Abstract
To better understand the emergence and transmission of antibiotic-resistant Streptococcus agalactiae, we compared phenotypic and genotypic characteristics of 52 human and 83 bovine S. agalactiae isolates. Serotypes found among isolates from human hosts included V (48.1%), III (19.2%), Ia and Ib (13.5% each), and II (5.8%). Among isolates from bovine hosts, molecular serotypes III and II were predominant (53 and 14.5%, respectively). Four and 21 different ribotypes were found among human and bovine isolates, respectively. A combination of ribotyping and serotyping showed that two bovine isolates were indistinguishable from human isolates. Resistance to tetracycline and erythromycin was more common among human (84.6% and 26.9%, respectively) than bovine (14.5% and 3.6%, respectively) isolates. tetM was found in all tetracycline-resistant human isolates, while tetO was the predominant resistance gene among bovine isolates. tet genes were found among various ribotypes. ermB, ermTR, and mefA were detected among erythromycin-resistant human isolates, while ermB was the only erythromycin resistance determinant among isolates from bovine hosts. For isolates from human hosts, erythromycin resistance genes appeared to be associated with specific ribotypes. We conclude that (i) human and bovine S. agalactiae isolates represent distinct populations; (ii) human host-associated S. agalactiae subtypes may occasionally be transmitted to bovines; (iii) while emergence of erythromycin and tetracycline resistance appears to largely occur independently among human and bovine isolates, occasional cross-species transfer of resistant strains or transmission of resistance genes between human- and bovine-associated subtypes may occur; and (iv) dissemination of antibiotic-resistant S. agalactiae appears to include both clonal spread of resistant strains as well as horizontal gene transfer.
Streptococcus agalactiae, also known as group B streptococcus (GBS), is an important pathogen in both humans and dairy cattle. First recognized as a pathogen causing bovine mastitis, the organism is distinguished from other pathogenic streptococci by the presence of the cell wall-associated group B carbohydrate (Lancefield group B) (28). Since the 1970s, S. agalactiae has emerged as an important human pathogen. Currently, S. agalactiae is the leading cause of bacterial sepsis, pneumonia, and meningitis in neonates in the United States and Europe, with a mortality rate of 4 to 6% (41, 42, 47). S. agalactiae is also an increasingly important cause of invasive infections in immunocompromised adults and the elderly (42, 48). In dairy cows, S. agalactiae is a major cause of mastitis, which is currently considered the economically most important disease affecting the dairy industry (8, 52).
Serotyping has been a traditional epidemiological tool for investigating S. agalactiae infections in humans. Based on capsular polysaccharide antigens, nine serotypes have been identified to date. Serotypes Ia, Ib, II, III, and V have been predominant in the United States among infants and pregnant women, while serotypes VI and VIII have been associated with colonization of humans in Japan (9, 31). S. agalactiae strains of capsular serotype V were first detected in 1985 and are currently the most common capsular serotype-associated with GBS infections among nonpregnant adults in the United States (6, 23). In addition to serotyping, genotyping methods such as randomly amplified polymorphic DNA and ribotyping have been used to characterize S. agalactiae isolates of human and bovine origin (11, 26, 34). Most subtyping studies have shown that S. agalactiae isolates from human and bovine hosts represent largely separate populations, even though identical subtypes have occasionally been isolated from humans and animals (34, 44). Application of a recently described multilocus sequence typing (MLST) scheme for S. agalactiae also provided evidence that a specific human hyperinvasive GBS clone appears to have emerged from a bovine ancestor (5).
S. agalactiae infections in both humans and bovines are treated by administration of antibiotics. Antibiotics are also applied prophylactically in pregnant women to prevent infections in newborns. In 1996, the U.S. Centers for Disease Control and Prevention issued specific recommendations for GBS prophylaxis, which led to a significant decline in the incidence of early-onset human neonatal infections (40, 42). Penicillin is the drug of choice for treatment of both human and bovine S. agalactiae infections (28, 40, 42). For penicillin-allergic individuals, erythromycin and clindamycin are recommended. The prevalence of resistance to erythromycin and clindamycin has been increasing in S. agalactiae (19, 35, 48, 49); recently, resistance to erythromycin has been associated with human S. agalactiae serotype III and V isolates (33, 49).
