Abstract
A novel denaturing high-performance liquid chromatography (DHPLC)-based technique allows rapid high-resolution analysis of PCR products. We used this technique for unequivocal molecular identification of seven Candida species. We show the application of this PCR/DHPLC approach for direct detection and identification of yeast species from blood cultures and for detection of Candida colonization in the gastrointestinal tract of allogeneic transplant patients.
The incidence of Candida infections has increased significantly over the past decades, being an important cause of morbidity and mortality in both critically ill and immunocompromised patients (1). Candida species colonize mucosal surfaces in humans and represent a major source for local and systemic endogenous infections. In addition, the ability of these yeasts to adhere to inert plastic surfaces (11) renders them important pathogens associated with indwelling catheters. Although Candida albicans is still the most prevalent species in clinical samples, the increasing incidence of non-C. albicans Candida species such as C. dubliniensis, C. krusei, C. glabrata, C. parapsilosis, C. tropicalis, C. guilliermondii, and C. lusitaniae has been reported (10, 13, 15, 19). Given the high lethality of invasive Candida infections, and due to various in vitro susceptibilities of different Candida species (10), rapid and reliable identification at the species level is required for correct treatment of these patients. Routine morphological and biochemical identification is time-consuming and laborious. Therefore, a number of PCR-based techniques have been developed for identification of medically important yeast species. The most popular targets for PCR amplification are the internal transcribed spacer regions ITS1 and ITS2 (2, 3, 6, 8, 16, 17). Methods used for identification of amplicons include hybridization with species-specific probes, direct sequencing, precise identification of the product length using a capillary sequencer, and characterization of restriction fragment length polymorphisms. However, most methods are inappropriate for the accurate analysis of clinical samples containing more than one yeast species. To overcome these limitations, we developed a novel technique based on PCR amplification of the variable ITS2 region and subsequent analysis of the amplicons by using the WAVE microbial analysis system (Transgenomic Ltd., Omaha, NE) (4, 5, 9, 18). This technique allows the analysis of complex samples containing more than one Candida species. Rapid verification of results can be done by direct sequencing of individual collected peaks.
MATERIALS AND METHODS
Yeast strains.
We used reference strains from Candida membranaefaciens (ATCC 201377), Candida tropicalis (DSMZ 5991), Candida parapsilosis (ATCC 90018), Candida magnoliae (ATCC 201379), Candida lusitaniae (DSMZ 70102), Candida dubliniensis (CBS 7987), Candida albicans (ATCC 90028), Candida inconspicua (DSMZ 70631), Candida krusei (ATCC 6258), Cryptococcus neoformans (ATCC 90112), Candida kefyr (DSMZ 11954), Candida glabrata (ATCC 90030), Trichosporon mucoides (ATCC 201383), Candida norvegensis (DSMZ 70760), and Saccharomyces cerevisiae (ATCC 9763). In addition, clinical isolates of C. albicans (25 isolates), C. glabrata (22 isolates), C. tropicalis (9 isolates), C. kefyr (7 isolates), C. krusei (7 isolates), C. parapsilosis (7 isolates), and C. dubliniensis (12 isolates) were included to study the reproducibility of the method. Strains were collected consecutively in the clinical mycology laboratory from April through July 2001 from inpatients of the Charité University Hospital. The collection of C. albicans and C. glabrata strains was discontinued once 25 and 22 isolates of these species were identified, respectively. Copy strains were excluded.
Isolation of DNA.
