M2 medium supplemented with 0.3% maltose, and dried, and the radioactivity was determined.
For inhibition of maltose transport with carbonyl cyanide 3-chlorophenylhydrazone (CCCP), CCCP was dissolved to 0.1 M in 5% acetic acid and then adjusted with 1 M NaOH to pH 7.1. Cells were incubated with 0.1 mM CCCP for 10 min at 30°C prior to the addition of [14C]maltodextrins.
Inhibition experiments with sodium arsenate (10 μM, 0.5 mM, and 1 mM) were performed in a 0.1 M Tris-HCl-0.1 mM MgCl2 buffer, pH 7.0. Cells were cultured in M2 medium supplemented with 0.3% maltose and then washed twice with the Tris-HCl-MgCl2 buffer to remove phosphate. The initial transport rate in this medium was half the rate in the M2 medium but reached the same value after 2 min.
HPLC analysis of maltodextrins.
The purity of the purchased nonradioactive maltodextrins was determined on a Shimadzu HPLC analyzer equipped with a Reprosil 100 NH2 column (5 μm; 250 × 4 mm), a SCL-10A controller, two LC-10At pumps, an SIL-10AXL autoinjector, a SPD-10AV UV/VIS spectrophotometric detector, and a C-R4 AX Chromatopac computer. Samples (20 μl) were injected and chromatographed with an isocratic solvent system consisting of double-distilled water and acetonitrile (35:65, vol/vol) with a flow rate of 1 ml per min. The eluted compounds were detected at 220 nm.
Sequence alignments.
The C. crescentus genome was analyzed and compared with other genomes using Gene versus All Alignment at http://www.tigr.org/tigr-scripts/CMR2/GenePage.spl?, http://www.ebi.ac.uk/fasta33/proteomes.html, and http://www.ebi.ac.uk/blast2. For the prediction of TonB-dependent outer membrane proteins, the program at http://www.ncbi.nlm.nih.gov./cgi-bin/COG/palox?COG1629 and HHpred (58) were used. The presence of signal sequences was analyzed with the program at http://expasy.org/cgi-bin/niceprot.pl?Q9A608.
RESULTS
Identification of a maltose-inducible outer membrane protein.
C. crescentus contains a quantitatively dominant protein, designated RsaA, at the cell surface that forms an S-layer (21). To avoid interference of RsaA with the identification of a nutrient-inducible outer membrane protein among the predicted TonB-dependent proteins, strain JS1003, which lacks RsaA, was used.
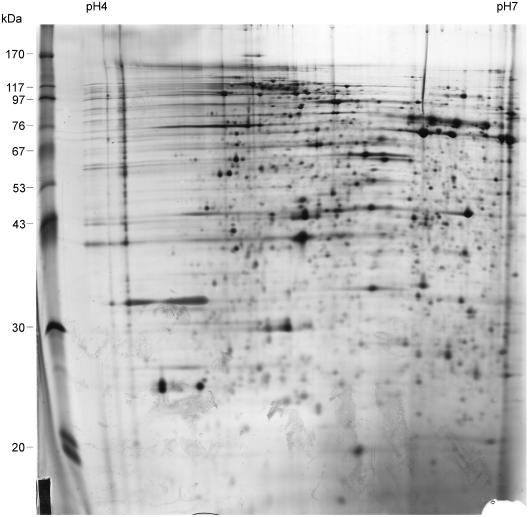
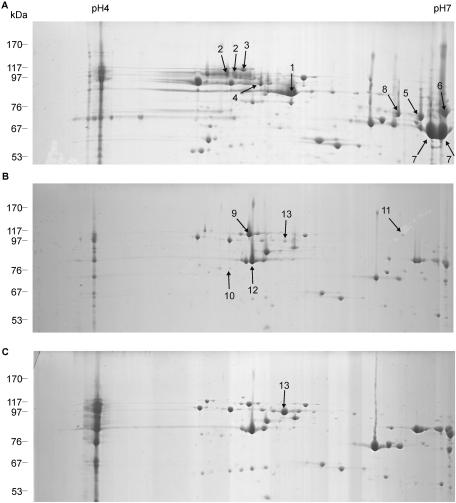
After growth of strain JS1003 on the rich PYE medium, the outer membrane fraction was isolated and various methods were evaluated for solubilization and subsequent separation of the outer membrane proteins by 2-D gel electrophoresis. Figure 1 shows a protein profile obtained with the method described in Materials and Methods. The outer membrane of C. crescentus contains many proteins, particularly in the 80- to 100-kDa range of the predicted TonB-dependent proteins. The high-molecular-mass section of the 2-D gels is shown in Fig. 2, where outer membrane proteins of cells grown in PYE medium (Fig. 2A), minimal M2 medium with glucose (Fig. 2B), and minimal M2 medium with maltose (Fig. 2C) as carbon sources were compared. Maltose was chosen as the substrate to be tested, since starch is a major carbon source in nature and E. coli contains the maltose-inducible LamB protein, which facilitates diffusion of maltodextrins across the outer membrane. We hypothesized that the diffusion facilitator of E. coli has in C. crescentus developed further to an energy-coupled transporter. The numbered protein spots were eluted and analyzed by MALDI-TOF mass spectrometry. The proteins numbered 1 to 13 belonged to the predicted TonB-dependent proteins (Table 2). The protein numbered 13 was the most highly expressed protein in cells grown with maltose and was studied further.
FIG. 1.
Two-dimensional gel electrophoresis of an outer membrane preparation (40 μg protein) of C. crescentus JS1003 grown in rich PYE medium. Proteins were separated in the first dimension by isoelectric focusing (pH 4 to 7) and in the second dimension according to the size of the proteins in the presence of 0.1% SDS. Protein spots were visualized by silver staining.
FIG. 2.
Two-dimensional gel electrophoresis of outer membrane proteins of C. crescentus grown in PYE rich medium (A), M2 minimal medium with glucose (B), and M2 minimal medium with maltose (C). Only the high-molecular-mass section of the 2-D gels is shown. The arrows indicate the spots analyzed by MALDI-TOF, and spot 13 is the protein induced by maltose. The gel was stained with Coomassie blue.
TABLE 2.
Identification of selected protein spots by MALDI-TOF
| Protein spot no.a | Enhanced spot on mediumb | Predicted protein | Gene | Protein mass (Da) |
|---|---|---|---|---|
| 1 | PYE | TonB-dependent receptor | cc0210 | 89,687 |
| 2 | PYE | TonB-dependent receptor | cc3013 | 121,153 |
| 3 | PYE | TonB-dependent receptor | cc0983 | 122,929 |
| 4 | PYE | TonB-dependent receptor | cc1099 | 108,856 |
| 5 | PYE | TonB-dependent receptor | cc2928 | 76,571 |
| 6 | PYE | TonB-dependent receptor | cc3146 | 88,772 |
| 7 | PYE | TonB-dependent receptor | cc3500 | 77,641 |
| 8 | PYE | TonB-dependent receptor | cc2194 | 77,507 |
| 9 | M2M, M2G | TonB-dependent receptor | cc0999 | 111,438 |
| 10 | M2M, M2G | TonB-dependent receptor | cc1666 | 80,843 |
| 11 | M2M, M2G | TonB-dependent receptor | cc2804 | 114,514 |
| 12 | M2M, M2G | TonB-dependent receptor | cc0185 | 85,815 |
| 13 | M2M | TonB-dependent receptor | cc2287 | 99,441 |
Numbers in Fig. 2.