While it is almost undisputed that extensive use of antibiotics in medicine and animal husbandry results in increased antibiotic resistance among bacterial populations (1, 53), evidence that antibiotic usage in farm animals contributes to the emergence of human antibiotic-resistant pathogens is more limited, even though several studies have suggested that antimicrobial use with animals influences development of antibiotic resistance among pathogens in humans (1, 53). While one study of antibiotic resistance patterns among S. agalactiae strains from human and bovine hosts isolated in Brazil has been recently published (18), to our knowledge, no similar data are available for S. agalactiae strains isolated in other countries. Data of this nature from other countries will help to define the transmission and emergence of antibiotic resistance genes in S. agalactiae and support rational and economical decisions on the application and use of specific antibiotics to treat infections. To better understand the prevalence, emergence, and transmission of antibiotic-resistant S. agalactiae among human and bovine host populations in the United States, we (i) expanded a previously described collection of 104 temporally and spatially matched S. agalactiae isolates from human and bovine hosts to include an additional 31 bovine isolates, including EcoRI ribotype characterization of these additional isolates; (ii) performed molecular and conventional serotyping of isolates; (iii) determined the antibiotic susceptibility patterns of all isolates; and (iv) screened for the presence of selected macrolide and tetracycline resistance genes among resistant isolates.
MATERIALS AND METHODS
Bacterial isolates.
A total of 135 S. agalactiae isolates were used in this study. A total of 104 isolates were previously described and represent geographically and temporally matched isolates from human (n = 52) and bovine (n = 52) hosts; EcoRI ribotypes for these isolates have also previously been reported (44). An additional 31 bovine S. agalactiae isolates were included in this study (yielding a total of 83 bovine isolates), since bovine isolates showed a lower prevalence of antibiotic resistance. Bovine isolates were obtained from 83 farms as previously described (44). From each farm, four isolates from four different cows were obtained; one isolate was randomly selected from each farm for inclusion in the isolate set. Human clinical isolates of S. agalactiae were obtained through the Emerging Infectious Diseases surveillance program of the New York State Department of Health (44). All human and bovine isolates were collected in New York State from the years 2000 to 2002. Bacterial isolates were stored at −80°C in brain heart infusion broth (Becton Dickinson, Sparks, MD) with 20% glycerol.
Serotyping.
All isolates were initially characterized by molecular serotyping using a combination of PCR assays and DNA sequencing of selected capsular polysaccharide (cps) genes, as described by Kong et al. (30) with some modifications. Bacterial lysates for PCR were prepared using lysozyme and proteinase K as described by Furrer et al. (21). Lysates were used to perform an initial set of six separate serotype-specific PCR assays, which allow classification of isolates into serotypes Ia, Ib, III, IV, V, and VI (30). Isolates which did not yield a PCR product with in any of these six PCR assays were characterized by sequencing a 790-bp region encompassing the 3′ end of cpsE, cpsF, and the 5′ end of cpsG (cpsEFG; positions 1437 to 2226; GenBank accession number AF332908); DNA sequence data for this fragment have been shown to differentiate serotypes Ia, Ib, II, III-1, III-2, III-3, III-4, IV, V, VI, and VII (30). These sequence data thus should allow classification into serotypes II or VII for isolates that did not yield serotype-specific PCR products by the initial PCR assay. Since sequence comparisons for some isolates yielded sequence matches with serotypes that should have yielded PCR products in the initial PCR assays (e.g., the cpsEFG sequence yielded a match with Ia and III, two serotypes which should have been identified by the initial PCR assays), additional DNA sequencing of the 5′ end of cpsD and the 3′ end of cpsE was performed in an attempt to clarify these inconsistent results. PCR amplification and sequencing were performed using primers cpsDS (5′ GCA AAA GAA CAG ATG GAA CAA AGT GG 3′) and cpsEA2 (5′ AAA MGC TTG ATC AAC AGT TAA GCA GG 3′); these primers amplify a 750-bp region encompassing the 3′ end of cpsD and the 5′ end of cpsE (cpsDE; positions 1 to 750; GenBank accession number AF332908). These two primer sequences were kindly provided by F. Kong (personal communication). PCR cycling parameters were 95°C for 30 s, 65°C for 30 s, and 72°C for 1 min for a total of 35 cycles, with final elongation at 72°C for 10 min. Sequence data for cpsEFG and cpsDE were compared to sequence data for the same gene fragments obtained for a panel of reference strains, as described by Kong et al. (30). While most sequences obtained in our study described here perfectly matched a reference strain sequence, a bootstrap cpsDE neighbor-joining consensus tree had to be constructed for some animal isolates to identify the closest sequence matches.
