Abstract
Mycobacterium peregrinum consists of two taxa: types I and II. We evaluated 43 clinical type II strains from throughout the United States. They were responsible for soft-tissue and bone infections, catheter-related infections, and possible pneumonitis. By carbohydrate utilization, they were indistinguishable from type I strains, being d-mannitol and trehalose positive. However, they had a distinct susceptibility pattern that included intermediate ciprofloxacin MICs but low clarithromycin and doxycycline MICs of ≤1 μg/ml. These features were also shared by reference isolates of Mycobacterium senegalense from African bovine cases of “farcy.” By 16S rRNA gene sequencing, the type II isolates shared 100% sequence identity with M. senegalense. Partial sequencing of the type II hsp65 gene (441 bp) revealed four sequevars showing ≥98.4% identity with each other and ≥98.6% identity with the sequence of five bovine strains of M. senegalense. There was ≤97.1% identity with M. peregrinum type I isolates and other Mycobacterium fortuitum group species. Sequencing of additional gene targets including the 16S-23S rDNA internal transcribed spacer region and the rpoB gene (partial sequence) revealed a similar phylogenetic grouping. DNA-DNA hybridization showed 76 to 99% relatedness between the bovine and human strains. These studies demonstrate that type II isolates are not isolates of M. peregrinum but represent human strains of M. senegalense. This study is the first to demonstrate this species as a human pathogen. Representative human M. senegalense strains include ATCC 35755 and newly submitted strains ATCC BAA-849, ATCC BAA-850, and ATCC BAA-851.
In 1962, Bojalil and colleagues described a new species of nonpigmented, rapidly growing mycobacteria which they named Mycobacterium peregrinum, a name derived from the Latin meaning “strange” or “foreign” (4). Presumably, the name was chosen based on the observation that the isolate was unusual or differed from the common Mycobacterium fortuitum. The type strain of the species was ATCC 14467T. A study by Rudolph Bönicke in 1966 was one of the first to note different biochemical groups within the species Mycobacterium fortuitum using carbohydrate studies (5). Bönicke noted that one group, which he called group A, was negative for carbohydrates; a second group, which he called group B, was positive for mannitol only; and a third group, which he called group C, was positive for mannitol and inositol.
A detailed study of rapidly growing mycobacteria by Kubica et al. and the International Working Group on Mycobacterial Taxonomy was published in 1972 (20). The authors noted that the biochemical results of Bojalil et al. with ATCC 14467T (4) could not be reproduced, likely because the authors of the earlier study utilized sugar fermentations which produce erratic results in taxa such as this one that produce complete sugar hydrolysis. They did observe that ATCC 14467T and related strains formed a unique taxonomic cluster with groupings similar to those of Bönicke. They positioned the three groups of Bönicke as biovariants of M. fortuitum. Bönicke's group B, which was positive for mannitol only among the commonly tested sugars, was named Mycobacterium fortuitum biovariant peregrinum. The other two groups became known as Mycobacterium fortuitum biovariant fortuitum (group A) and M. fortuitum third-biovariant complex (group C).
In 1981, Silcox et al. described laboratory criteria for the three biovariants and noted recovery of all three taxa from patient sources highly suggestive of clinical disease (32). Subsequent DNA-DNA homology studies (2, 22, 24) resolved the issue of the status of Bojalil's ATCC 14467T; they established this strain and Bonicke's original group B as a species. The original name of M. peregrinum proposed by Bojalil et al. (4) remains its accepted designation (22). The first complete and accurate 16S rRNA gene sequence of the type strain of M. peregrinum was submitted to GenBank in 2000 and updated in 2002 (accession number AF130308).
Compared to the other two Bönicke groups recognized within M. fortuitum (now also species, with the third biovariant comprising multiple species), little has been documented in the literature about M. peregrinum. It is known to be a human pathogen, but no large study of clinical disease has been reported, and isolates described prior to the 1980s were from sputum and of unknown clinical significance. Silcox et al. reported nine isolates in 1981 associated with cavitary lung disease and wounds of the sternum and foot (32). In 1983 Wallace et al. described 64 cases of human disease due to the M. fortuitum group, 3 of which were due to M. peregrinum and associated with skin or soft-tissue disease (52). One pseudo-outbreak of respiratory disease due to a contaminated ice machine (23) and one cluster of sternal wound infections in Hong Kong (54) due to M. peregrinum have been described since that time. No detailed laboratory investigation of the species has been reported since the species description by Kusunoki and Ezaki in 1992 (22).
The possibility that the species M. peregrinum might include more than one taxonomic group was first noted in 1985. Wallace and colleagues evaluated the β-lactamase patterns of M. peregrinum using polyacrylamide gels (49, 55). Two major patterns were seen, one represented by ATCC 14467T and the other represented by ATCC 35755. In a disk diffusion study of the susceptibility of the M. fortuitum group to an early quinolone known as pipemidic acid, Steele and Wallace (36) noted that clinical strains and the same two ATCC reference strains of M. peregrinum could be divided into two groups: one susceptible and one resistant to pipemidic acid. The resistant group represented 3.1% of 162 consecutive clinical isolates of the M. fortuitum group and was comparable in prevalence to the susceptible group. Subsequent studies of isoenzymes (3), mycobactin patterns (6), ribotyping (54), and PCR restriction enzyme analysis (PRA) patterns for M. peregrinum using the groEl heat shock protein gene (25) showed similar groupings, though the relationship to pipemidic acid susceptibility was not noted.
In a 1995 description of the use of PRA of the 441-bp Telenti fragment of the hsp65 gene (40) for identification of rapidly growing mycobacteria, Steingrube et al. noted that two restriction fragment length polymorphism patterns were present among isolates that met the current phenotypic definition of M. peregrinum published by Silcox et al. (32, 37). These two groups corresponded to the pipemidic acid-susceptible and -resistant groups and also differed in other characteristics, including growth at 45°C and use of acetamide as a carbon source (36, 37). These two groups then became known as the pipemidic acid-susceptible and -resistant groups, or M. peregrinum type I and type II. The pipemidic acid-susceptible group (type I) included the ATCC type strain 14467T of Bojalil et al. (4), while the pipemidic acid-resistant group (type II) included ATCC 35755.
In 1999, Ringuet et al. compared results of sequencing of the Telenti fragment (40) of the hsp65 gene among members of the M. fortuitum complex (30). Fourteen clinical isolates of M. peregrinum were sequenced, of which 11 (79%) were identical or within 2 bp of ATCC 14467T, while 3 strains (21%) had very distant sequences that were intermediate between M. peregrinum and M. fortuitum. The relationship of the latter group to pipemidic acid susceptibility or to PRA patterns was not reported, and strain ATCC 35755 was not studied.
While the 16S rRNA gene of the type strain of M. peregrinum has been sequenced and is present in GenBank as well as in other database systems such as RIDOM (17), strain ATCC 35755 has not, to our knowledge, been sequenced previously.
Molecular analysis has become an essential component of accurate mycobacterial species identification (34). We have collected a large series of the type II isolates of M. peregrinum, and here we present a polyphasic phenotypic and molecular analysis of this group with a proposal for its recognition as human isolates of the bovine species Mycobacterium senegalense.
MATERIALS AND METHODS
Isolates.
PRA of the hsp65 gene has been used routinely in the Mycobacteria/Nocardia Laboratory at the University of Texas Health Center for identification of rapidly growing mycobacteria since 1995. For the current study, PRA records were screened for isolates that had the PRA pattern corresponding to the pipemidic acid-resistant group of M. peregrinum (type II) described by Steingrube et al. (37). Upon analysis of these isolates, it became apparent that M. peregrinum type II isolates had a unique susceptibility pattern that allowed their recognition among isolates of the M. fortuitum group which had been submitted specifically for susceptibility testing without species identification. Two hundred fifty-five consecutive isolates of the M. fortuitum group that had been submitted for susceptibility testing and not identification (and that therefore had not undergone hsp65 PRA analysis) between 2001 and 2004 were then screened for the drug susceptibility pattern of the type II isolates. These isolates, with the type II drug pattern, were then taken from stocks frozen at −70°C, subcultured, and subjected to hsp65 PRA and biochemical analysis. Isolates confirmed by PRA as type II were included in the study. Approximately 10% of the M. peregrinum type II strains used in the study were initially recognized at the Mayo Clinic Laboratories or ARUP Laboratories based on partial sequencing of the 16S rRNA gene.