Media are described in Materials and Methods. M2M and M2G, M2 minimal medium supplemented with maltose and glucose, respectively.
Predicted structure of the MalA protein.
Spot number 13 was assigned to locus cc2287 of the C. crescentus genome sequence and designated MalA. MalA has a molecular mass of 99,501 Da and contains a predicted signal peptide of 32 residues (Fig. 3). The sequence EEVVIT starts at residue 14 of the mature protein; this sequence resembles E. coli TonB box sequences, for example, EETVIV of FhuE. Computer-assisted sequence analyses with the HHpred program clearly predict a 141-residue N-terminal sequence that resembles the folding of the cork (plug, hatch) of the five outer membrane transporters with known crystal structures, including the conserved four-stranded β-sheet (hβ1-hβ4 and the two amphipathic α-helices [hα1 and hα2]). A strong propensity for α-helix formation is predicted for residues 26 to 33 which are close to the so-called switch helix of FhuA and FecA and seen there only in the proteins unloaded with substrates (24, 25, 38, 62). In addition, 22 antiparallel β-strands with short turns connecting the β-strands in the periplasm and large loops above the cell surface are predicted (Fig. 3). The largest loop between β-strands 7 and 8 encompasses 79 residues, and the second largest loop between β-strands 9 and 10 encompasses 61 residues. For comparison, the largest loop in the outer membrane transporters with known crystal structures is in FepA with 37 residues (34). The larger size of MalA (100 kDa) compared to the 80 kDa of the ferric siderophore transporters seems to be caused by the larger surface loops. Among the outer membrane transporters with available crystal structures the highest sequence identity is to the cork of BtuB (25%) and the barrel of FecA (25%). Further sequence analysis revealed similarities to functionally uncharacterized proteins; CC1754 of C. crescentus (40.5% sequence identity); XAC2600, FecA, and FiuA of Xanthomonas axonopodis (37.8, 30.4, and 28.1% identity); CirA of Xylella fastidiosa (31.1%); and FhuA of Xanthomonas campestris (32.8%). MalA exhibits no sequence similarity to LamB of E. coli.
FIG. 3.
Genome-derived amino acid sequence of the MalA protein. The predicted signal sequence is shown in italic type, the TonB box is shown in bold type, the cork domain is underlined, and the 22 antiparallel β-strands are indicated by numbered lines above the sequence. Note the large cell surface loops between β-strands 5 and 6, 7 and 8, 9 and 10, 13 and 14, and 15 and 16.
Predicted maltodextrin transport locus.
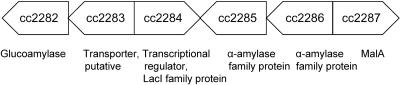
The malA gene is located near a predicted gene encoding a transport system across the cytoplasmic membrane (Fig. 4) with sequence similarities to sugar transporters in prokaryotes and sucrose transporters in plants. Also in this region of the chromosome are found one gene assigned to the family of lacI regulatory genes, two genes encoding α-amylases, and one gene encoding a glucoamylase with a predicted signal sequence. The transcription polarity of the mal genes suggests a divergent promoter upstream of cc2286 and malA which controls transcription of cc2283 to cc2286 and in the opposite direction transcription of malA. It is likely that these genes are all involved in the uptake and metabolism of maltodextrins, similar to the periplasmic α-amylase of E. coli, which degrades larger maltodextrins, and α-amylase and glucophosphorylase, which cleave maltose in the cytoplasm.
FIG. 4.
Open reading frames identified in the genome of C. crescentus adjacent to the malA gene and predicted to be related to maltose utilization. The arrows indicate the direction of transcription.
MalA-dependent growth on maltodextrins.
To determine the function of MalA in maltodextrin transport, a mutant with a cassette insertion mutation in the malA gene was constructed. Growth of the resulting strain, C. crescentus HB2003 malA, was tested on M2 minimal agar plates containing no carbon source. Maltodextrins were applied to paper disks placed on the agar. Growth of mutant HB2003 was supported by maltose, maltotriose, and maltotetraose, but not by maltoheptaose and maltohexaose (Table 3). Since the growth zone around the paper disk with maltotetraose was relatively poor, the exclusion limit of the outer membrane lacking MalA was between the size of maltotetraose and maltopentaose. Transformation of the malA mutant with the wild-type malA gene restored growth on maltopentaose and maltohexaose (Table 3). MalA supported uptake of the larger maltodextrins across the outer membrane of C. crescentus.
TABLE 3.
Growth promotion of C. crescentus strains and E. coli KB419 by maltodextrins
| Maltodextrin | Growth zone of straina:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| JS1003 malA+ | HB2003 malA | HB2003 malA/pMR2287 | HB2004 tonB | HB2007 exbBD | HB2007 exbBD/pMRexb1 | HB2006 tonB exbBD | KB419 lamB | |
| Maltose | 3 | 3 | 3 | 3 | 3 | 2.5 | 3 | 2.5 |
| Maltotriose | 3 | 2.2 | 3 | 3 | 2.5 | 2.5 | 2.4 | 1.3 |
| Maltotetraose | 3 | 0.9 | 3 | 3 | 1.3 | 2.5 | 1.4 | - |
| Maltopentaose | 3 | - | 3 | 3 | - | 2.5 | - | - |
| Maltohexaose | 3 | - | 2.8 | 3 | - | 2.5 | - | - |
The growth zones are given in centimeters including the filter paper disk (0.6 cm). -, no growth.
MalA-dependent maltodextrin transport.