For sequencing, PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Inc., Chatsworth, CA). Sequencing of PCR products was performed at Cornell University's BioResource Center using the ABI 3700 automated sequencing unit (Perkin-Elmer Biosystems, Foster City, CA).
All of the bovine isolates and two of the human isolates from each serotype were also serotyped by conventional methods as previously described (51) at the National Centre for Streptococcus (Alberta, Canada). Briefly, antisera for types Ia, Ib, II, III, IV, V, VI, VII, and VIII and for c and x protein antigens were prepared in rabbits by using recognized type stains. Antisera were used to test Lancefield hot-acid extracts of test and control strains using Ouchterlony immunodiffusion.
Ribotyping.
EcoRI ribotyping was performed as previously described (44) using the restriction enzyme EcoRI and the RiboPrinter (Qualicon, Inc., Wilmington, DE) system. Similarity coefficients for ribotype patterns from different isolates were calculated using the RiboPrinter's proprietary algorithm. Similarity coefficients and visual evaluation were used to define distinct ribotypes.
Phenotypic susceptibility testing.
Kirby-Bauer disk diffusion tests were performed for each of the 135 isolates according to the methods recommended by CLSI (formerly NCCLS) (37). Isolates were tested for susceptibility to cephalosporin, tetracycline, gentamicin, erythromycin, and clindamycin. Classification as “susceptible,” “intermediate,” or “resistant” was based on the CLSI-recommended breakpoints for inhibition zone diameters.
Detection of tetracycline and erythromycin resistance genes.
The presence of selected tetracycline and macrolide antibiotic resistance genes was determined using previously described PCR conditions and primers for tetM (39), tetO (38), ermA (45), ermB (45), ermC (45), ermTR (10), and mefA (13). All S. agalactiae isolates were examined for the presence of tetM and tetO, which encode resistance to tetracycline. All isolates classified as resistant or with intermediate resistance to erythromycin, as well as four erythromycin-sensitive isolates, were examined for the presence of ermA, ermB, ermC, ermTR, and mefA as described previously (10, 13, 45).
To confirm the specificity of the PCRs, tetO, ermB, ermTR, and mefA PCR products from two randomly selected resistant isolates were sequenced. In addition, the complete tetM coding region was sequenced for one human tetracycline-resistant isolate and one bovine tetracycline-resistant isolate and for two human isolates with intermediate resistance. PCR and sequencing was performed with four primer pairs. In addition to three previously published primers (38, 39), five new primers were designed, based on a published tetM sequence (GenBank accession number X90939). Primer pairs used included (i) forward 1 (5′ TGC TTT GTA TGC CTA TGG TTA TGC 3′) (this study) and reverse 1 (5′ CGG TAA AGT TCG TCA CAC AC 3′) (38), (ii) forward 2 (5′ TGG AAT TGA TTT ATC AAC GG 3′) (39) and reverse 2 (5′ CTC GCA AAT GCA GTA CGC CAC TAT 3′) (this study), (iii) forward 3 (5′ TAT TCA TCA ACA CAT CGA GGT C 3′) (this study) and reverse 3 (5′ TTC CAA CCA TAC AAT CCT TG 3′) (39), and (iv) forward 4 (5′ GTG CCG CCA AAT CCT TTC TGG 3′) (this study) and reverse 4 (5′ TCC TCC TTT ACA CTT TAA TTC AAA TC 3′) (this study). Sequence analyses and alignments were performed using Seqman and Megalign (DNAStar, Madison, WI).