Reference strains used for comparison included the type strains of M. peregrinum (ATCC 14467T), M. houstonense (ATCC 49403T), M. fortuitum (ATCC 6841T), and M. neworleansense (ATCC 49404T), as well as all 10 strains of M. peregrinum currently in the ATCC: ATCC 35755 (formerly TMC 1545), ATCC 700686, ATCC 23001, ATCC 23015, ATCC 23017, ATCC 23020, ATCC 23022, ATCC 23041, ATCC 23047, and ATCC 23049. Also evaluated were M. senegalense ATCC 13781 and ATCC 35796T (both bovine strains) and M. farcinogenes ATCC 35753T. Additional bovine strains of M. senegalense were obtained from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany): DSM 43658, DSM 43661, DSM 43663, and DSM 43664.
Information including geographic location, source of infection, and type of disease was obtained. For some patients, follow-up from consultation with their physician on the use of drug therapy was also obtained. Chart reviews were not performed. Thus, informed consent was not obtained.
Phenotypic species identification.
Organisms were identified as rapidly growing mycobacteria on the basis of growth within 7 days on both Middlebrook 7H10 or 7H11 agar and Trypticase soy agar and on the basis of typical colony morphology. These isolates had also been identified as rapidly growing mycobacteria or as members of Mycobacterium species before their submission to our laboratory by the referring laboratory. Isolates were identified as presumptive members of the Mycobacterium fortuitum group on the basis of the absence of pigmentation and typical drug susceptibility patterns that included disk diffusion susceptibility to polymyxin B and amikacin (51), susceptible MICs to amikacin (38), ciprofloxacin (43), and sulfamethoxazole, and intermediate or susceptible MICs to cefoxitin (38, 44) and imipenem (see the next section for methods and breakpoints) (39, 44).
Isolates within the M. fortuitum group were identified as M. peregrinum by the pattern of utilization of carbohydrates as sole carbon sources according to Silcox et al. (32) and Tsukamura (41), which included being positive for d-mannitol and negative for i-myo-inositol, d-glucitol (sorbitol), and citrate (14-day incubation). These studies had not included M. senegalense, while the study by Chamoiseau showed this rapidly growing mycobacterium to have a similar carbohydrate utilization pattern (12).
Isolates were also identified as M. peregrinum by PRA of the 441-bp Telenti fragment of the hsp65 gene according to Steingrube et al. (37). By using PRA and selected growth and biochemical tests, the isolates were grouped as M. peregrinum type I on the basis of pipemidic acid susceptibility (36), failure to grow at 45°C, inability to utilize acetamide as a sole carbon source, and an hsp65 gene PRA pattern of BstEII fragments of 235 and 210 bp and HaeIII fragments of 145, 140, and 100 bp. The previous study by Steingrube et al. (37) had shown M. peregrinum ATCC 14467T of Bojalil et al. to belong to this group. Isolates were grouped as M. peregrinum type II if they were pipemidic acid resistant by the disk diffusion method (36), grew at 45°C, were able to utilize acetamide as a sole carbon source, and had an hsp65 gene PRA pattern with BstEII fragments of 235, 115, and 80 bp and with HaeIII fragments of 140, 125, and 60 bp or 140, 125, and 100 bp. The reference strain for this group was ATCC 35755.
Susceptibility testing.
Broth microdilution MIC testing for susceptibility to 12 antimicrobial agents was performed according to standard methods (9, 27, 38, 39). Drugs tested included amikacin, ciprofloxacin, sulfamethoxazole, imipenem, cefoxitin, tobramycin, and doxycycline (group A). More-recent isolates were also tested for susceptibility to five newer agents: linezolid (47), levofloxacin, gatifloxacin, meropenem, and clarithromycin (group B) (10, 11). Susceptible and resistant breakpoints were those recently recommended by the Clinical Laboratory Standards Institute (CLSI, formerly NCCLS) except for meropenem, levofloxacin, and gatifloxacin, which have not been addressed by the CLSI (27). For these drugs, the breakpoints used were those of bacterial species that grow aerobically (26). Quality control organisms utilized were Staphylococcus aureus ATCC 29213 and M. peregrinum ATCC 700686, as recommended by CLSI guidelines (27).
Disk diffusion testing for susceptibility to cephalothin (30 μg) (50), polymyxin B (300 U) (51), pipemidic acid (30 μg) (36), and amikacin (30 μg) (51) was also performed using commercial disks as previously described. The pipemidic acid disks are no longer commercially available, and consequently they were tested only against early isolates.
HPLC.
High-performance liquid chromatography (HPLC) of cell wall mycolic acids was performed as previously described using fluorescence detection (8, 46, 53). Reference strains used for comparison included M. peregrinum ATCC 14467T of Bojalil, M. senegalense ATCC 35796T, M. fortuitum ATCC 6841T, M. houstonense ATCC 49403T, and M. farcinogenes ATCC 35753T.
MALDI-TOF MS.
A previous study suggested that intact cell matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS), which detects ions derived from cell wall components, could be used for mycobacterial species and/or strain identification (M. L. Pignone, K. M. Greth, and J. Tang, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. I-77, 2003; K. M. Greth, M. I. Pignone, and J. Tang, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. C-237, 2003). Selected mycobacterial strains were chosen for comparison by this method. Details of the method are included in the supplemental material.
Reference strains used included M. fortuitum ATCC 6841T, M. porcinum ATCC 33776T, M. farcinogenes ATCC 35753T, M. houstonense ATCC 49403T, M. senegalense ATCC 35796T and ATCC 13781, M. chelonae ATCC 35752T, Mycobacterium chelonae subsp. niacinogenes ATCC 19237T, and M. abscessus ATCC 19977T.
Partial sequencing of the hsp65 gene.
DNA from isolates of M. peregrinum type II (23 isolates, including ATCC 35755) and the bovine strains of M. senegalense (6 isolates) underwent sequencing of the 441-bp Telenti fragment using primers TB11 and TB12 (40) and an ABI PRISM 310 Genetic Analyzer capillary sequencer (Applied Biosystems). Sequences were compared to those in GenBank or sequenced in-house. Strains for hsp65 sequencing were the same as those for 16S rRNA gene extended or complete sequencing. Pairwise alignments were performed using ClustalW and phylogenetic analyses using the neighbor-joining method in the MEGA 2.1 software (21).
PRA.
PRA was performed on the 441-bp Telenti fragment of the hsp65 gene as described for mycobacteria (40) and modified by Steingrube et al. for rapidly growing mycobacteria (37). The amplicon was digested with BstEII and HaeIII and separated on 3% metaphor agarose gels. Fragment sizes in base pairs were estimated on a computerized Bio Image System (Millipore, Bedford, Mass.).
Sequencing of the 16S rRNA gene.
Genomic DNAs from selected isolates were sequenced using the MicroSeq 500 16S rDNA Bacterial Identification System (Applied Biosystems, Foster City, Calif.), which sequences the first 500 bp at the 5′ end (13, 15, 28) and includes hypervariable regions A and B (19, 33). Data analysis was performed using MicroSeq version 1.4 (Applied Biosystems) at the Mayo Clinic (Rochester, Minn.) or ARUP (Salt Lake City, Utah).
This fragment of the 16S rRNA gene cannot differentiate between several rapidly growing species of mycobacteria, and additional sequencing in the latter part of the 16S rRNA gene was performed as described by Turenne et al. (42) on selected clinical isolates with the type II pattern (n = 22), M. peregrinum ATCC strains 35755, 23001, and 23015, and the bovine strains of M. senegalense from the DSMZ. Analysis spanned at least Escherichia coli bp 970 to 1390, a region that includes differentiating bases within species that are identical in the first 500 bp of the gene. To our knowledge, no mycobacterial species that are identical in these two regions present variations outside this range.
Complete 16S rRNA sequencing (E. coli bp 8 to 1508) was performed for clinical strains MF-1868, MF-548, and MF-1796 and was previously performed for reference strains M. senegalense ATCC 35796T, M. farcinogenes ATCC 35753T, and M. peregrinum ATCC 14467T, ATCC 23001, and ATCC 23015 (42). Sequence databases used for comparison included an in-house database (42), RIDOM (http://www.ridom-rdna.de) (17, 18), and GenBank. Phylogenetic analyses were performed using MEGA2 as described above.
16S-23S ITS1 region sequencing.