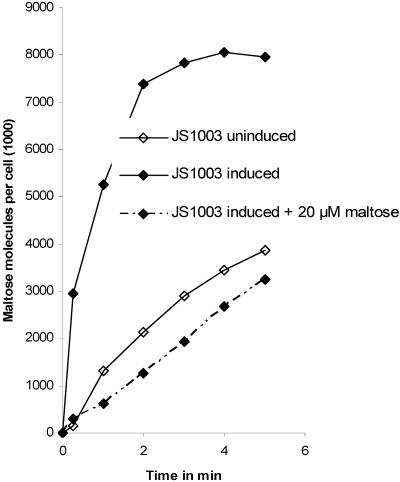
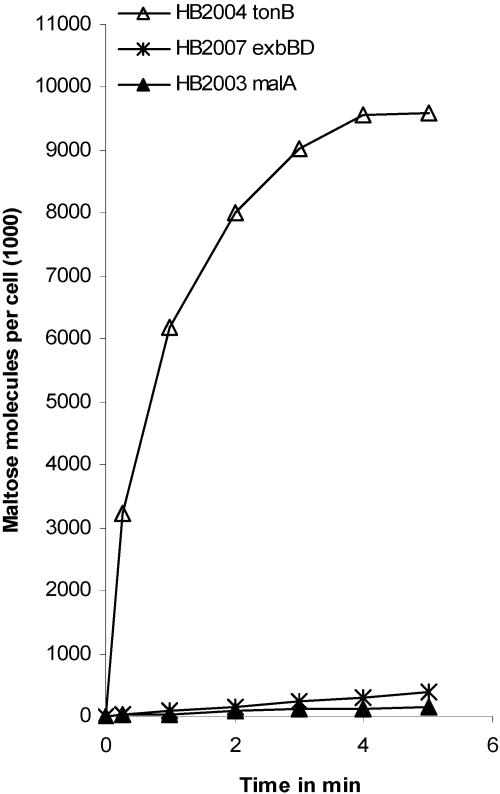
The use of maltopentaose and maltohexaose as a carbon source required MalA, in contrast to maltose, on which the malA mutant could grow. Since even low uptake rates (time period of minutes) could be sufficient to support growth (time period of a day), the uptake rate of [14C]maltose into wild-type and mutant cells were compared. C. crescentus JS1003 grown on maltose as a carbon source rapidly took up [14C]maltose (Fig. 5). After 1-min incubation, 70% of the final value was attained. A 100-fold excess of unlabeled maltose added together with the radioactive maltose reduced the transport rate to 14% of the uninhibited level (Table 4). Maltopentaose nearly abolished [14C]maltose transport. A 15-fold excess of maltose over [14C]maltose reduced the 1-min transport value to 23% of the uninhibited level compared to 9% attained with maltotetraose. The stronger inhibition of maltotetraose and maltopentaose may reflect a tighter binding of the larger maltodextrins to MalA (see later) and possibly the cytoplasmic membrane transporter than the binding of maltose. Strain JS1003 grown on glucose showed a maltose transport rate 29% of that of cells grown on maltose (Fig. 5 and Table 4). The data indicate that maltose is transported by C. crescentus, maltose transport is inducible by growth on maltose, and maltodextrins inhibit uptake of labeled maltose.
FIG. 5.
[14C]maltose transport into wild-type C. crescentus JS1003 grown in M2 minimal medium supplemented with maltose (induced), glucose (uninduced), and induced in the presence of 20 μM unlabeled maltose added immediately prior to the addition of 0.2 μM [14C]maltose.
TABLE 4.
[14C]maltose transport rates related to maltose-induced C. crescentus JS1003 malA+ or E. coli W3110 wild type
| C. crescentus strain | % Transport |
|---|---|
| JS1003 induced | 100 |
| JS1003 uninduced | 29 |
| JS1003 induced + maltosea | 14 |
| HB2003 malA | 1.3 |
| HB2007 exbB exbD | 2.2 |
| HB2004 tonB | 100 |
20 μM maltose was added immediately prior to the addition of 0.2 μM [14C]maltose. The 2-min transport values were used for comparison.
C. crescentus HB2003 malA transported [14C]maltose very slowly (Fig. 6). Maltose transport was linear and after 2 min reached 1.3% of the level of transport into strain JS1003 (Table 4) and after 25 min reached 7% of the transport into strain JS1003 (data not shown). The low maltose uptake rate into mutant HB2003 suffices to support growth, and apparently the slow uptake was determined by slow diffusion of maltose across the outer membrane. The diffusion-controlled uptake of [14C]maltose into mutant HB2003 was not inhibited by maltose (data not shown).
FIG. 6.
[14C]maltose transport into C. crescentus HB2003 malA, HB2007 exbB exbD, and HB2004 tonB. Cells were grown in M2 minimal medium supplemented with maltose.
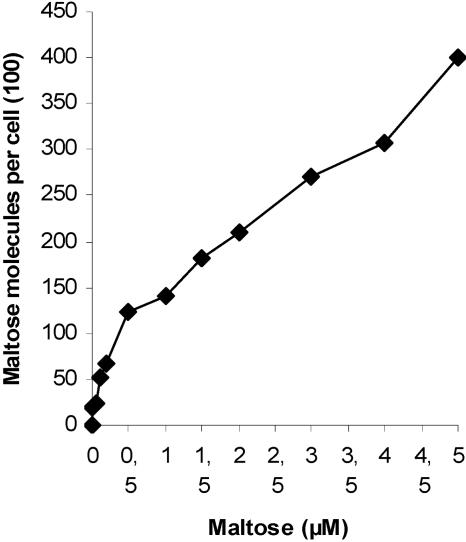
Concentration-dependent maltose transport showed biphasic kinetics, a fast rate between 0.1 and 0.5 μM maltose and a slow rate between 1 and 5 μM maltose (Fig. 7). The Kd value derived from the fast rate was 0.22 μM, and the Kd derived from the slow rate was 5.4 μM. It is assumed that the low Kd value mainly reflects maltose binding to MalA and the higher Kd value mainly reflects binding to the cytoplasmic membrane transporter encoded by cc2283.
FIG. 7.
Concentration-dependent [14C]maltose transport into C. crescentus. Cells were grown in maltose-containing medium, and transport was measured for 12 s at the indicated maltose concentrations.
Dependence of MalA transport activity on ExbB ExbD.
To test whether maltodextrin uptake through MalA of C. crescentus occurs by facilitated diffusion as through E. coli LamB or by energy-coupled transport, mutants with Ω or Gmr cassette insertion mutations in C. crescentus genes homologous to E. coli tonB (cc2327) and exbB exbD (cc2336 cc2335) were constructed. The C. crescentus TonB, ExbB, and ExbD proteins show 26.1, 40, and 46.4% identity to the E. coli TonB, ExbB, and ExbD proteins. The homologous ExbB proteins share a stretch of 57 identical residues, interrupted in C. crescentus at eight sites by single conservative substitutions. The overall similarity is reduced by an N-terminal 67-residue segment in C. crescentus that is lacking in E. coli ExbB and 16 C-terminal residues in E. coli ExbB that are lacking in C. crescentus ExbB. The homologous ExbD proteins share a stretch of 33 identical residues, interrupted in C. crescentus at four sites by single conservative substitutions.