Total DNA was isolated by phenol-chloroform extraction from 1 g of feces, 200 μl of blood culture fluid, or 200 μl of aqueous suspensions of yeast strains according to modified protocols (20). Samples were homogenized in 1 ml lysis buffer (500 mM Tris, pH 9.0, 20 mM EDTA, 10 mM NaCl, 1% sodium dodecyl sulfate) with 10 μl proteinase K (10 U/ml) and incubated at 56°C. After 1 h, 150 μl of phenol and 0.2 g of zirconium beads (0.1 mm; BioSpec) were added, and cells were disintegrated by shaking three times for 30 s in a FastPrep FP120 (Bio101) device. The aqueous phase was transferred to a fresh sterile tube, and DNA was further purified by chloroform/isoamyl alcohol (24:1) extraction. After centrifugation at 16,000 × g for 5 min, the aqueous phase was transferred to a fresh sterile tube, and DNA extraction was repeated with chloroform/isoamyl alcohol (24:1). The aqueous phase was transferred to a fresh sterile tube, and DNA was precipitated on ice with 0.1 volume of sodium acetate (3 M) and 2.5 volumes of 99% (vol/vol) ethanol. After an additional centrifugation at 16,000 × g for 20 min, the supernatant fluid was discarded. The DNA pellet was washed twice with 70% (vol/vol) ethanol, dried in a Speed Vac, and dissolved in 100 μl of Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 8.0). Extraction of pure lysis buffer was performed in parallel as a negative control.
Conditions for PCR amplification.
ITS2 amplification primers (MycoSeq, catalog no. 79018/19) were obtained from Transgenomic (Omaha, NE). The PCR mixture (total volume, 50 μl) consisted of 1 μl of template, 0.5 μM of forward and reverse primers, 5 μl of 10× Optimase buffer (Transgenomic, Omaha, NE), 400 μM of each deoxyribonucleotide triphosphate (Roche Diagnostics, Mannheim, Germany), and 2.5 U of Optimase DNA polymerase (Transgenomic, Omaha, NE). PCR products were generated using a T3 thermocycler (Biometra, Goettingen, Germany) with the following touchdown parameters: a denaturation step at 95°C for 2 min and 14 cycles at 95°C for 30 s, 61.3°C for 30 s with a −0.5°C increment every 30 s (reaching 54.3°C after 14 cycles), and 72°C for 40 s, followed by 19 cycles at 95°C for 30 s, 54.3°C for 30 s, and 72°C for 40 s and a final extension step at 72°C for 5 min. PCR products were verified on 2% (wt/vol) agarose gel in 1× Tris-borate-EDTA buffer after ethidium bromide staining.
DHPLC analysis.
PCR products were analyzed by ion-pair reverse-phase denaturing high-performance liquid chromatography (DHPLC) on the WAVE microbial analysis system (Transgenomic, Omaha, NE). Basically, 1 μl of each PCR product was loaded onto the autosampler of the system. PCR products were separated on a DNASep HT column cartridge (catalog no. 993710; Transgenomic, Omaha, NE). Separation conditions were adapted by temperature, flow rate, and gradient. The optimal separation conditions are listed in Table 1. PCR products were stained by using an intercalating stain (HS staining solution I, catalog no. 553440; Transgenomic, Omaha, NE) and visualized with an HSX-3500 fluorescence detector. In some experiments, unstained PCR products were analyzed using a UV detector. All buffer solutions were obtained from Transgenomic (Omaha, NE). Results were analyzed using Navigator software version 1.5.4 (Build 23).
TABLE 1.
Optimized conditions for the identification of yeasts using the WAVE microbial analysis systema
| Gradient | Time (min) | % A | % B |
|---|---|---|---|
| Loading | 0 | 52 | 48 |
| Start gradient | 0.1 | 45 | 55 |
| Stop gradient | 18.1 | 36 | 64 |
| Start clean | 18.2 | 52 | 48 |
| Stop clean | 18.3 | 52 | 48 |
| Start equilibrate | 18.4 | 52 | 48 |
| Stop equilibrate | 19.9 | 52 | 48 |
The program was run at 60°C and at a flow rate of 0.5 ml/min. A, 0.1 M triethylammonium acetate; B, 0.1 M triethylammonium acetate in 25% acetonitrile.
Sequence analysis.