RESULTS
Serotype characterization of isolates from bovine and human hosts.
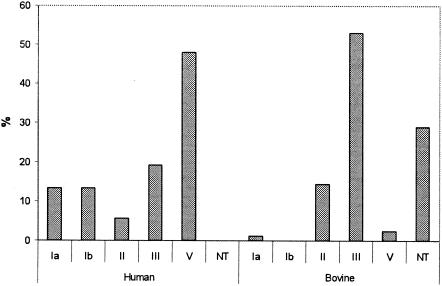
PCR primer sets for identification of serotypes Ia, Ib, III, IV, V, and VI, as described by Kong et al. (30), allowed serotype classification for 49 of the 52 human isolates. Sequencing of a 790-bp cpsEFG fragment allowed for classification of the remaining three human isolates as molecular serotype II. The most common serotype among human isolates was V (48.1%), followed by III (19.2%). Serotypes Ia and Ib were less commonly identified (13.5% each). Only three isolates (5.8%) were serotype II (Fig. 1). Accuracy of the molecular serotyping results was confirmed by conventional serotyping of two randomly selected human isolates for each of the five serotypes identified; molecular and conventional serotyping results matched for all 10 isolates.
FIG. 1.
Distribution of molecular serotypes among S. agalactiae isolates from human (n = 52) and bovine (n = 83) hosts. NT, not typeable.
Among the 83 bovine isolates, a total of 47 could be identified to serotype level by using the PCR primers specific for serotypes Ia, Ib, III, IV, V, and VI; the vast majority of isolates (n = 44) (Table 1) were identified as serotype III. Sequencing of the 790-bp cpsEFG fragment for the 36 isolates that could not initially be identified to serotype by the PCR assay yielded a PCR product for 33 isolates. Sequence similarity analyses (30) yielded matches for serotypes “Ia/III-3” and “II/III-4” for 21 and 12 isolates, respectively. Classification as “Ia/III-3” was deemed inconclusive, since isolates with this sequence should have yielded a PCR product by either the serotype Ia- or the serotype III-specific PCR assay. All 33 bovine isolates that yielded matches for serotypes “Ia/III-3” and “II/III-4” were thus further characterized by sequencing of a 750-bp cpsDE fragment. Most (17/21) isolates with matches for serotypes “Ia/III-3” yielded a match for serotype “III-3,” based on partial cpsDE sequences and were thus classified as nontypeable (NT), since none of these isolates yielded serotype III when characterized by conventional serotyping. The remaining four isolates with matches for serotypes “Ia/III-3” in the cpsEFG sequencing also yielded inclusive results in the cpsDE sequencing and were thus also classified as NT. All 12 bovine isolates with matches for serotypes “II/III-4” based on partial cpsEFG sequences were classified as molecular serotype II by partial cpsDE sequencing (30). In summary, molecular serotyping classified 44 bovine isolates (53%) as serotype III, 12 isolates as serotype II, 2 isolates as serotype V, 1 isolate as serotype Ia, and 24 bovine isolates as nontypeable (Fig. 1). All three human isolates that were classified as molecular serotype II based on partial cpsDE sequencing showed cpsDE sequences distinct from the sequences for the 12 bovine isolates classified as molecular serotype II.
TABLE 1.
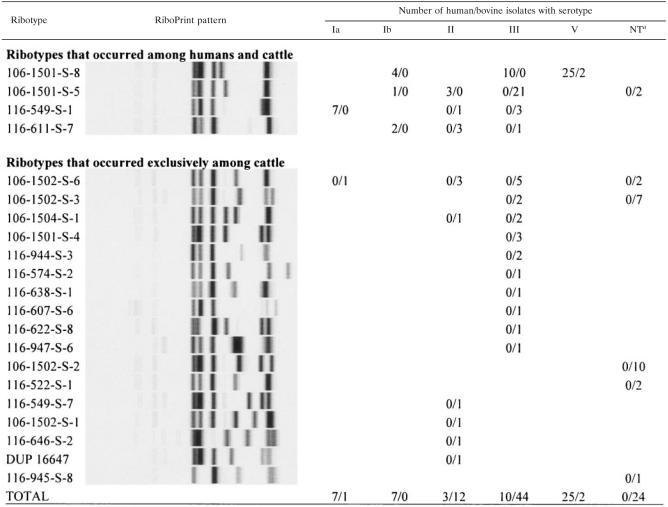
Distribution of S. agalactiae ribotypes and molecular serotypes among human and bovine isolates
NT, nontypeable.