Growth of C. crescentus HB2004 tonB on maltodextrins was not impaired (Table 3), and [14C]maltose transport in strain HB2004 was as high as in strain JS1003 (Fig. 6 and Table 4). In the tonB mutant, the tonB deletion was verified by Southern blotting of BamHI and BamHI-NotI fragments of chromosomal DNA and hybridization with tonB was amplified by PCR from plasmid pETextonB. Wild-type JS1003 yielded the expected 3.5-kb BamHI fragment and the 2-kb and 0.8-kb BamHI-NodI fragments, whereas these fragments were lacking in HB2004 (data not shown). In addition, PCR with one of the primers binding in tonB yielded the expected fragment with the wild-type strain and no fragment with the tonB mutant. Growth of the HB2007 exbBD double mutant was reduced on maltotriose and maltotetraose and abolished on maltopentaose and maltohexaose. Growth of the HB2007 exbBD mutant on the maltodextrins was largely restored after transformation with plasmid pMRexb1, which carries the wild-type exbBD genes (Table 3). [14C]maltose transport into HB2007 exbBD was very low (Fig. 6 and Table 4). Treatment of cells with 0.1 mM CCCP reduced the residual maltose transport to zero, suggesting that transport into the exbBD mutant resulted from diffusion through the outer membrane and transport across the cytoplasmic membrane. The latter was inhibited by CCCP. The tonB exbB exbD triple mutant HB2006 displayed growth properties similar to those of the exbBD double mutant (Table 3). Transformation of the triple mutant with pETextonB (tonB exbB exbD) restored growth on maltopentaose and maltohexaose and transport of the maltose (data not shown).
To examine whether the failure of the exbBD mutant to transport maltose was caused by a lack of MalA in the outer membrane, 2-D electrophoresis was performed with an isolated outer membrane fraction of strain HB2007. The MalA spot was observed in amounts similar to those shown in Fig. 2C for JS1003, and MALDI-TOF mass spectrometry identified the spot as MalA (data not shown).
Transport of larger maltodextrins.
To relate the absence of growth of the malA and exbBD mutants on the larger maltodextrins to the lack of transport, [14C]maltotriose, [14C]maltotetraose, and [14C]maltopentaose were incubated with strain JS1003 under the same conditions as in the maltose transport assays. The maltodextrins were transported at rates similar to the transport rate of maltose. The 15-s transport values were more than twofold higher than those of maltose, which probably reflects the enhanced binding of the maltodextrins to MalA. In the malA mutant [14C]maltotriose was transported at a lower rate than the transport rate of maltose, which probably reflected the lower diffusion rate through porins. No transport of [14C]maltotetraose and [14C]maltopentaose occurred into the malA mutant, supporting the strict requirement of MalA for entry of the larger maltodextrins. Transport into the exbBD mutant was abolished, whereas transport into the tonB mutant was the same as transport into wild-type cells (data not shown).
Maltodextrin binding to MalA.
In intact cells, the FhuA ferrichrome, FepA ferric enterobactin, and BtuB vitamin B12 transporter bind their substrates with binding constants close to the nanomolar (5, 20) and subnanomolar (11, 53) range. To estimate maltose binding, inhibition of maltose transport was determined from 0.1 to 20 μM maltose. The initial rate of transport (15-s value) was reduced to half the value of the uninhibited transport by 0.4 μM maltose. Nonradiolabeled maltotetraose supplied at 0.1 μM reduced the initial rate of [14C]maltose uptake to 50%. Although these values may reflect binding to not only MalA but also partially to the transporter in the cytoplasmic membrane, it shows that binding to MalA is in the micromolar range and not as strong as substrate binding to the TonB-dependent E. coli outer membrane transporters.
Energy depletion of cells should allow determination of maltose binding to MalA in intact cells, provided binding was not influenced by energization of MalA. A 10 μM concentration of CCCP reduced maltose transport to 65%, 50 μM CCCP reduced maltose transport to 36%, and 100 μM CCCP reduced maltose transport to 1%. The CCCP concentrations are higher than are required for E. coli, where 10 μM CCCP completely inhibits proton motive force-dependent transport, but the same (100 μM) in Pseudomonas aeruginosa (17). Using the transport assay, the 15-s [14C]maltose value of CCCP-treated cells was 0.6% of the non-CCCP-treated cells (200 molecules per cell), whereas the [14C]maltotriose value was 6%, the [14C]maltotetraose value was 9%, and the [14C]maltopentaose value was 15%. About the same low level of [14C]maltose stayed associated with the exbBD mutant and to cells incubated on ice. It seems that maltodextrin binding to MalA is energy dependent. Arsenate (1 mM) did not inhibit [14C]maltose transport. No specific [14C]maltose binding was observed to isolated outer membranes, since the very low levels were the same in outer membrane preparations from wild-type cells and malA mutant cells. As will be shown later, binding of maltose to isolated MalA could not be determined, since no native MalA was obtained.
Iron uptake by the tonB mutant and the exbB exbD mutant.
Lack of maltose transport and growth of mutant HB2007 exbB exbD on maltopentaose and maltoheptaose pointed toward an energy-dependent maltodextrin transport through MalA. Because Fe3+ siderophore transport depends on ExbB ExbD, whether iron uptake by C. crescentus required ExbB ExbD was tested. Strain JS1003 did not grow on M2G minimal medium without the addition of FeSO4; 5 μM was sufficient. At this iron concentration, the growth curve of mutant HB2007 exbB exbD showed a longer lag phase than strain JS1003, but the cell density reached after 22 h was similar to that of the wild type. Whether strain JS1003 uses Fe3+ siderophores to achieve growth in M2 medium supplemented with 0.2% Casamino Acids to which no iron was added was tested. Addition of the iron-complexing compounds dipyridyl (50 μM) and ethylenediamine dihydroxyphenylacetic acid (EDDA) (10 μM), which cannot be used by C. crescentus, prevented growth. Of the siderophores tested on plates containing dipyridyl and EDDA, only rhodotorulic acid and desferral supported growth, whereas ferrichrome, ferricrocin, ferrichrome A, citrate, fusigen, arthrobactin, schizokinen, coprogen, rhizoferrin, staphyloferrin, and dihydroxybenzoic acid failed to support growth. In contrast to strain JS1003, growth of HB2007 exbB exbD was not supported by rhodotorulic acid, and growth on desferral was reduced from 25- to 15-mm growth around filter paper disks to which 10 μl of a 1 mM solution of the siderophore had been added. Mutant HB2004 tonB showed growth properties similar to those of strain JS1003. These results support the conclusion that ExbB and ExbD are required for Fe3+-rhodotorulate uptake.