ITS2 amplicons from selected peak fractions were collected with the FCW 200 DNA fragment collector. The volume of each fraction varied from 100 to 200 μl depending upon the peak size and height. An aliquot was applied as a template for reamplification by PCR using the same primers and conditions described above. Purified PCR products (PCR purification kit; QIAGEN, Hilden, Germany) served as a template for sequencing reactions using the chemicals and protocols from Beckman Coulter (Fullerton, CA). ITS2 sequences were determined with the CEQ8000 DNA sequencing system (Beckman Coulter, Fullerton, CA) as recommended by the vendor. The sequences were compared with entries in public databases by using the BLAST software (BlastN algorithm) provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
Conditions for blood culture analysis.
Growth-positive BacT/ALERT FA (bioMérieux) blood culture bottles were analyzed by Gram stain. Microscopically positive materials (yeast cells in Gram stain) were subcultured on Sabouraud agar and CHROMagar (Mast Diagnostics) and incubated at 30°C and 37°C, respectively. Yeasts grown for 2 to 7 days were subcultured and identified using phenotypical and molecular methods as described previously (7, 8, 12, 14, 16). For some experiments, blood culture bottles from our clinical diagnostic microbiology laboratory that did not show growth after 7 days of incubation were spiked with 2 × 105 yeast cells (final concentration, 5 × 103 yeast cells/ml) and incubated under routine conditions. When growth was indicated by the system, yeasts were identified as described above. Aliquots of each blood culture were used for DNA isolation and subsequent PCR/DHPLC analysis.
Collection and treatment of fecal samples.
Stool samples were taken from allogeneic stem cell recipients. Fresh fecal samples (2 to 3 g) were aliquoted, and part (0.5 to 1 g) was transferred to 1.5 ml nonselective liquid culture medium (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl) containing 70% (vol/vol) glycerin and immediately frozen at −80°C for cultural analysis. For culture, aliquots of 100 μl each of the stool samples were plated onto Sabouraud agar and CHROMagar (Mast Diagnostics) and incubated at 30°C and 37°C, respectively. In addition, 4 ml Sabouraud medium was inoculated with 200 μl of the stool sample and incubated for 2 days at 30°C. One hundred microliters of this suspension was then spread on Sabouraud agar and CHROMagar (Mast Diagnostics) and incubated at 30°C and 37°C, respectively. Identification of yeasts was done using phenotypic tests as described above. Remaining portions (1 to 1.5 g) were transferred to sterile plastic tubes containing 10 ml lysis buffer for subsequent DNA extraction.
RESULTS
Universal fungus-specific primers flanking the ITS2 region complementary to conserved regions of the 5.8S and 28S rRNA genes, respectively, were used to generate PCR products ranging from ∼200 to ∼300 bp.
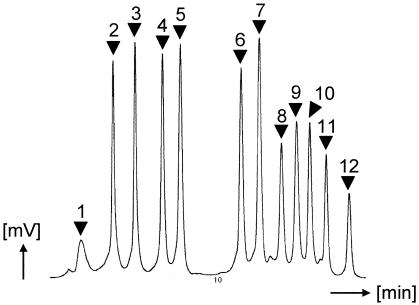
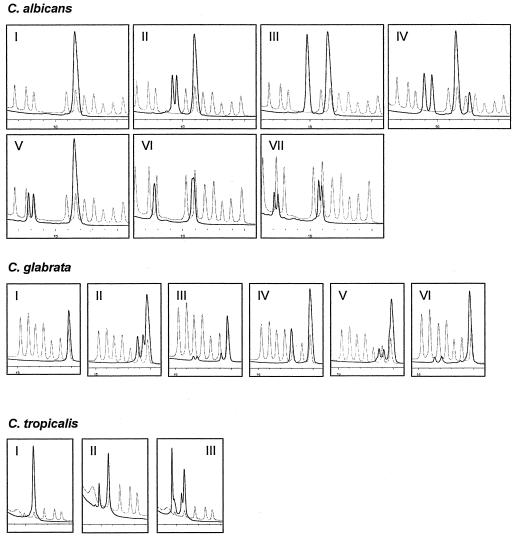
The PCR/DHPLC approach with ITS2 as the target region was evaluated using reference strains from Candida membranaefaciens, Candida tropicalis, Candida parapsilosis, Candida magnoliae, Candida lusitaniae, Candida dubliniensis, Candida albicans, Candida inconspicua, Candida krusei, Cryptococcus neoformans, Candida kefyr, and Candida glabrata. Both single and mixed ITS2 amplicons were injected into the WAVE microbial analysis system (Fig. 1). Resulting peak profiles showed unique signals for each reference strain. Hence, a mixture of these PCR products was used as a marker for the identification of fungal DNA (Fig. 1). Reference strains from Trichosporon mucoides, Candida