By conventional serotyping 26 (31.3%) and 4 (4.8%) isolates from bovine hosts were classified as capsular polysaccharide serotypes III and II, respectively. One isolate was classified as serotype V and 62.6% of the isolates were NT. While all isolates classified as serotype III by conventional serotyping were also assigned serotype III by the PCR-based serotype III-specific assay, 18 isolates classified as serotype III by PCR were nontypeable by classical serotyping. While our data show that serotyping methods developed for human S. agalactiae isolates perform poorly with bovine isolates, molecular serotyping overall classified fewer isolates as nontypeable (28.9%) than conventional serotyping (62.6%). Molecular serotyping results were used in the rest of the paper for comparison purposes.
Association of serotypes with ribotypes.
As previously reported (44), S. agalactiae isolates from human hosts only represented four different EcoRI ribotypes (Table 1). While two ribotypes were predictive for molecular serotypes among human isolates (e.g., all human isolates with ribotype 116-549-S-1 were serotype Ia) (Table 1), human isolates with ribotype 106-1501-S-8 represented three different serotypes (Ib, III, and V). Ribotype 106-1501-S-5 represented two different serotypes (Ib and II).
The 83 isolates from bovine hosts represented 21 different ribotypes (Table 1), 4 more than the 17 ribotypes previously identified among the 52 isolates described by Sukhnanand et al. (44), which were also included in this study. Serotype III bovine isolates classified into 13 ribotypes, with none identified as ribotype 106-1501-S-8, which was the only ribotype among human serotype III isolates. Eight different ribotypes were found among 12 serotype II bovine isolates. Both of the serotype V bovine isolates ribotyped as 106-1501-S-8. The single serotype Ia bovine isolate had a different ribotype than that of the seven human serotype Ia isolates. While four ribotypes were found among both human and animal isolates, only one ribotype-molecular serotype combination (ribotype 106-1501-S-8, serotype V) was found among both human and animal isolates.
Antimicrobial susceptibility profiles.
Overall, all of the isolates from human hosts and 77 of 83 S. agalactiae isolates from bovine hosts showed resistance to at least one of the antibiotics tested. All isolates were fully susceptible to cephalosporin. Resistance to tetracycline was more common among S. agalactiae isolates from humans (84.6%) than among isolates from cattle (14.5%) (Table 2). Similarly, resistance to erythromycin was more common among human isolates (26.9%) than among bovine isolates (3.6%). Clindamycin resistance was rare among isolates from both bovine and human hosts (7.8% and 3.6%, respectively). All clindamycin-resistant isolates were also resistant to erythromycin. All human isolates and 91.6% of the bovine isolates were found to be resistant to gentamicin.
TABLE 2.
Antibiotic susceptibility patterns of human and bovine S. agalactiae isolatesa
| Antibiotic | % of human isolates classified as:
|
% of bovine isolates classified as:
|
||||
|---|---|---|---|---|---|---|
| S | I | R | S | I | R | |
| Tetracycline | 3.8 | 11.5 | 84.6 | 83.1 | 2.4 | 14.5 |
| Erythromycin | 55.8 | 17.3 | 26.9 | 81.9 | 14.5 | 3.6 |
| Cephalosporin | 100 | 0 | 0 | 100 | 0 | 0 |
| Gentamicin | 0 | 0 | 100 | 3.6 | 4.8 | 91.6 |
| Clindamycin | 74.5 | 17.6 | 7.8 | 94 | 2.4 | 3.6 |
Antibiotic susceptibilities were classified as susceptible (S), intermediate (I), or resistant (R), based on inhibition zone sizes, following CLSI guidelines.
Detection of tetracycline and erythromycin resistance genes.