The TolQ TolR proteins of C. crescentus cannot functionally replace the ExbB ExbD proteins.
exbB exbD mutants of E. coli can partially be suppressed by the homologous tolQ tolR genes (8). A search of the C. crescentus genome sequence revealed the tolQ and tolR homologs cc3233 and cc3232, respectively. Additional open reading frames belonging to the E. coli tol locus were found at the same site: cc3229, homologous to the PAL gene; cc3230, homologous to tolB; and cc3231, homologous to tolA. Clustering of these genes in the same order and transcriptional polarity, tolQ tolR tolA tolB pal, as in E. coli (12, 13) suggests that this is indeed the tol locus of C. crescentus. The clear phenotype of the exbB exbD mutants indicates that in C. crescentus, tolQ tolR cannot functionally replace exbB exbD in MalA-mediated maltodextrin transport and rhodotorulic acid-mediated Fe3+ transport. Nevertheless, we attempted to isolate a chromosomal tolQ tolR deletion mutant. A Gmr insertion and an Ω insertion between the genes flanking tolQ tolR were constructed (plasmids pHB141 and pHB142, respectively), but double recombination into the C. crescentus genome failed. It is possible that the tol locus plays an essential role in C. crescentus, e.g., in the assembly of the outer membrane, as has been suggested for E. coli (13).
MalA does not complement a LamB mutant.
LamB facilitates diffusion of maltodextrins across the outer membrane of E. coli. To examine whether malA complements an E. coli lamB mutant, E. coli KB419 was transformed with plasmid pBAD2287 malA. A second KB419 transformant contained, in addition to malA, the C. crescentus tonB exbB exbD genes on plasmid pETextonB in case MalA functions as an energy-coupled transporter and requires the homologous TonB, ExbB, and ExbD proteins. The transformants were suspended in soft agar containing M2 minimal medium lacking a carbon source, which was then poured onto M2 minimal medium plates supplemented with 0.2% arabinose to induce malA transcription. This concentration of arabinose supported slow growth which did not interfere with the growth stimulation assay around paper disks onto which 10 μl of a 40% solution of glucose, maltose, or maltodextrins was applied. Growth of mutant KB419 lamB and its transformants was stimulated by glucose, maltose, and maltotriose, but not by maltotetraose, which indicated that MalA was not active in E. coli.
Isolation of MalA from an E. coli malA transformant.
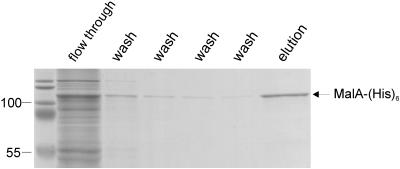
There were several reasons to purify MalA from E. coli. (i) Purified MalA could not be isolated in sufficient amounts from C. crescentus. The level of MalA was low, and there were too many outer membrane proteins with similar properties. (ii) The properties of isolated MalA from E. coli may provide hints for its inactivity in E. coli. (iii) Isolated MalA was required for raising MalA antiserum. (iv) Isolated MalA may be used to study pore-forming properties in artificial lipid bilayer membranes. E. coli porins and LamB maltoporin exhibit single-channel conductance when incorporated into artificial lipid bilayer membranes (2-4). In contrast, TonB-dependent outer membrane transporters form closed channels that do not support flux of ions (34). To facilitate purification, a His6 tag was added to the C terminus of MalA (MalAHis6). The malA gene was cloned into E. coli omp8 lamB ompC ompF ompA, which lacks the major porins which, as contaminants, might interfere with MalA conductance. Cloning of malA in the medium-copy-number vector pT7-7, which provides an ideal ribosome binding site for high levels of malA translation, resulted in transformants that lysed. Surviving colonies had mutations in malA. Apparently, large amounts of MalA are lethal to E. coli. Therefore, the malA gene was cloned under the tight control of the arabinose operon, and MalA synthesis was induced by 0.002% l-arabinose. MalA formed a prominent band in an SDS-polyacrylamide gel at an electrophoretic position corresponding to100 kDa (data not shown). MalAHis6 in isolated outer membranes was solubilized by overnight incubation at 12°C in 0.1% SDS, whereas 1.5% dodecyl-N,N-dimethylamine-N-oxide (LDAO), 5% LDAO-0.6 M NaCl, 1.5% octylglycoside, and 1.5% CHAPS did not solubilize the protein. MalAHis6 in SDS was bound to Ni2+-NTA agarose and eluted with buffer containing 250 mM imidazole (Fig. 8). Outer membrane proteins are usually soluble in LDAO buffers from which they were crystallized. Resistance of MalA to mild solubilization suggests that it was not integrated with its native conformation in the outer membrane of E. coli and may even form inclusion bodies which cofractionate with the outer membrane. This conclusion was supported by ion flux measurements in artificial lipid bilayer membranes which were performed according to the published procedures (2-4). SDS-solubilized MalAHis6 added to one of two compartments containing 1 M KCl resulted in an ion flux. The conductance increased more or less steadily and did not show distinct single conductance steps as is characteristic for outer membrane porins. The histogram of the probability of the occurrence of certain conductivity steps showed a broad range of values from 0.25 to 4.75 G/nS. A control sample isolated exactly in the same way from untransformed E. coli omp8 did not show any conductance increase indicating the absence of outer membrane porins (data not shown). If MalA contains a binding site for maltodextrins in the pore, addition of maltodextrins might inhibit conductance, as has been shown for LamB (4). Only very high concentrations of maltose (up to 0.35 M) reduced conductance by 70% of that of the untreated MalA sample. The 50% saturation constant derived from the inhibition data was about 220 mM (mean of four titration experiments). Inhibition by maltopentaose revealed a 50% saturation constant of about 80 mM. Inhibition similar to maltose was observed with glucose, but not with fructose. It is concluded that the data reveal the pore-forming potential of MalA. Apparently, the isolated sample was not in the native state, which resulted in KCl conductance and very low affinities for the natural substrates maltose and maltoheptaose.
FIG. 8.
Purification of the MalAHis6 protein on a Ni2+-NTA agarose column. MalAHis6 was extracted from E. coli BL21 omp8 with 1% SDS and purified by affinity chromatography. The eluted fractions were subjected to SDS-polyacrylamide gel electrophoresis and stained with Coomassie blue.
DISCUSSION
Growth of C. crescentus on maltose as the sole carbon source induced the synthesis of the MalA protein. MalA was necessary for growth on maltopentaose and maltohexaose but was dispensable for growth on maltose, maltotriose, and maltotetraose, which indicated the presence of other proteins in the outer membrane that support diffusion or facilitated diffusion of the smaller maltodextrins. Lack of MalA reduced growth of C. crescentus on maltotetraose, whereas lack of the LamB protein reduced growth of E. coli already on maltotriose. The MalA-independent pores in the outer membrane of C. crescentus are somewhat larger than the LamB-independent pores in the outer membrane of E. coli.
To examine whether MalA forms a maltose-specific pore or an energy-dependent transporter, C. crescentus tonB and exbB exbD mutants were isolated. The exbB exbD mutants could not grow on maltopentaose and maltohexaose but grew on the smaller maltodextrins. Uptake of [14C]maltose into the exbBD mutant was very low (Table 4) but sufficed to support growth. The larger maltodextrins were not transported. The relationship of ExbBD dependence to the energy requirement was supported by the ExbB ExbD dependence of iron transport mediated by rhodotorulic acid. These results indicated that MalA is a transporter and not a maltoporin. The smaller maltodextrins pass through the outer membrane by diffusion most likely not mediated by MalA, but rather by other, unidentified proteins. This hypothesis is based on the properties of the iron, ferric siderophore, and heme transporters across the outer membrane, which do not form diffusion pores unless energized by interaction with TonB.