The same strains for which hsp65 sequencing was performed also underwent sequencing of the 16S-23S internal transcribed spacer (ITS1) region using primers 16S-1511f (5′-AAG TCG TAA CAA GGT ARC CG-3′) and 23S-23r (5′-TCG CCA AGG CAT CCA CC-3′) (14). Sequences were compared to available sequences currently in GenBank and RIDOM (17, 18). Phylogenetic analyses were performed using MEGA2 as described above.
Partial sequencing of the rpoB gene.
Sequence analysis of a 723-bp region of the rpoB gene was performed as described by Adékambi et al. (1), using primers MycoF and MycoR for amplification and primers MycoseqF and MycoseqR for sequencing of the PCR product. Sequences were compared with those of Adékambi et al. (1), available in GenBank, and were retrieved for phylogenetic analyses using MEGA2 as described above.
DNA-DNA hybridization.
Total-DNA relatedness studies were performed on a set of nine strains. The methods used to cultivate cells and to prepare, isolate, and purify labeled and unlabeled DNA, as well as the methods used for DNA reassociation and the separation of single-stranded and double-stranded DNA on hydroxyapatite, have been described previously (31). DNA relatedness was determined at the optimal (75°C) reassociation temperature. Percent divergence was calculated to the nearest 0.5% (7).
Nucleotide sequence accession numbers and newly submitted strains.
Sequences of the partial hsp65 gene (accession numbers AY684045 to AY684049), the 16S-23S internal transcribed spacer regions (AY684050 to AY684055), and the partial rpoB gene (AY684056 to AY684063) recognized in this study have been deposited in GenBank.
Human isolates of M. senegalense have been deposited in the American Type Culture Collection as ATCC BAA-849, BAA-850, and BAA-851.
RESULTS
Isolates.
By routine PRA of clinically submitted isolates, 24 isolates of M. peregrinum type II were identified. Among the 255 consecutive clinical isolates of the M. fortuitum group submitted for susceptibility testing between 2001 and 2004, 15 had the susceptibility pattern suggestive of M. peregrinum type II and were screened by phenotypic testing and PRA. Of these, 14 (5.5%) were M. peregrinum type II. Five additional isolates were recognized based on partial 16S rRNA gene sequences.
Overall, a total of 43 clinical isolates of M. peregrinum type II were identified and studied in more detail. The majority of the 42 isolates with available information were from skin and soft-tissue infections (25/42 [60%]), with infections following surgical procedures or accidental trauma. At least five cases were known to be associated with osteomyelitis. Other sources included catheter- or pacemaker-related infections (7/42 [17%]) and respiratory sites (8/42 [19%]) whose clinical significance was generally unknown (see Table S1 in the supplemental material).
The 41 isolates for which geographic sites were known were from 16 states and the Dominican Republic. The majority of the isolates (22/41 [54%]) were from Texas, Florida, and North Carolina, and 26/41 (63%) were from Southern coastal states.
Phenotypic identification.
All of the isolates of M. peregrinum type II grew in less than 7 days on Trypticase soy agar and Middlebrook 7H10 or 7H11 agar, were nonpigmented, and had morphology typical of the M. fortuitum group. On Mueller-Hinton agar, used for disk diffusion susceptibility testing, the isolates tended to show a fine growth and were often unreadable at 72 h, which is relatively unusual for members of the M. fortuitum group.
The results of growth and biochemical testing are listed in Table S2 in the supplemental material. The isolates were similar to other members of the M. fortuitum group including M. peregrinum type I (ATCC 14467T) and previous groupings that did not recognize the two types. Selected tests were performed against all isolates (group A). These revealed that all isolates were nonpigmented and grew at 30 and 35°C and that 78% grew at 45°C. Like isolates of M. peregrinum type I, including ATCC 14467T, the isolates were able to utilize d-mannitol as a sole carbon source but not citrate, i-myo-inositol, or d-sorbitol (20, 32, 37). Isolates were also negative for l-rhamnose and positive (90%) for d-trehalose (but generally positive late [at 2 weeks], unlike the pattern for other carbohydrates, for which isolates were positive after 3 days of incubation). Unlike isolates of M. peregrinum type I, the type II isolates were positive for acetamide, and most grew at 45°C, as previously noted (37).
Some traditional tests were performed against a minority of the type II isolates (group B). They showed that the type II isolates were arylsulfatase positive at 3 days, reduced nitrate, grew on MacConkey agar, and were iron uptake positive (20, 32).
The two bovine ATCC reference strains of M. senegalense had the same biochemical reactions as the M. peregrinum type II isolates, except that both isolates of the former were d-trehalose negative. The single available ATCC strain of M. peregrinum type II (ATCC 35755) was also d-trehalose negative (see Table S2 in the supplemental material).
M. farcinogenes ATCC 35753T was also studied, because it is the closest relative by 16S rRNA gene sequencing to M. senegalense, along with M. houstonense ATCC 49403T, with which M. farcinogenes shares 100% 16S rRNA gene sequence identity. M. farcinogenes grew slowly, had a low semiquantitative catalase reaction (10 mm), was Tween positive, was negative for 3-day arylsulfatase, nitrate, iron uptake, urease, and d-mannitol, and did not grow on 5% NaCl. These results are similar to those listed in the catalogue of the Trudeau Mycobacterial Culture Collection (TMC) (25a), where the isolate was stored prior to being transferred to the ATCC.
Susceptibility testing.
Susceptibility testing was performed using the CLSI-recommended broth microdilution method for rapidly growing mycobacteria (27). The type II isolates gave a strikingly uniform pattern of susceptibility that was readily separable from those of other clinical species within the M. fortuitum group, including the M. peregrinum type I isolates. In general they were highly drug susceptible (Table 1). For all the type II isolates, clarithromycin MICs were ≤1 μg/ml, much lower than those for most other recognized species (10, 46, 48). In keeping with the resistance of type II isolates to pipemidic acid (an early quinolone) (36), ciprofloxacin MICs were in the upper limits of the CLSI susceptible category (27). The type II isolates were all doxycycline susceptible, with MICs of ≤0.5 μg/ml, which makes this the first M. fortuitum group taxon whose members are uniformly susceptible to the tetracyclines (27, 38, 39, 45, 46). The type II isolates also had low susceptible MICs to amikacin, were uniformly susceptible to linezolid, imipenem, and sulfamethoxazole, and were susceptible or intermediate to cefoxitin (27).
TABLE 1.
Susceptibility results of Mycobacterium peregrinum type II clinical isolates (including two designated reference strains) and two reference strains of M. senegalense
| Isolates tested and drug | MIC (μg/ml) for the indicated ATCC reference strain
|
Type II clinical isolates
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Type II BAA-849 | M. peregrinum 35755 |
M. senegalense
|
No. tested | MIC (μg/ml)a
|
|||||
| 13781 | 35796T | Range | 50% | 90% | Mode | ||||
| Group A (all isolates) | |||||||||
| Amikacin | 0.5 | ≤0.25 | ≤2 | ≤1 | 43 | ≤0.25-4 | ≤1 | ≤2 | ≤1 |
| Cefoxitin | 16 | 8 | 16 | 16 | 43 | 4-32 | 16 | 32 | 16 |
| Ciprofloxacin | 0.25 | ≤0.063 | 1 | 0.5 | 43 | 0.25-1 | 0.5 | 1 | 0.5 |
| Doxycycline | 1 | ≤0.25 | ≤0.12 | ≤0.12 | 42 | ≤0.12-1 | ≤0.25 | 0.5 | ≤0.12 |
| Imipenem | 2 | ≤0.5 | 4 | 2 | 42 | 0.25-4 | 1 | ≤4 | 1 |
| Sulfamethoxazole | 64 | ≤1 | 16 | 16 | 42 | ≤1-64 | 8 | 32 | 8 |
| Tobramycin | 8 | 2 | 4 | 8 | 43 | 1-8 | 4 | 8 | 4 |
| Group B (selected isolates) | |||||||||
| Clarithromycin | 0.5 | ≤0.12 | 0.5 | 0.25 | 30 | ≤0.12-2 | 0.25 | 1 | ≤0.25 |
| Levofloxacin | 0.5 | ≤0.12 | 0.5 | 0.5 | 23 | ≤0.12-1 | 0.5 | 1 | 0.5 |
| Linezolid | 4 | ≤2 | 4 | ≤2 | 30 | 1-8 | 2 | 8 | ≤2 |
| Gatifloxacin | 0.25 | ≤0.06 | 0.25 | 0.12 | 24 | ≤0.12-0.5 | ≤0.12 | 0.25 | ≤0.12 |
| Meropenem | 1 | 2 | 8 | 4 | 24 | ≤0.5-4 | 2 | 4 | 1, 2, 4 |
50% and 90%, MICs at which 50 and 90% of isolates are inhibited, respectively.