norvegensis, and Saccharomyces cerevisiae showed unique peak profiles but were excluded from the marker mixture to limit the complexity of the marker set (data not shown). Reproducibility was studied using clinical isolates of C. albicans (25 isolates), C. glabrata (22 isolates), C. tropicalis (9 isolates), C. kefyr (7 isolates), C. krusei (7 isolates), C. parapsilosis (7 isolates), and C. dubliniensis (12 isolates). Although major peaks were consistent with the marker, some additional side peaks were observed (Fig. 2). Thus, in addition to strains exhibiting one major peak only (type I), six variant profiles of C. albicans (types II to VII), five additional profiles of C. glabrata (types II to VI), and two additional profiles of C. tropicalis (types II and III) could be distinguished (Fig. 2).
FIG. 1.
DHPLC peak profile of mixed fungal amplicons. Shown are mixtures of PCR-amplified ITS2 DNA fragments from C. membranaefaciens (peak 1), C. tropicalis (peak 2), C. parapsilosis (peak 3), C. magnoliae (peak 4), C. lusitaniae (peak 5), C. dubliniensis (peak 6), C. albicans (peak 7), C. inconspicua (peak 8), C. krusei (peak 9), C. neoformans (peak 10), C. kefyr (peak 11), and C. glabrata (peak 12).
FIG. 2.
Peak profiles obtained from Candida variants. Profiles consisting of only one major peak were designated type I. Dashed line, marker profile containing type I reference peaks and the type II variant of C. tropicalis. (A) Besides type I, six additional Candida albicans variants (types II to VII) were identified by analyzing clinical isolates and blood culture samples. (B) Candida glabrata variants (types I to VI). (C) Three variants were observed for Candida tropicalis (types I to III). Note that the marker profile contains types I and II of C. tropicalis.
To simulate diagnostic procedures for candidemia, a total of 14 negative blood cultures was spiked in duplicate with pure cultures of reference strains of C. parapsilosis, C. tropicalis, C. albicans, C. glabrata, C. dubliniensis, C. krusei, and a mixture of C. albicans and C. dubliniensis. This panel of blood cultures was blinded for subsequent DHPLC analysis. Total DNA was extracted from positive blood cultures and PCR amplified. Resulting DHPLC peaks of the panel were assigned to the corresponding reference peaks of the Candida marker set. All Candida species tested were unequivocally identified. In a second set of experiments, the assay was applied to real clinical specimens. Positive blood cultures (yeast cells in Gram stain) consecutively collected in the clinical mycology laboratory of our institution from November 2004 through June 2005 were analyzed using the PCR/DHPLC approach. The 66 blood culture samples studied comprised 26 copy strains, e.g., the second or third isolates from independent blood samples of the same patient. Results were in agreement with those achieved by culture analysis. Most samples (34 out of 66 samples) were positive for C. albicans. Other samples contained C. dubliniensis (1 sample), C. parapsilosis (2 samples), C. glabrata (12 samples), C. krusei (8 samples), and C. tropicalis (7 samples). Moreover, in one blood culture, the WAVE system identified a mixed infection with C. albicans and C. glabrata. One Trichosporon asahii infection showed a major peak in DHPLC that could not be resolved since T. asahii was not included in the marker used as a reference for species identification. While identification of major peaks was simple by using the marker mixture as a reference, side peaks were observed in some samples. All profiles could be assigned to variants shown in Fig. 2. The distribution of DHPLC variants in the strains or samples studied is given in Table 2.