Unexpectedly, the tonB mutant could grow on maltopentaose, maltohexaose, and Fe3+-rhodotorulate, and transported [14C]maltose at the wild-type rate. The predicted TonB protein shows 26.1% sequence identity to E. coli TonB, contains an N-proximal transmembrane segment with a histidine residue at a position equivalent to that of the functionally important His-20 of E. coli TonB (59) but lacks the functionally important Ser-16 (36). It also contains a number of proline residues that are thought to play a role in spanning the periplasmic space (29, 37). However, the sequence around Gln-160, which in E. coli TonB interacts with outer membrane transporters (27, 52), is very different in C. crescentus TonB. It is therefore questionable whether this TonB homolog can interact with the MalA TonB box, and this is consistent with the finding that lack of TonB does not affect MalA activity.
C. crescentus, belonging to the α-Proteobacteria, is not very closely related to E. coli, which belongs to the γ-Proteobacteria. No protein other than the TonB protein encoded by the C. crescentus genome is similar in size and sequence to the E. coli TonB protein; there is, however, a larger protein of 401 amino acid residues that is 41% identical to C. crescentus TonB. This protein might function as TonB or substitute for the examined TonB in maltopentaose and maltoheptaose transport when TonB is lacking. Several TonB proteins which can completely or partially functionally substitute for each other have been found in various other organisms, e.g., Serratia marcescens (42), Vibrio cholerae (54), and Pseudomonas aeruginosa (29, 63). The function of the 401-residue protein is currently being investigated.
Further evidence for a transporter function of MalA in contrast to a pore activity was obtained from transport kinetics and maltodextrin binding studies. Concentration-dependent maltose transport showed biphasic kinetics, a fast initial rate and a slower second rate. The Kd value derived from the fast rate was 0.22 μM, and the Kd derived from the slower rate was 5.4 μM. Similar biphasic kinetics were first observed in the energy-coupled vitamin B12 transport into E. coli in which the initial rapid phase was energy independent and showed saturation kinetics, whereas the second slower phase was energy dependent. It was concluded that the first phase reflects loading of a membrane carrier and the second phase reflects release from the membrane binding site into the interior of the cells (20). Subsequent studies identified the BtuB energy-coupled transporter in the outer membrane that binds vitamin B12 with a Kd of 5 nM and an ABC transporter in the cytoplasmic membrane (14, 15, 20, 61). In contrast to BtuB and the iron siderophore outer membrane transporters, binding of maltose displayed a higher Kd value. Nonradioactive maltose at a concentration of 0.4 μM reduced radiolabeled maltose transport to 50%, which indicates a Kd of 0.4 μM. This value was similar to the Kd of 0.22 μM derived from the initial rapid transport rate in the maltose concentration-dependent transport assay. Maltotetraose reduced maltose transport at a concentration of 0.1 μM which reflects the higher affinity of MalA to the larger maltodextrins. Although the Kd of maltose binding to MalA is 100-fold larger than the Kd values of the vitamin B12 and iron transporters, it is 1,000-fold lower than the Km of p-nitrophenyl-α-d-maltohexaoside binding, 0.13 mM, to the LamB porin (26). Stability constants determined in vitro for LamB incorporated into artificial lipid bilayer membrane range from 10 mM for maltose to 0.067 mM for maltohexaose (3, 4).
Unexpectedly, cells deprived of energy by treatment with CCCP or kept on ice, as well as exbBD mutants, showed very low maltose binding which increased somewhat with the larger maltodextrins. This could mean that binding of maltodextrins to MalA requires energy that is used to induce a binding-competent MalA conformation. In contrast, TonB-coupled transporters do not require energy for substrate binding but do require substrate release from the transporters (15, 17, 42, 51, 60). Since C. crescentus is very distinct from E. coli and related γ-proteobacteria, it is not unexpected that the transport mechanism across the outer membrane differs in details.
MalA was able to integrate into artificial lipid bilayer membranes and form pores through which KCl ions moved. This was unexpected for an energy-coupled outer membrane transporter that does not form permanently open pores (14, 15, 34, 35, 46). We assume that solubilization of MalA with 0.1 or 1% SDS resulted in a partially denatured protein that formed a permanently open pore that is not present in the native protein. Milder detergents did not solubilize MalA. It is also possible that MalA did not assume its native conformation in E. coli from which it was isolated. Cloning of malA on a medium-copy-number plasmid in C. crescentus in order to increase the concentration of MalA for isolation from its natural membrane was not successful. The requirement of much higher maltose concentrations to inhibit conductance than are required to inhibit in vivo maltose transport also points toward an artificial pore. Glucose inhibited the flux of KCl, which supports the notion of an artificial pore; however, binding wasnot completely unspecific, since fructose did not cause inhibition.
The results demonstrated the potential of MalA to form a pore. We propose that under natural conditions in the C. crescentus membrane, MalA forms a closed pore that opens upon interaction with TonB. Opening of the pore and release of the maltodextrins from their binding site require energy, which is harvested from the transmembrane potential of the cytoplasmic membrane by the predicted TonB-ExbB-ExbD protein complex. The prediction of a TonB box, plug, and β-barrel supports the proposed active MalA transporter. An alternative, less likely model proposes a permanently open pore in MalA through which maltose and maltotriose slowly diffuse. Such a pore would be too small for diffusion of maltopentaose and maltohexaose, or tight binding to MalA results in a diffusion that is too slow to support growth. At low maltodextrin concentrations with a low MalA substrate occupancy, energy input is important and changes the conformation of the maltodextrin binding site, and maltodextrins are released and diffuse through the pore into the periplasm. In addition, energization may widen the pore to allow a faster diffusion.
Experiments aimed at providing further support for an energy requirement of MalA-mediated transport by using inhibitors of the proton motive force, such as CCCP, were hampered by the likely involvement of a proton or sodium gradient in the transport of maltodextrins across the cytoplasmic membrane. Analysis of the C. crescentus genome sequence revealed a gene encoding a putative transporter close to malA that resembles the Na+-melibiose cotransporter and H+-sugar cotransporters in prokaryotes and in particular sucrose transporters of plants. In the systems studied, substrates such as Fe3+, Fe3+-siderophores, heme, and vitamin B12, which are translocated across the outer membrane by proton motive force-coupled transporters, are translocated across the cytoplasmic membrane by ATP-dependent ABC transporters. In the case of maltodextrin transport of C. crescentus, inhibitors of the transmembrane potential of the cytoplasmic membrane would inhibit transport across the outer membrane and the cytoplasmic membrane as well. Up to now all substrates that are ExbBD-dependent transported across the outer membrane are transported across the cytoplasmic membrane by ABC transporters. ABC transporters require a periplasmic binding protein which is not encoded in the C. crescentus mal locus. The C. crescentus Mal system is the first which most likely uses the electrochemical potential for transport across the cytoplasmic membrane.