The two ATCC bovine strains of M. senegalense (ATCC 35796T and ATCC 13781) gave the same susceptibility pattern as did the type II clinical isolates (see Table 1).
HPLC.
By fluorescence HPLC, M. peregrinum type II isolates gave mycolic acid patterns that were generally indistinguishable from those of other members of the M. fortuitum-M. smegmatis group, including M. fortuitum, M. porcinum, the bovine strain of M. senegalense, M. houstonense, and M. peregrinum ATCC 14467T of Bojalil (type I). The reference strain of M. farcinogenes, ATCC 35753T, had a late major peak that differed from the peaks of these isolates (see Fig. S3 in the supplemental material).
Mass spectroscopy.
The 22 isolates tested were grouped into four clusters that showed intracluster variations of 0.12 or less (see Fig. S4 in the supplemental material). Six of the seven ATCC strains identified as M. peregrinum type I formed a cluster along with the M. peregrinum type strain, ATCC 14467T. The three isolates of M. chelonae/abscessus formed the second cluster. The third cluster consisted of both isolates of M. porcinum and three out of four strains of M. peregrinum type II, while the fourth cluster consisted of one strain each of M. peregrinum type I and type II, the two bovine strains of M. senegalense, M. fortuitum, M. houstonense, and M. farcinogenes. Overall, the clustering of strains by MALDI-TOF MS was similar to that by molecular methods.
Partial sequencing of the hsp65 gene.
Four sequevars (sqv), which diverged from each other by 1 to 7 bp, or 0.2 to 1.6%, were identified among the type II isolates. These were referred to as sqv II through V (Table 2). These sequences, along with the sequences for the bovine strains of M. senegalense, were submitted to GenBank and were assigned accession numbers AY684045 to AY684049.
TABLE 2.
Sequevar designations of 8 reference strains and 19 clinical strains for which sequencing of the partial hsp65 and rpoB genes and the complete 16S-23S ITS1 region was performed
| Strain(s) | Sequevar by sequencing of:
|
||
|---|---|---|---|
| hsp65 | 16S-23S rRNA ITS1 | rpoB | |
| M. senegalense ATCC 35796T, DSM 43661, DSM 43663, DSM 43664 | I | I | I |
| M. peregrinum type II | |||
| MF-278, MF-417, MF-421, MF-528, MF-532, MF-548, MF-593, MF-620, MF-1756, MF-1787, MF-1788, MF-1796, MF-1868, MF-2132 | II | IIb | IIc |
| MF-263 (ATCC BAA-849) | II | VI | VI |
| ATCC 35755 | III | IIIa | IIIa |
| MF-378 | III | IIIa | IIIb |
| MF-495 | III | IIIa | II |
| MF-386 (ATCC BAA-851) | III | No datad | IIIa |
| MF-125a | III | IIIb | IIIa |
| MF-1738 (ATCC BAA-850) | IV | IV | IVa |
| MF-1816 | IV | IV | IVb |
| MF-1925 | V | No data | V |
Strain MF-125 presented with a single-base-pair variation in the 16S rRNA gene from the sequence of M. senegalense at bp 1246 in the E. coli sequence.
Strains MF-593, MF-1788, and MF-1796 were not tested.
Strains MF-1788 and MF-1796 were not tested.
No data, evidence of the presence of two different ribosomal operons having different 16S-23S sequences; analysis could not be performed.
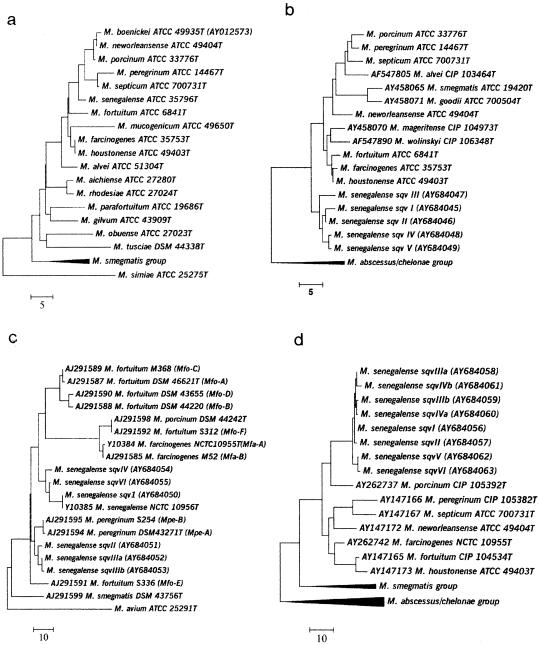
The two ATCC bovine strains of M. senegalense, ATCC 13781 and ATCC 35796T, and the three DSM bovine strains of M. senegalense had the same hsp65 sequence, sqv I, and exhibited 2-, 6-, 4-, and 5-bp differences (0.5 to 1.4% divergence) from sequevars II, III, IV, and V of the M. peregrinum type II group, respectively (Table 2). The most prevalent sequevar, sqv II, was seen in 15 of 23 (65%) type II isolates and differed by only 2 bp from sqv I at positions 168 (T to C) and 342 (A to G) of the 441-bp fragment. Interestingly, these two variations are present in all sequevars analyzed against the animal strains of M. senegalense and also in closely related taxa such as M. farcinogenes, M. houstonense, M. neworleansense, M. fortuitum, and M. peregrinum (type I) (see Table S5 in the supplemental material). The largest divergence (1.6%) was seen between sqv III and V, with 7 variations. The intraspecies variation exhibited by the different type II sequevars was comparable to the variation seen with the bovine reference strains of M. senegalense (≤1.4%), and all clustered together by phylogenetic analyses (Fig. 1b).
FIG. 1.
Phylogenetic analyses of M. senegalense by 16S rRNA gene sequencing (1,427 bp) (a) and of its various sequevars by sequencing of the partial hsp65 gene (441 bp) (b), the 16S-23S ITS1 region (c), and the partial rpoB gene (687 bp) (d). Analyses were determined using the neighbor-joining method. Bars, numbers of base pair differences.
These sequences were then compared to the closest relatives based on the 16S rRNA gene sequence, mostly strains within the M. fortuitum group and M. farcinogenes, and against hsp65 sequences submitted to GenBank. The species established as closest by 16S rRNA analysis exhibited a 6- to 10-bp hsp65 sequence divergence from the four type II sequevars and M. senegalense. These species (strains) included the two species with identical 16S rRNA gene sequences, M. houstonense (ATCC 49403T) and M. farcinogenes (ATCC 35753T), and M. fortuitum (ATCC 6841T) (see Table S5 in the supplemental material).
The remaining species within the M. fortuitum group exhibited a higher degree of hsp65 sequence divergence, with differences of ≥12 bp (≥2.7%) from the type II gene sequences. This included all isolates of the M. fortuitum third-biovariant sorbitol-negative group (M. neworleansense ATCC 49404T, M. porcinum ATCC 33776T, M. boenickei ATCC 49935T, M. septicum ATCC 700731T), as well as M. peregrinum type I (ATCC 14467T). The latter differed from the four type II sequevars by 13, 13, 14, and 15 bp, respectively, or ≥3.2%. All nucleotide variations seen among the type II sequevars, the bovine reference strains of M. senegalense, and their closest relatives were synonymous.
PRA of the hsp65 partial gene sequence.
The four hsp65 gene sequevars among the clinical isolates of M. peregrinum type II gave one major and two minor hsp65 gene PRA patterns (PRA types II to IV). The majority of isolates (40/43 [93%]) gave fragments of 235, 115, and 85 bp with BstEII and 140, 125, and 60 bp with HaeIII (PRA type II). These selected isolates (including ATCC 35755) were subsequently identified as having hsp65 gene sequevars II and III as described above. A second group (2/43 [5%]) (PRA type III) had the same BstEII restriction fragments but gave HaeIII restriction fragments of 140, 125, and 100 bp. Sequencing showed that these isolates lacked one of the six HaeIII restriction sites present in the 441-bp sequence of sequevars II and III due to a C→T transition at base 402 (bp 546 of the complete gene sequence). This pattern identified M. peregrinum type II hsp65 sequevar IV. A third group (1/43 [2%]) had the same HaeIII restriction fragments as PRA type III but gave a BstEII fragment of 125 (PRA type IV) instead of 115 bp, resulting from a T→C transition at base 327 (bp 471 of the complete hsp65 gene), and identified hsp65 sequevar V.