TABLE 2.
Distribution of Candida DHPLC subtypes in clinical samplesa
| Source |
C. albicans
|
C. glabrata
|
C. tropicalis
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | I | II | III | IV | V | VI | I | IIb | III | |
| Blood cultures | 21 | 1 | 3 | 2 | 5 | 3 | 1 | 1 | 3 | |||||||
| Various clinical samples | 18 | 1 | 2 | 1 | 3 | 14 | 2 | 4 | 1 | 1 | 5 | 4 | ||||
Copy strains were not considered.
The C. tropicalis type II profile was only found in the reference strain.
In addition, this novel method was applied for detecting yeast populations in human feces to test its ability for future monitoring of fungal colonization of mucosal surfaces. Fecal samples from four hospitalized patients were tested by PCR/DHPLC, and results were similar to those obtained by cultural analysis. Two samples were positive for C. glabrata. Furthermore, in one stool sample, the WAVE system identified mixed colonization with C. albicans and C. kefyr. Again, this result was confirmed by culture analysis. One sample gave divergent molecular and cultural results. While C. albicans. was detected by both methods, colonization by C. glabrata was found by DHPLC only. In contrast, Geotrichum candidum was cultivated from this sample but not found by PCR/DHPLC.
DISCUSSION
Results demonstrate that the PCR/DHPLC method is suited to identify fungal species in clinical specimens, e.g., blood culture and fecal samples. All strains tested, mainly Candida spp., were characterized by unique peak profiles. Amplicons from 12 fungal strains were combined and used as a marker for the identification of yeasts in clinical sample material. Seventy-nine of a total of 80 blood culture samples studied (14 spiked and 66 clinical samples) were correctly identified. Side peaks observed in some cases did not affect the identification of the respective Candida species. Rare ambiguities could be resolved by collecting and PCR/sequencing of the respective DHPLC peak. The more complex profiles found in three species allowed the identification of subtypes, an observation that may be of importance for epidemiological studies. PCR/DHPLC variants were observed in C. albicans, C. glabrata, and C. tropicalis. In C. albicans and C. glabrata, the single-peak profile (type I) is much more prevalent than any PCR/DHPLC profile with side peaks (Table 2). While PCR/DHPLC subtypes of Candida spp. have not been analyzed in detail, we speculate that variants may occur due to microheterogeneities between various rRNA gene clusters of a genome.
Taken together, we conclude that this method may replace the time-consuming and sometimes challenging culture-based identification of yeast species. Assuming that DNA extraction takes 4 hours, PCR takes 3 hours, and DHPLC on the WAVE system takes 30 minutes, results can be obtained in 1 working day. Processing of blood culture samples may be further accelerated by using a commercial genomic DNA blood kit.
The applications of the WAVE microbial analysis system presented here capitalize on the ability of the system to unequivocally identify more than one yeast species in a given sample. Therefore, in addition to rapid and simple identification of yeasts from positive blood cultures, the system enables fast and reliable detection of yeasts in complex human microbiota, e.g., intestinal microflora or skin, and should be helpful for epidemiological studies.
Acknowledgments
We thank Gernot Reifenberger for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (grant GO 363/10-1 to U.B.G.).