The Mal system of C. crescentus differs from the Mal system of E. coli not only with respect to the transport mechanisms across the outer membrane and the cytoplasmic membrane but also with regard to its regulation. Analysis of the genome sequence predicts a transcriptional repressor of the LacI family, whereas the maltose regulon of E. coli is regulated by the MalT transcriptional activator and the MalK ATPase of the ABC transporter (32). There are also similarities. The glucoamylase encoded close to malA contains a signal peptide and might degrade larger maltodextrins in the periplasm to smaller maltodextrins, as does MalS in E. coli. The two α-amylases encoded close to malA might degrade maltodextrins in the cytoplasm as do the E. coli MalP maltodextrin phosphorylase and MalQ amylomaltase. We found no evidence of secretion of the glucoamylase and the two α-amylases into the culture medium; starch did not serve as a substrate.
Previous 2-D electrophoretic analysis of outer membrane proteins and MALDI-TOF analysis identified 16 putative TonB-dependent outer membrane proteins in C. crescentus (44). In another 2-D, MALDI-TOF study, 18 putative TonB-dependent outer membrane proteins were identified in the stalk of C. crescentus (30). The protein composition varied when cells were grown in minimal and rich media (28, 44). Xylose induced expression of nine putative TonB-dependent receptors (28). These studies did not relate the use of a nutrient to a particular TonB-dependent receptor. A protein of Bacteroides thetaiotaomicron, SusC, which showed sequence similarity to known TonB-dependent receptors of other gram-negative bacteria, has been related to the utilization of maltooligosaccharides (49). The dependence of TonB, ExbB, and ExbD transport on growth on substrates was not determined in any of these studies.
In conclusion, this study relates for the first time uptake of nutrients other than iron and vitamin B12 to an ExbB/ExbD-dependent transporter in a gram-negative cell. Maltodextrins derived from abundant starch in nature serve as nutrients. At low concentrations, as they probably occur in the habitats of C. crescentus, energy-coupled transport of the maltodextrins across the outer membrane is required. At higher concentrations, maltose and maltotriose can diffuse through the outer membrane in the absence of MalA. The diffusion pores exclude larger compounds, such as maltopentaose and maltohexaose.
Acknowledgments
We thank R. Benz and E. Maier, Biotechnology, University of Würzburg, for MalA conductance measurements; W. Boos, University of Konstanz, for providing radiolabeled maltodextrins and helpful discussions; Andrei Lupas for MalA sequence analysis; M. Valdebenito for HPLC analysis; K. A. Brune for critical reading of the manuscript; and K. Hantke for fruitful comments.
This work was supported by the Deutsche Forschungsgemeinschaft (Graduiertenkolleg “Infektionsbiologie,” Forschergruppe Bakterielle Zellhülle: Synthese, Funktion und Wirkort and Be 865/10) grant 120/11-1 to A.N. and the Proteom Centrum Tübingen and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Alexeyev, M. F., I. N. Shokolenko, and T. P. Croughan. 1995. New mini-Tn5 derivatives for insertion mutagenesis and genetic engineering in gram-negative bacteria. Can. J. Microbiol. 41:1053-1055. [DOI] [PubMed] [Google Scholar]
- 2.Benz, R. 1988. Structure and function of porins from gram-negative bacteria. Annu. Rev. Microbiol. 42:359-393. [DOI] [PubMed] [Google Scholar]
- 3.Benz, R., and F. Orlik. 2004. Functional reconstitution of specific porins, p. 183-212. In R. Benz (ed.), Bacterial and eukaryotic porins. Wiley-VCH, Weinheim, Germany.
- 4.Benz, R., A. Schmid, and G. H. Vos-Scheperkeuter. 1987. Mechanism of sugar transport through the sugar-specific LamB channel of Escherichia coli outer membrane. J. Membr. Biol. 100:21-29. [DOI] [PubMed] [Google Scholar]
- 5.Bradbeer, C. 1993. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 175:3146-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Lett. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V. 2003. Iron uptake by Escherichia coli. Front. Biosci. 8:s1409-s1421. [DOI] [PubMed] [Google Scholar]
- 8.Braun, V., and C. Herrmann. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261-268. [DOI] [PubMed] [Google Scholar]
- 9.Braun, V., and H. J. Krieger-Brauer. 1977. Interrelationship of the phage lambda receptor protein and maltose transport in mutants of Escherichia coli K12. Biochim. Biophys. Acta 469:89-98. [DOI] [PubMed] [Google Scholar]
- 10.Braun, V., K. Hantke, and W. Köster. 1998. Bacterial iron transport: mechanisms, genetics, and regulation. Met. Ions Biol. Syst. 35:67-145. [PubMed] [Google Scholar]
- 11.Cao, Z., P. Warfel, S. M. C. Newton, and P. Klebba. 2003. Spectroscopic observations of ferric enterobactin transport. J. Biol. Chem. 278:1022-1028. [DOI] [PubMed] [Google Scholar]
- 12.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cascales, E., R. Lloubes, and J. N. Sturgis. 2001. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42:795-807. [DOI] [PubMed] [Google Scholar]
- 14.Chimento, D. P., A. K. Mohanty, R. J. Kadner, and M. C. Wiener. 2003. Substrate-induced transmembrane signalling in the cobalamin transporter BtuB. Nat. Struct. Biol. 10:394-401. [DOI] [PubMed] [Google Scholar]
- 15.Chimento, D. P., R. J. Kadner, and M. C. Wiener. 2005. Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins 59:240-251. [DOI] [PubMed] [Google Scholar]
- 16.Clarke, T. E., L. W. Tari, and H. J. Vogel. 2001. Structural biology of iron uptake systems. Curr. Top. Med. Chem. 1:7-30. [DOI] [PubMed] [Google Scholar]
- 17.Clement, E., P. J. Mesini, F. Pattus, and I. J. Schalk. 2004. The binding mechanism of pyoverdin with the outer membrane receptor FpvA in Pseudomonas aeruginosa is dependent on its iron-loaded status. Biochemistry 43:7954-7965. [DOI] [PubMed] [Google Scholar]
- 18.Cobessi, D., H. Celia, N. Folschweiller, I. J. Schalk, M. A. Abdallah, and F. Pattus. 2005. The crystal structure of the pyoverdine membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 Å resolution. J. Mol. Biol. 347:121-134. [DOI] [PubMed] [Google Scholar]
- 19.Cornelissen, C. N. 2003. Transferrin-iron uptake by gram-negative bacteria. Front. Biosci. 8:d836-d847. [DOI] [PubMed] [Google Scholar]
- 20.Di Girolamo, P. M., and C. Bradbeer. 1971. Transport of vitamin B12 in Escherichia coli. J. Bacteriol. 106:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards, P., and J. Smith. 1991. A transducing bacteriophage for Caulobacter crescentus uses the paracrystalline surface array. J. Bacteriol. 173:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 23.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 26.Freundlieb, S., U. Ehmann, and W. Boos. 1988. Facilitated diffusion of p-nitrophenyl-α-D-maltohexaoside through the outer membrane of Escherichia coli. J. Biol. Chem. 263:314-320. [PubMed] [Google Scholar]