The 10 reference strains of M. peregrinum in the ATCC were subjected to PRA. Of these 10 isolates, 1 produced the common PRA pattern of M. senegalense (ATCC 35755), 1 exhibited the pattern of M. porcinum (ATCC 23041), and 5 showed patterns of M. peregrinum type I (ATCC 14467T, 23001, 23020, 23047, and 23049). Three isolates (ATCC 23015, 23017, and 23022) yielded unrecognized PRA patterns.
A restriction digest pattern identical to that of the hsp65 PRA type II was observed with three other species: the M. fortuitum third-biovariant d-sorbitol-negative species M. neworleansense (ATCC 49404T), the M. fortuitum third-biovariant d-sorbitol-positive species M. houstonense (ATCC 49403T), and M. farcinogenes ATCC 35753T. However, these species were readily separated from the type II isolates by drug susceptibilities, carbohydrate utilization, and growth rates.
The two bovine reference strains of M. senegalense (ATCC 13781 and ATCC 35796T) gave a different and unique PRA pattern (PRA type I) that consisted of the same BstEII fragments but only two HaeIII fragments, of 185 and 140 bp. Sequencing of the hsp65 gene fragment showed that the bovine strains also lack a single HaeIII cutting site present in the other closely related taxa, a result of a C→T substitution at position 168 of the 441-bp fragment. This sequence was designated sequevar I. This profile was also observed for the three bovine strains of M. senegalense from the German culture collection: DSM 43661, DSM 43663, and DSM 43664. Thus, despite the fact that for M. peregrinum type II sequevar II, there was only a 2-bp difference from the bovine M. senegalense sequevar I, the two sequevars gave different PRA patterns by use of HaeIII.
Partial 16S rRNA gene sequencing.
A total of 34 M. peregrinum type II isolates were subjected to 500-bp 16S rRNA gene sequencing using MicroSeq (16, 28). All resulted in 100% sequence identity to the type strains of M. senegalense, M. farcinogenes, and M. houstonense.
A total of 25 of the type II isolates and representatives from each hsp65 PRA type were subjected to more-complete 16S rRNA gene sequencing. Two belonged to other species and were excluded. Of the remaining 23 isolates (including ATCC 35755), 22 had 100% sequence identity with M. senegalense, while isolate MF-125 showed a single-base-pair difference from M. senegalense at E. coli position 1246. Overall, all 43 clinical isolates underwent partial (500-bp) and/or more-complete (903-bp) 16S rRNA gene sequencing.
Of the animal reference strains from the DSMZ, three were identified as M. senegalense with 100% identity (DSM 43661, DSM 43663, and DSM 43664) while DSM 43658 was identified as M. farcinogenes/M. houstonense (species which cannot be distinguished from each other by 16S rRNA genes) and was therefore excluded from the study.
By using the complete 16S rRNA gene sequence, the closest species to M. senegalense are M. farcinogenes, M. houstonense, and M. porcinum, each with 4-bp differences, followed by M. neworleansense (5 bp), M. boenickei (6 bp), M. septicum (7 bp), M. fortuitum (8 bp), and both M. alvei and M. peregrinum with 11-bp differences. The taxonomic placement of M. senegalense among its closest mycobacterial relatives can be seen in Fig. 1a.
Two additional ATCC strains of M. peregrinum (type I) were available for testing, and 16S rRNA gene analysis revealed 100% sequence identity with M. peregrinum ATCC 14467T of Bojalil.
16S-23S ITS1 region.
Rapidly growing mycobacteria, with the exception of M. abscessus and M. chelonae, carry two ribosomal operons in which interoperon variability in spacer regions can render sequence analysis unsuccessful if the PCR product is used directly for sequencing. However, since ITS1 sequence analysis was previously successful for bovine strains of M. senegalense (16), 20 type II clinical isolates were subjected to such analysis, along with M. senegalense ATCC 35796T and the 3 DSM animal strains identified as M. senegalense by 16S rRNA gene analysis. Of these isolates, two (MF-386 and MF-1925) showed two different sequences and could not be analyzed. The remaining 18 type II strains and the bovine M. senegalense strains revealed six sequevars: sqv I, II, IIIa, IIIb, IV, and VI. As much as possible, these were in correlation with the sequevar numbers assigned to hsp65 gene sequevars to avoid confusion. No sequevar V was assigned, since the one isolate in this group, strain MF-1925, could not be analyzed by the current PCR-based sequencing method. All four bovine strains shared 100% sequence identity, as previously reported (16) (sqv I). Sqv IIIa and IIIb differed from each other by 1 bp. While our analysis of the type strain M. senegalense ATCC 35796T revealed 100% identity with type strains DSM 43656T in RIDOM and NCTC 10956T in GenBank, none of the five M. peregrinum type II ITS1 sequevars corresponded with any close similarity with other sequences in public databases (Fig. 1c). However, organisms with top scores included members of the M. fortuitum group, including M. senegalense. As with the hsp65 gene, the ITS1 sequences of the bovine M. senegalense strains had a similar but unique sequence compared with the clinical M. peregrinum type II isolates. Six examples of these type II sequences were submitted to GenBank as sequences AY684050 to AY684055.
Partial sequencing of the rpoB gene.
Partial rpoB gene sequencing (687 bp) was performed for 21 clinical strains of M. peregrinum type II, M. senegalense bovine strains ATCC 35796T, DSM 43661, and DSM 43663, and M. farcinogenes TMC 805T (ATCC 35753T). Eight closely related sequevars were identified. These were designated sqv I, II, IIIa, IIIb, IVa, IVb, V, and VI. As with ITS1 sequences, these were in correlation with the sequevar numbers assigned to hsp65 gene sequevars to avoid confusion (Table 2). The seven type II sequevars differed from each other by 1 to 7 bases, with 99.0 to 99.9% identity. The type II sequevars differed by only 1 to 4 bases from the bovine sequence (sqv I) of M. senegalense (99.4 to 99.9% identity). These sequences were submitted to GenBank and were assigned accession numbers (AY684056 to AY684063). As with the sequevars of hsp65, the base pair changes with rpoB were all synonymous. However, this was also true when the rpoB sequences were compared to that of M. porcinum CIP 105392T (AY262737). rpoB sequences of Mycobacterium species are limited in GenBank, yet sequences of most members of the non- or late-pigmenting rapidly growing species are available for comparison (1). The closest match for the eight sequevars was the rpoB sequence of M. porcinum CIP 105392T (AY262737), with 96.8 to 97.2% identities. While partial rpoB gene sequence determination revealed a larger number of sequevars among the type II group, there is a significant jump in the number of base pair differences when the type II isolates are compared by rpoB sequencing to their closest relative, M. porcinum (Table 3).
TABLE 3.
Hamming distancesa among M. senegalense rpoB sequevar I (region V), the seven sequevars of M. peregrinum type II, and the type strains of the closest speciesb
| Sequevar or strainc | No. of base pair differences between the indicated sequevars or strains
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | IIIa | IIIb | IVa | IVb | V | VI | M. porcinum | M. farcinogenes | M. neworleansense | M. fortuitum | M. houstonense | M. septicum | |
| II | 1 | |||||||||||||
| IIIa | 1 | 2 | ||||||||||||
| IIIb | 2 | 3 | 1 | |||||||||||
| IVa | 1 | 2 | 2 | 1 | ||||||||||
| IVb | 3 | 4 | 2 | 3 | 4 | |||||||||
| V | 3 | 4 | 4 | 5 | 4 | 6 | ||||||||
| VI | 4 | 5 | 5 | 6 | 5 | 7 | 1 | |||||||
| M. porcinum | 17 | 18 | 18 | 19 | 18 | 20 | 20 | 19 | ||||||
| M. farcinogenes | 21 | 22 | 22 | 23 | 22 | 24 | 24 | 23 | 24 | |||||
| M. neworleansense | 26 | 27 | 27 | 28 | 27 | 29 | 28 | 27 | 24 | 19 | ||||
| M. fortuitum | 27 | 26 | 28 | 29 | 28 | 30 | 30 | 29 | 26 | 12 | 23 | |||
| M. houstonense | 31 | 32 | 32 | 33 | 32 | 34 | 34 | 33 | 30 | 16 | 27 | 6 | ||
| M. septicum | 35 | 36 | 36 | 37 | 36 | 38 | 38 | 37 | 35 | 24 | 25 | 25 | 28 | |
| M. peregrinum type I | 37 | 38 | 38 | 39 | 38 | 40 | 40 | 39 | 31 | 24 | 25 | 30 | 34 | 19 |
Numbers of base pair differences.