REFERENCES
- 1.Chen, Y. C., S. C. Chang, C. C. Sun, L. S. Yang, W. C. Hsieh, and K. T. Luh. 1997. Secular trends in the epidemiology of nosocomial fungal infections at a teaching hospital in Taiwan, 1981 to 1993. Infect. Control Hosp. Epidemiol. 18:369-375. [DOI] [PubMed] [Google Scholar]
- 2.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. LaFe, U. Bui, A. P. Limaye, and B. T. Cookson. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J. Clin. Microbiol. 39:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Baere, T., G. Claeys, D. Swinne, G. Verschraegen, A. Muylaert, C. Massonet, and M. Vaneechoutte. 2002. Identification of cultured isolates of clinically important yeast species using fluorescent fragment length analysis of the amplified internally transcribed rRNA spacer 2 region (ITS2). BMC Microbiol. 2:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domann, E., G. Hong, C. Imirzalioglu, S. Turschner, J. Kuhle, C. Watzel, T. Hain, H. Hossain, and T. Chakraborty. 2003. Culture-independent identification of pathogenic bacteria and polymicrobial infections in the genitourinary tract of renal transplant recipients. J. Clin. Microbiol. 41:5500-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frueh, F. W., and M. Noyer-Weidner. 2003. The use of denaturing high-performance liquid chromatography (DHPLC) for the analysis of genetic variations: impact for diagnostics and pharmacogenetics. Clin. Chem. Lab. Med. 41:452-461. [DOI] [PubMed] [Google Scholar]
- 6.Fujita, S., B. A. Lasker, T. J. Lott, E. Reiss, and C. J. Morrison. 1995. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J. Clin. Microbiol. 33:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graf, B., T. Adam, E. Zill, and U. B. Göbel. 2000. Evaluation of the VITEK 2 system for rapid identification of yeasts and yeast-like organisms. J. Clin. Microbiol. 38:1782-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graf, B., A. Trost, J. Eucker, U. B. Göbel, and T. Adam. 2004. Rapid and simple differentiation of C. dubliniensis from C. albicans. Diagn. Microbiol. Infect. Dis. 48:149-151. [DOI] [PubMed] [Google Scholar]
- 9.Hurtle, W., D. Shoemaker, E. Henchal, and D. Norwood. 2002. Denaturing HPLC for identifying bacteria. BioTechniques 33:386-388, 390-391. [DOI] [PubMed] [Google Scholar]
- 10.Kao, A. S., M. E. Brandt, W. R. Pruitt, L. A. Conn, B. A. Perkins, D. S. Stephens, W. S. Baughman, A. L. Reingold, G. A. Rothrock, M. A. Pfaller, R. W. Pinner, and R. A. Hajjeh. 1999. The epidemiology of candidemia in two United States cities: results of a population-based active surveillance. Clin. Infect. Dis. 29:1164-1170. [DOI] [PubMed] [Google Scholar]
- 11.Klotz, S. A., D. J. Drutz, and J. E. Zajic. 1985. Factors governing adherence of Candida species to plastic surfaces. Infect. Immun. 50:97-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreger-Van Rij, N. J. W. 1984. The yeasts: a taxonomic study. Elsevier, Amsterdam, The Netherlands.
- 13.Malani, P. N., S. F. Bradley, R. S. Little, and C. A. Kauffman. 2001. Trends in species causing fungaemia in a tertiary care medical centre over 12 years. Mycoses 44:446-449. [DOI] [PubMed] [Google Scholar]
- 14.Pinjon, E., D. Sullivan, I. Salkin, D. Shanley, and D. Coleman. 1998. Simple, inexpensive, reliable method for differentiation of Candida dubliniensis from Candida albicans. J. Clin. Microbiol. 36:2093-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swinne, D., M. Watelle, C. Suetens, K. Mertens, P. A. Fonteyne, and N. Nolard. 2004. A one-year survey of candidemia in Belgium in 2002. Epidemiol. Infect. 132:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trost, A., B. Graf, J. Eucker, O. Sezer, K. Possinger, U. B. Göbel, and T. Adam. 2004. Identification of clinically relevant yeasts by PCR/RFLP. J. Microbiol. Methods 56:201-211. [DOI] [PubMed] [Google Scholar]
- 17.Turenne, C. Y., S. E. Sanche, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 1999. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 37:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao, W., and P. J. Oefner. 2001. Denaturing high-performance liquid chromatography: a review. Hum. Mutat. 17:439-474. [DOI] [PubMed] [Google Scholar]
- 19.Yang, C. W., T. M. Barkham, F. Y. Chan, and Y. Wang. 2003. Prevalence of Candida species, including Candida dubliniensis, in Singapore. J. Clin. Microbiol. 41:472-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoetendal, E. G., A. D. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]