The tetracycline resistance gene tetM was identified in all 44 tetracycline-resistant human isolates. Among six human isolates with intermediate tetracycline resistance, two also bore tetM, while four bore neither tetM nor tetO. In one of the two isolates from human hosts with intermediate tetracycline resistance that carried tetM, the insertion sequence IS1381 was detected in tetM; the second isolate had a 26-bp deletion in tetM. In bovine isolates, tetO was the predominant tetracycline resistance gene. Specifically, among 12 tetracycline-resistant bovine isolates, 10 bore tetO while 2 carried tetM. One of the two bovine isolates with intermediate tetracycline resistance also bore tetO. tetM sequence analysis revealed that the tetM sequence for the bovine isolate was identical to that found in the human isolate for which tetM was sequenced. While one tetM-carrying bovine isolate was ribotype 106-1501-S-5, the ribotype most common among the bovine isolates in this study, the other tetM-positive bovine isolate showed isolate characteristics (ribotype 106-1501-S-8 and serotype V) that were otherwise typical for isolates associated with human hosts (Table 1). As it was unusual to isolate human-associated strains from cattle, we examined three additional S. agalactiae isolates obtained from different cows in the same farm; all four isolates bore the same characteristics (ribotype106-1501-S-8, serotype V, and tetM positive).
All three erythromycin-resistant bovine isolates and 1 of the 12 bovine isolates with intermediate erythromycin resistance were PCR positive for ermB. Among human isolates, erythromycin resistance was associated with the presence of ermB (three isolates), ermTR (seven isolates), or mefA (three isolates), including one human isolate that carried both ermB and ermTR. ermB presence was associated with either the presence of tetO (in bovine isolates) or the presence of tetM (in human isolates). Except for one ermB-bearing isolate, which was only resistant to erythromycin, isolates carrying ermB were resistant to both erythromycin and clindamycin, while isolates carrying ermTR or mefA were resistant to erythromycin but sensitive to clindamycin. In one erythromycin-resistant human isolate, we were not able to detect any of the erythromycin resistance determinants examined (ermA, ermB, ermC, ermTR, and mefA); this isolate was highly resistant to erythromycin but susceptible to clindamycin. Although not currently reported in S. agalactiae, single-nucleotide changes in 23S rRNA or in ribosomal proteins conferring macrolide resistance have been reported for many bacteria, including Streptococcus pneumoniae (46, 50). It is thus possible that a ribosomal mutation is responsible for the resistance in this specific isolate. Another human isolate resistant to erythromycin and clindamycin yielded an unspecific (larger-than-predicted) ermTR PCR product.
Distribution of antimicrobial resistance genes among serotypes and ribotypes.
Tetracycline resistance genes were identified in isolates representing various ribotypes and serotypes (Table 3). Among human isolates, erythromycin resistance genes appeared to be associated with specific serotypes and ribotypes (Table 3). Specifically, mefA was found only among serotype Ia isolates; overall, three of the seven serotype Ia isolates bore mefA. ermB was found only among serotype V human isolates. In bovine isolates, however, ermB was found in two different serotypes (Table 3). Three of the four ermB-carrying bovine isolates were ribotype 106-1502-S-6.
TABLE 3.