The closest species are M. porcinum, M. farcinogenes, M. neworleansense, M. fortuitum, M. houstonense, M. septicum, and M. peregrinum type I.
Sqv II through VI are sequevars of M. peregrinum type II.
DNA-DNA hybridization.
Mycobacterium senegalense ATCC 35796T was found to be related at the species level to the human M. peregrinum type II strains MF-263 (ATCC BAA-849), MF-125, MF-417, MF-1738 (ATCC BAA-850), MF-386 (ATCC BAA-851), MF-1925, and ATCC 35755 (76 to 99% relatedness, with divergence ranging from 1 to 2%). DNA hybridization data indicated that M. senegalense ATCC 35796T was unrelated to M. peregrinum ATCC 14467T (47% relatedness at 75°C) (Table 4).
TABLE 4.
DNA relatedness of representative strains labeled with Mycobacterium senegalense ATCC 35796T
| Source of unlabeled DNA | hsp65 sequevar | DNA-DNA bindinga (divergence)b |
|---|---|---|
| M. senegalense ATCC 35796T | I | 100 |
| M. peregrinum type II MF-263 (ATCC BAA-849) | II | 99 (1.5) |
| M. peregrinum type II MF-125 | III | 96 (1.5) |
| M. peregrinum type II MF-417 | II | 90 (1.5) |
| M. peregrinum type II MF-1738 (ATCC BAA-850) | IV | 89 (2.0) |
| M. peregrinum type II MF-386 (ATCC BAA-851) | III | 83 (1.5) |
| M. peregrinum type II (ATCC 35755) | III | 76 (1.0) |
| M. peregrinum type I ATCC 14467T | 47 (8.0) |
Relative binding ratio [(percentage of heterologous DNA bound to hydroxyapatite/percentage of homologous DNA bound by hydroxyapatite) × 100] at 75°C. Values were means for at least two hybridization reactions.
Divergence is the decrease in thermal stability (in degrees Celsius) of heterologous DNA duplexes compared with those of homologous DNA duplexes.
DISCUSSION
In this study, the bovine strains of M. senegalense and the human strains of M. peregrinum type II exhibited minimal differences and were closely clustered by polyphasic molecular and phenotypic properties. Their 16S rRNA genes exhibited 100% sequence identity, and the most common sequevar of the hsp65 gene found among the type II clinical isolates (sequevar II) differed by only 2 bp from the sequence for the bovine strains of M. senegalense (99.55% identity), although this difference affected a HaeIII cutting site, resulting in different PRA patterns. The most common sequevar of the rpoB gene among the type II isolates differed by only 1 bp from M. senegalense (99.9% identity). Similarly minimal differences were seen with the sequevar groupings of the ITS1 sequence, with a closely clustered relationship for all three sequences that, for other rapidly growing species, has been seen only among strains of the same species (1, 48). The degree of relatedness or difference for housekeeping genes that define the same or different species has not been established, however, and may vary depending on the species or the gene (sequence). Some guidelines for the rpoB sequence have been suggested, however. Adekambi et al. studied 20 type strains and 59 clinical strains of rapidly growing mycobacteria and observed an interspecies variability for the 723-bp rpoB fragment of >3%, with an intraspecies variability of <1.7% (1). The type II M. peregrinum strains and the bovine M. senegalense strains exhibited a variability of 0.1% to 1.0% among eight sequevars.
The bovine and human strains had identical drug susceptibility patterns that were unique relative to those of other members of the M. fortuitum complex in that all isolates had the combination of doxycycline susceptibility, higher ciprofloxacin MICs, and clarithromycin MICs of ≤1 μg/ml. They also had the same biochemical pattern except that they may differ in their abilities to utilize d-trehalose as a sole carbon source. Finally, DNA-DNA hybridization showed the bovine and human strains to have >70% homology, with 5/7 human strains exhibiting ≥89% homology with the bovine M. senegalense type strain. The obvious conclusion drawn from the highly similar or identical phenotypic and molecular profiles of the bovine strains of M. senegalense from Senegal and the human “M. peregrinum” type II isolates from the United States is that such similarity is possible only if they belong to the same species. DNA-DNA hybridization studies confirmed this hypothesis (35).
The fact that the sequevar of the hsp65 gene, the ITS1 region, and the rpoB gene found in the bovine strains of M. senegalense was not found among clinical isolates shows that the two groups are not identical. Interestingly, similar differences (i.e., 100% identical complete 16S rRNA gene sequence but minor hsp65 gene sequence variance) was demonstrated between porcine strains of M. porcinum from Japan and human strains of M. porcinum from the United States (48), and we have observed similar differences in the simian type strain of M. simiae (from India or Africa) and human clinical isolates (R. Wallace, Y. Zhang, and C. Turenne, unpublished data). As with the current M. senegalense study, however, DNA-DNA relatedness studies comparing the porcine and human M. porcinum strains proved them to be one species (31). In the situations involving M. porcinum and M. simiae, as well as M. senegalense, there is also a major geographic separation between the origins of the animal and human strains, and this may also be a factor in the differences observed.
A curious observation was presented here with regard to M. farcinogenes. Interestingly, both M. senegalense and M. farcinogenes were originally described in the same paper by Chamoiseau as causes of bovine lymphadenitis (“farcy”) in Senegal; hence the names of the two species (12). Although there is 100% 16S rRNA gene sequence identity between M. farcinogenes and the nonpigmented, rapidly growing third-biovariant complex species M. houstonense (31), M. farcinogenes was originally described as a yellow-pigmented, slow-growing species (12). In our hands, M. farcinogenes was demonstrated to be a nonpigmented slow grower, growing best on Middlebrook 7H11 agar and Löwenstein-Jensen agar, not at all on Trypticase soy agar, unlike other rapidly growing species. In a 1983 study of 15 strains by Ridell and Goodfellow (29), the species was not pigmented and required more than 5 days to grow. It was negative for 3-day arylsulfatase and catalase and generally clustered with slow growers. It should be noted that characterization of the type strain (ATCC 35753T) while it was housed in the Trudeau Mycobacterial Culture Collection (TMC 805) showed it to be negative for arylsulfatase at 3 days, negative for nitrate, and negative for iron uptake; M. peregrinum type II, M. senegalense, and other members of the M. fortuitum group are positive by all of these tests (25a). We obtained similar results with our testing. Given that the species grows and behaves biochemically as a slow grower, it is remarkable that it has a 16S rRNA gene sequence identical to that of another, rapidly growing species, that its hsp65 sequence shows >99% identity with that of a rapidly growing species, and that the HPLC pattern of its mycolic acids and the mass spectroscopy pattern of cell wall proteins are indistinguishable from those of members of the M. fortuitum group; thus, it behaves like a chimera that is part rapid grower and part slow grower.
In this study, we have evaluated the ITS1 regions and the partial hsp65 and rpoB genes of 23 strains of M. peregrinum type II. These sequences were heterogeneous, exhibiting several closely related sequevars. This should not be unexpected, since protein-coding genes are anticipated to exhibit more base pair divergence than the 16S rRNA gene. In a study by Ringuet et al. (30), the 441-bp Telenti fragment was sequenced for multiple species of rapidly growing mycobacteria, all of which exhibited some degree of intraspecies DNA polymorphism. As previously noted, these investigators studied 14 clinical isolates of M. peregrinum, 11 of which exhibited three sequences which were identical to or closely aligned with those of ATCC 14467T. A recent study of M. porcinum showed it to consist of five closely related hsp65 gene sequevars, one of which was unique to animals (pigs) (48).
Description of human isolates of M. senegalense.