- 27.Heller, K. J., R. J. Kadner, and K. Günther. 1988. Suppression of the btuB451 mutation by mutations in the tonB gene suggests a direct interaction between TonB and TonB-dependent receptor proteins in the outer membrane of Escherichia coli. Gene 64:147-153. [DOI] [PubMed] [Google Scholar]
- 28.Hottes, A. K., M. Meewan, D. Yang, N. Arana, P. Romero, H. H. McAdams, and C. Stephens. 2004. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 186:1448-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang, B., K. Ru, Z. Yuan, C. B. Whitchurch, and J. S. Mattick. 2004. tonB3 is required for normal twitching motility and extracellular assembly of type IV pili. J. Bacteriol. 186:4387-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ireland, M. M., J. A. Karty, E. M. Quardokus, J. P. Reilly, and Y. V. Brun. 2002. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol. Microbiol. 45:1029-1041. [DOI] [PubMed] [Google Scholar]
- 31.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 17:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joly, N., A. Bohm, W. Boos, and E. Richet. 2004. MalK, the ATP-binding cassette component of the Escherichia coli maltodextrin transporter, inhibits the transcriptional activator MalT by antagonizing inducer binding. J. Biol. Chem. 279:33123-33130. [DOI] [PubMed] [Google Scholar]
- 33.Kaback, H. R. 1968. The role of the phosphoenolpyruvate-phosphotransferase system in the transport of sugars by isolated membrane preparations of Escherichia coli. J. Biol. Chem. 234:3711-3724. [PubMed] [Google Scholar]
- 34.Killmann, H., R. Benz, and V. Braun. 1993. Conversion of the FhuA transport protein into a diffusion channel through the outer membrane of Escherichia coli. EMBO J. 12:3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klebba, P. E. 2003. Three paradoxes of ferric enterobactin uptake. Front. Biosci. 8:s1422-s1436. [DOI] [PubMed] [Google Scholar]
- 36.Larsen, R. A., and K. Postle. 2001. Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J. Biol. Chem. 276:8111-8117. [DOI] [PubMed] [Google Scholar]
- 37.Larsen, R. A., G. E. Wood, and K. Postle. 1993. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol. Microbiol. 10:943-953. [DOI] [PubMed] [Google Scholar]
- 38.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771-778. [DOI] [PubMed] [Google Scholar]
- 39.McAdams, H. H., and L. Shapiro. 2003. A bacterial cell-cycle regulatory network operating in time and space. Science 301:1874-1877. [DOI] [PubMed] [Google Scholar]
- 40.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, et al. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pajatsch, M., A. Bock, and W. Boos. 1998. Enzymatic preparation of radiolabeled linear maltodextrins and cyclodextrins of high specific activity from [14C] maltose using amylomaltase, cyclodextrin glucosyltransferase and cyclodextrinase. Carbohydr. Res. 307:375-379. [DOI] [PubMed] [Google Scholar]
- 42.Paquelin, A., J. M. Ghigo, S. Bertin, and C. Wandersman. 2001. Characterization of HasB, a Serratia marcescens TonB-like protein specifically involved in the haemophore-dependent haem acquisition system. Mol. Microbiol. 42:995-1005. [DOI] [PubMed] [Google Scholar]
- 43.Perkins, D. N., D. J. C. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 44.Phadke, N. D., M. P. Molloy, S. A. Steinhoff, P. J. Ulintz, P. C. Andrews, and J. R. Maddock. 2001. Analysis of the outer membrane proteome of Caulobacter crescentus by two-dimensional electrophoresis and mass spectrometry. Proteomics 1:705-720. [DOI] [PubMed] [Google Scholar]
- 45.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 47.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 48.Prilipov, A., P. S. Phale, P. Van Gelder, J. P. Rosenbusch, and R. Koebnik. 1998. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol. Lett. 163:65-72. [DOI] [PubMed] [Google Scholar]
- 49.Reeves, A. R., J. N. D'Elia, J. Frias, and A. A. Salyers. 1996. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J. Bacteriol. 178:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, H., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Schalk, I. J., W. W. Yue, and S. K. Buchanan. 2004. Recognition of iron-free siderophores by TonB-dependent iron transporters. Mol. Microbiol. 54:14-22. [DOI] [PubMed] [Google Scholar]
- 52.Schöffler, H., and V. Braun. 1998. Transport across the outer membrane of Escherichia coli K12 via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol. Gen. Genet. 217:378-383. [DOI] [PubMed] [Google Scholar]
- 53.Scott, D. C., Z. Cao, Z. Qi, M. Bauler, J. D. Igo, S. M. C. Newton, and P. E. Klebba. 2001. Exchangeability of N-termini in the ligand-gated porins of Escherichia coli. J. Biol. Chem. 276:13025-13033. [DOI] [PubMed] [Google Scholar]
- 54.Seliger, S. S., A. R. Mey, A. M. Valle, and S. M. Payne. 2001. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39:801-812. [DOI] [PubMed] [Google Scholar]
- 55.Shapiro, L., H. H. McAdams, and R. Losick. 2002. Generating and exploiting polarity in bacteria. Science 298:1942-1946. [DOI] [PubMed] [Google Scholar]
- 56.Shevchenko, A., I. Chernushevich, W. Ens, K. G. Standing, B. Thomson, M. Wilm, and M. Mann. 1997. Rapid ‘de novo’ peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 11:1015-1024. [DOI] [PubMed] [Google Scholar]
- 57.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 58.Söding, J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951-960. [DOI] [PubMed] [Google Scholar]
- 59.Traub, I., S. Gaisser, and V. Braun. 1993. Activity domains of the TonB protein. Mol. Microbiol. 8:409-423. [DOI] [PubMed] [Google Scholar]
- 60.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources. From siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 61.White, J. C., P. M. Di Girolamo, M. L. Fu, Y. A. Preston, and C. Bradbeer. 1973. Transport of vitamin B12 in Escherichia coli. Location and properties of the initial B12-binding site. J. Biol. Chem. 248:3978-3986. [PubMed] [Google Scholar]
- 62.Yue, W. W., S. Grizot, and S. K. Buchanan. 2003. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J. Mol. Biol. 332:353-368. [DOI] [PubMed] [Google Scholar]
- 63.Zhao, Q., and K. Poole. 2002. Mutational analysis of the TonB1 energy coupler of Pseudomonas aeruginosa. J. Bacteriol. 184:1503-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]