The organism is acid fast and grows readily in 3 to 4 days on Middlebrook 7H10 or 7H11 agar as well as on Trypticase soy agar. It is nonpigmented and produces smooth to rough colonies. It is positive for 3-day arylsulfatase, acetamide, iron uptake, nitrate reduction, d-mannitol, and d-trehalose as a sole carbon source (14 days) and generally grows at 45°C. It is negative for l-rhamnose, d-sorbitol, i-myo-inositol, and citrate. It contains typical mycobacterial mycolic acids, and by HPLC its pattern is not distinguishable from that of other members of the M. fortuitum/smegmatis group. The isolates are uniformly susceptible to doxycycline, clarithromycin, amikacin, imipenem, and sulfamethoxazole by use of current CLSI guidelines (27). They have relatively high MICs for ciprofloxacin (0.5 to 1.0 μg/ml) compared to M. fortuitum or M. peregrinum type I and are susceptible or intermediate to cefoxitin. The organism is a recognized cause of human disease, including skin and soft-tissue infections, posttraumatic or postsurgical osteomyelitis, catheter-related infections, and possibly pulmonary infections. Its complete 16S rRNA gene sequence (bp 8 to 1500) shows 100% identity with that of the bovine strain of M. senegalense ATCC 35796T. Its hsp65 gene exhibits four closely related sequence variants and gives three PRA patterns with the 441-bp Telenti fragment and restriction enzymes BstEII and HaeIII. The most common PRA pattern is not distinguishable from the pattern seen with M. farcinogenes, M. houstonense, and M. neworleansense but is distinguishable from the bovine sequevar of M. senegalense and from M. peregrinum type I (ATCC 14467T). The hsp65 sequence observed for the bovine reference strains of M. senegalense (ATCC 35796T and ATCC 13781) and its associated PRA pattern with HaeIII of fragments of 185 and 140 bp were not observed among the human isolates. The human strains exhibited five closely related ITS1 sequevars, while a few strains showed evidence of two different spacer sequences. They also exhibited seven closely related sequevars of the rpoB gene partial sequence. By DNA-DNA reassociation, they exhibited >70% relatedness with the bovine strain from Senegal ATCC 35796T. The strains chosen as species reference strains are ATCC 35755 (a sputum isolate from Texas, formerly TMC 1545); ATCC BAA-849, which was recovered from a case of foot osteomyelitis in Florida; ATCC BAA-850, which was recovered from an abdominal wound in Florida; and ATCC BAA-851, which was recovered from a thigh infection in Texas. Examples of the different sequevars have been deposited in GenBank as sequences AY684045 to AY684049 (hsp65), AY684056 to AY684063 (rpoB), and AY684050 to AY684055 (16S-23S ITS1 region).
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Adékambi, T., P. Colson, and M. Drancourt.2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baess, I.1982. Deoxyribonucleic acid relatedness among species of rapidly-growing mycobacteria. Acta Pathol. Microbiol. Scand. Sect. B 90:371-375. [DOI] [PubMed] [Google Scholar]
- 3.Blom-Potar, M.-C., H. L. David, and N. Rastogi.1989. Isoenzymes as tools to discriminate various subdivisions in the Mycobacterium fortuitum complex. Acta Leprol. 7:39-43. [PubMed] [Google Scholar]
- 4.Bojalil, L. F., J. Cerbón, and A. Trujillo.1962. Adansonian classification of mycobacteria. J. Gen. Microbiol. 28:333-346. [DOI] [PubMed] [Google Scholar]
- 5.Bönicke, R.1966. The occurrence of atypical mycobacteria in the environment of man and animal. Bull. Int. Union Tuberc. Lung Dis. 37:361-368. [Google Scholar]
- 6.Bosne, S., and V. V. Lévy-Frébault.1992. Mycobactin analysis as an aid for the identification of Mycobacterium fortuitum and Mycobacterium chelonae subspecies. J. Clin. Microbiol. 30:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner, D. J., F. W. Hickman-Brenner, J. V. Lee, A. G. Steigerwalt, G. R. Fanning, D. G. Hollis, J. J. Farmer III, R. E. Weaver, S. W. Joseph, and R. J. Seidler.1983. Vibrio furnissii (formerly aerogenic biogroup of Vibrio fluvialis), a new species isolated from human feces and the environment. J. Clin. Microbiol. 18:816-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, B. A., B. Springer, V. A. Steingrube, R. W. Wilson, G. E. Pfyffer, M. J. Garcia, M. C. Menendez, B. Rodriguez-Salgado, K. C. Jost, S. H. Chiu, G. O. Onyi, E. C. Böttger, and R. J. Wallace, Jr.1999. Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov., two new rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 49:1493-1511. [DOI] [PubMed] [Google Scholar]
- 9.Brown, B. A., J. M. Swenson, and R. J. Wallace, Jr.1992. Broth microdilution MIC test for rapidly growing mycobacteria, p. 5.11.1. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook. American Society for Microbiology, Washington, D.C.
- 10.Brown, B. A., R. J. Wallace, Jr., G. Onyi, V. DeRosas, and R. J. Wallace III.1992. Activities of four macrolides including clarithromycin against Mycobacterium fortuitum, Mycobacterium chelonae, and Mycobacterium chelonae-like organisms. Antimicrob. Agents Chemother. 36:180-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown-Elliott, B. A., R. J. Wallace, Jr., C. J. Crist, L. Mann, and R. W. Wilson.2002. Comparison of in vitro activities of gatifloxacin and ciprofloxacin against four taxa of rapidly growing mycobacteria. Antimicrob. Agents Chemother. 46:3283-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chamoiseau, G.1979. Etiology of farcy in African bovines: nomenclature of the causal organisms Mycobacterium farcinogenes Chamoiseau and Mycobacterium senegalense (Chamoiseau) comb. nov. Int. J. Syst. Bacteriol. 29:407-410. [Google Scholar]
- 13.Cloud, J. L., H. Neal, R. Rosenberry, C. Y. Turenne, M. Jama, D. R. Hillyard, and K. C. Carroll.2002. Identification of Mycobacterium spp. by using a commercial 16S ribosomal DNA sequencing kit and additional sequencing libraries. J. Clin. Microbiol. 40:400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dostal, S., E. Richter, and D. Harmsen.2003. Concise guide to mycobacteria and their molecular differentiation, p. 133-138. Ridom Press, Würzburg, Germany.
- 15.Hall, L., K. A. Doerr, S. L. Wohlfiel, and G. D. Roberts.2003. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J. Clin. Microbiol. 41:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamid, M. E., A. Roth, O. Landt, R. M. Kroppenstedt, M. Goodfellow, and H. Mauch.2002. Differentiation between Mycobacterium farcinogenes and Mycobacterium senegalense strains based on 16S-23S ribosomal DNA internal transcribed spacer sequences. J. Clin. Microbiol. 40:707-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmsen, D., J. Rothganger, M. Frosch, and J. Albert.2002. RIDOM: ribosomal differentiation of medical micro-organisms database. Nucleic Acids Res. 30:416-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmsen, D., S. Dostal, A. Roth, S. Niemann, J. Rothgänger, M. Sammeth, J. Albert, M. Frosch, and E. Richter.2003. RIDOM: comprehensive and public sequence database for identification of Mycobacterium species. BMC Infect. Dis. 3:26. [Online.] http://www.biomedcentral.com/1471-2334/3/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirshner, P., P. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Bottger.1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubica, G. P., I. Baess, R. E. Gordon, P. A. Jenkins, J. B. G. Kwapinski, C. McDurmont, S. R. Pattyn, H. Saito, V. Silcox, J. L. Stanford, K. Takeya, and M. Tsukamura.1972. A co-operative numerical analysis of rapidly growing mycobacteria. J. Gen. Microbiol. 73:55-70. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei.2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 22.Kusunoki, S., and T. Ezaki.1992. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int. J. Syst. Bacteriol. 42:240-245. [DOI] [PubMed] [Google Scholar]
- 23.Laussucq, S., A. L. Baltch, R. P. Smith, R. W. Smithwick, B. J. Davis, E. K. Desjardin, V. A. Silcox, A. B. Spellacy, R. T. Zeimis, H. M. Gruft, R. C. Good, and M. L. Cohen.1988. Nosocomial Mycobacterium fortuitum colonization from a contaminated ice machine. Am. Rev. Respir. Dis. 138:891-894. [DOI] [PubMed] [Google Scholar]
- 24.Lévy-Frébault, V., F. Grimont, P. A. D. Grimont, and H. L. David.1986. Deoxyribonucleic acid relatedness study of the Mycobacterium fortuitum-Mycobacterium chelonae complex. Int. J. Syst. Bacteriol. 36:458-460. [Google Scholar]
- 25.Lungu, O., P. Della Latta, I. Weitzman, and S. Silverstein.1994. Differentiation of Nocardia from rapidly growing Mycobacterium species by PCR-RFLP analysis. Diagn. Microbiol. Infect. Dis. 18:13-18. [DOI] [PubMed] [Google Scholar]
- 25a.National Institutes of Health.1980. Catalogue of the Trudeau Mycobacterial Culture Collection. National Institutes of Health publication 80-289. National Institutes of Health, Bethesda, Md.
- 26.NCCLS.2002. Performance standards for antimicrobial susceptibility testing. Twelfth informational supplement. NCCLS document M100-S12. NCCLS, Wayne, Pa.
- 27.NCCLS.2003. Susceptibility testing of mycobacteria, nocardia and other aerobic actinomycetes. Approved standard. NCCLS document M24-A. NCCLS, Wayne, Pa. [PubMed]
- 28.Patel, J. B., D. G. B. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nachamkin.2000. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J. Clin. Microbiol. 38:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ridell, M., and M. Goodfellow.1983. Numerical classification of Mycobacterium farcinogenes, Mycobacterium senegalense and related taxa. J. Gen. Microbiol. 129:599-611. [DOI] [PubMed] [Google Scholar]
- 30.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier.1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Saubolle, M. A., T. E. Kiehn, M. H. White, M. F. Rudinsky, and D. Armstrong.1996. Mycobacterium haemophilum: microbiology and expanding clinical and geographic spectra of disease in humans. Clin. Microbiol. Rev. 9:435-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schinsky, M. F., R. E. Morey, A. G. Steigerwalt, M. P. Douglas, R. W. Wilson, M. M. Floyd, W. R. Butler, M. I. Daneshvar, B. A. Brown-Elliott, R. J. Wallace, Jr., M. M. McNeil, D. J. Brenner, and J. M. Brown.2004. Taxonomic variation in the Mycobacterium fortuitum third-biovariant complex: description of Mycobacterium boenickei sp. nov., Mycobacterium houstonense sp. nov., Mycobacterium neworleansense sp. nov., Mycobacterium brisbanense sp. nov., and recognition of Mycobacterium porcinum from human clinical isolates. Int. J. Syst. Evol. Microbiol. 54:1653-1667. [DOI] [PubMed] [Google Scholar]
- 32.Silcox, V. A., R. C. Good, and M. M. Floyd.1981. Identification of clinically significant Mycobacterium fortuitum complex isolates. J. Clin. Microbiol. 14:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Springer, B., E. C. Böttger, P. Kirschner, and R. J. Wallace, Jr.1995. Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int. J. Syst. Bacteriol. 45:262-267. [DOI] [PubMed] [Google Scholar]
- 34.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Böttger.1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kampfer, M. C. J. Maiden, X. Nesme, R. Rossella-Mora, J. Swings, H. G. Truper, A. Vauterin, A. C. Ward, and W. B. Whitman.2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 36.Steele, L. C., and R. J. Wallace, Jr.1987. Ability of ciprofloxacin but not pipemidic acid to differentiate all three biovariants of Mycobacterium fortuitum from Mycobacterium chelonae. J. Clin. Microbiol. 25:456-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steingrube, V. A., J. L. Gibson, B. A. Brown, Y. Zhang, R. W. Wilson, M. Rajagopalan, and R. J. Wallace, Jr.1995. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J. Clin. Microbiol. 33:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swenson, J. M., C. Thornsberry, and V. A. Silcox.1982. Rapidly growing mycobacteria: testing of susceptibility to 34 antimicrobial agents by broth microdilution. Antimicrob. Agents Chemother. 22:186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swenson, J. M., R. J. Wallace, Jr., V. A. Silcox, and C. Thornsberry.1985. Antimicrobial susceptibility of five subgroups of Mycobacterium fortuitum and Mycobacterium chelonae. Antimicrob. Agents Chemother. 28:807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer.1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsukamura, M.1984. Identification of mycobacteria, p. 54-58. Mycobacteriosis Research Laboratory of the National Chubu Hospital, Obu, Aichi, Japan.
- 42.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani.2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallace, R. J., Jr., G. Bedsole, G. Sumter, C. V. Sanders, L. C. Steele, B. A. Brown, J. Smith, and D. R. Graham.1990. Activities of ciprofloxacin and ofloxacin against rapidly growing mycobacteria with demonstration of acquired resistance following single-drug therapy. Antimicrob. Agents Chemother. 34:65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallace, R. J., Jr., B. A. Brown, and G. Onyi.1991. Susceptibilities of Mycobacterium fortuitum biovar fortuitum and the two subgroups of Mycobacterium chelonae to imipenem, cefmetazole, cefoxitin, and amoxicillin-clavulanic acid. Antimicrob. Agents Chemother. 35:773-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace, R. J., Jr., B. A. Brown, V. A. Silcox, M. Tsukamura, D. R. Nash, L. C. Steele, V. A. Steingrube, J. Smith, G. Sumter, Y. Zhang, and Z. Blacklock.1991. Clinical disease, drug susceptibility, and biochemical patterns of the unnamed third biovariant complex of Mycobacterium fortuitum. J. Infect. Dis. 163:598-603. [DOI] [PubMed] [Google Scholar]
- 46.Wallace, R. J., Jr., B. A. Brown-Elliott, L. Hall, G. Roberts, R. W. Wilson, L. B. Mann, C. J. Crist, S. H. Chiu, R. Dunlap, M. J. Garcia, J. T. Bagwell, and K. C. Jost, Jr.2002. Clinical and laboratory features of Mycobacterium mageritense. J. Clin. Microbiol. 40:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallace, R. J., Jr., B. A. Brown-Elliott, S. C. Ward, C. J. Crist, L. B. Mann, and R. W. Wilson.2001. Activities of linezolid against rapidly growing mycobacteria. Antimicrob. Agents Chemother. 45:764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallace, R. J., Jr., B. A. Brown-Elliott, R. W. Wilson, L. Mann, L. Hall, Y. Zhang, K. C. Jost, Jr., J. M. Brown, A. Kabani, M. F. Schinsky, A. G. Steigerwalt, C. J. Crist, G. D. Roberts, Z. Blacklock, M. Tsukamura, V. Silcox, and C. Turenne.2004. Clinical and laboratory features of Mycobacterium porcinum. J. Clin. Microbiol. 42:5689-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace, R. J., Jr., D. R. Nash, T. Udou, V. A. Steingrube, L. C. Steele, J. M. Swenson, and V. A. Silcox.1985. Isoelectric focusing of beta-lactamases in Mycobacterium fortuitum: association of a single enzyme pattern with cefoxitin resistance. Am. Rev. Respir. Dis. 132:1093-1097. [DOI] [PubMed] [Google Scholar]
- 50.Wallace, R. J., Jr., V. A. Silcox, M. Tsukamura, B. A. Brown, J. O. Kilburn, W. R. Butler, and G. Onyi.1993. Clinical significance, biochemical features, and susceptibility patterns of sporadic isolates of the Mycobacterium chelonae-like organism. J. Clin. Microbiol. 31:3231-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallace, R. J., Jr., J. M. Swenson, V. A. Silcox, and R. C. Good.1982. Disk diffusion testing with polymyxin and amikacin for differentiation of Mycobacterium fortuitum and Mycobacterium chelonei. J. Clin. Microbiol. 16:1003-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallace, R. J., Jr., J. M. Swenson, V. A. Silcox, R. C. Good, J. Tschen, and M. S. Stone.1983. Spectrum of disease due to rapidly growing mycobacteria. Rev. Infect. Dis. 5:657-679. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, R. W., V. A. Steingrube, E. C. Böttger, B. Springer, B. A. Brown-Elliott, V. Vincent, K. C. Jost, Jr., Y. Zhang, M. J. Garcia, S. H. Chiu, G. O. Onyi, H. Rossmoore, D. R. Nash, and R. J. Wallace, Jr.2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks, and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int. J. Syst. Evol. Microbiol. 51:1751-1764. [DOI] [PubMed] [Google Scholar]
- 54.Yew, W., P. Wong, H. Woo, C. Yip, C. Chan, and F. Cheng.1993. Characterization of Mycobacterium fortuitum isolates from sternotomy wounds by antimicrobial susceptibilities, plasmid profiles, and ribosomal ribonucleic acid gene restriction patterns. Diagn. Microbiol. Infect. Dis. 17:111-117. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y., R. J. Wallace, Jr., V. A. Steingrube, B. A. Brown, D. R. Nash, V. A. Silcox, and M. Tsukamura.1992. Isoelectric focusing patterns of β-lactamases in the rapidly growing mycobacteria. Tuber. Lung Dis. 73:337-344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.