Distribution of antimicrobial resistance genes among S. agalactiae serotypes and ribotypes
| Antimicrobial resistance gene (no. of isolates) | Molecular serotype (no. of isolates) | Ribotype (no. of isolates) |
|---|---|---|
| Human | ||
| ermB (3) | V (3) | 106-1501-S-8 (3) |
| ermTR (7) | V (6), III (1) | 106-1501-S-8 (7) |
| mefA (3) | Ia (3) | 116-549-S-1 (3) |
| tetM (46) | Ia (6), Ib (6), II (2), III (9), V (23) | 106-1501-S-8 (36), 106-1501-S-5 (3), 116-549-S-1 (6), 116-611-S-7 (1) |
| Bovine | ||
| ermB (4) | NT (2), III (2) | 106-1502-S-6 (3), 106-1502-S-2 (1) |
| tetM (2) | III (1), V (1) | 106-1501-S-8 (1), 106-1501-S-5 (1) |
| tetO (11) | Ia (1), II (1), III (3), NT (6) | 116-522-S-1 (1), 106-1501-S-5 (1), 106-1502-S-2 (1), 106-1502-S-3 (2), 106-1502-S-6 (6) |
DISCUSSION
Through molecular and phenotypic characterization of 135 S. agalactiae isolates from human and bovine hosts, we provide evidence supporting that (i) human and bovine isolates represent distinct populations; (ii) human host-associated subtypes may occasionally be transmitted to bovine hosts; (iii) while emergence of erythromycin and tetracycline resistance appears to largely occur independently among human and animal isolates, occasional cross-species transfer of resistant strains or transmission of resistance genes between human and cattle host-associated subtypes may occur; and (iv) spread of antibiotic resistance determinants in S. agalactiae appears to include clonal spread of antibiotic-resistant strains, as well as horizontal gene transfer. We conclude that human and bovine S. agalactiae strains represent largely separate populations and that emergence of antibiotic resistance may largely occur independently between these two populations. Nevertheless, occasional S. agalactiae transmission between human and bovine hosts, as well as antibiotic resistance gene transfer between human- and bovine-associated strains, may occur.
Human and bovine isolates represent distinct populations.
Molecular subtyping data reported here further support that human and bovine S. agalactiae isolates represent distinct molecular subtype populations, as also shown in a previous report (44), which used a smaller set of 104 human and bovine isolates (including all human isolates characterized here, as well as 52 of the 83 bovine isolates characterized here). Specifically, serotype characterization of the 135 isolates characterized here further supports, by an independent subtyping approach, that human and bovine S. agalactiae isolates represent distinct populations. While the majority of bovine isolates were either serotype II or III or nontypeable, the majority of human isolates were serotype V; in addition, serotypes Ia and Ib were exclusively or almost exclusively found among human isolates. Our data are consistent with other reports on serotypes typically associated with human and bovine S. agalactiae infections. While serotypes I, II, and III were the most prevalent serotypes among isolates from human neonates with early-onset GBS infections in the 1970s (3), more recent studies have shown serotypes Ia and III are the most common serotypes currently associated with GBS infections among neonates (2, 32). Serotype V strains emerged as a cause of human infections in 1985; these strains have been shown to currently cause the majority of GBS infections among nonpregnant adults in the United States (6, 23). Similar to our data, previous studies have also found a predominance of serotypes II and III or a nontypeable status among bovine mastitis S. agalactiae isolates (17, 34). In addition, one recent study of bovine mastitis and human S. agalactiae isolates collected in Brazil also indicated that bovine- and human-associated subtypes may differ in the distribution of specific virulence-associated genes (bca, lmb, and scpB) (18).
Interestingly, associations between EcoRI ribotypes and serotypes further support that S. agalactiae isolates from human and bovine hosts represent distinct populations. For human isolates, serotypes generally showed a clear association with ribotypes, such that all isolates with a given serotype showed the same ribotype and only isolates with serotype Ib represented three ribotypes. Bovine isolates classified as serotype III, on the other hand, represented 13 different ribotypes. Although serotype III was common among both human and bovine isolates, human and bovine serotype III isolates had different ribotypes, which suggests that S. agalactiae isolated from human and bovine origins represent different clonal groups that bear the same capsular polysaccharide, a hypothesis consistent with findings by a variety of groups (5, 7, 24, 36) that serotype III isolates represent at least two distinct clusters and/or evolutionary lineages. Interestingly, similar to our ribotyping data, others have also found serotype III isolates, even within a given lineage or cluster, may show considerable molecular subtype diversity (as determined by randomly amplified polymorphic DNA and/or MLST) (5, 34). Possible explanations for genetic heterogeneity within the same serotype include the horizontal spread of capsular polysaccharide genes (e.g., between ancestors of the different serotype II lineages) and diversification within a given lineage or cluster. While we were unable to find any published reports providing evidence for the horizontal transfer of capsular genes in S. agalactiae, the horizontal spread of capsular genes has been reported with two closely related organisms, Streptococcus suis and S. pneumoniae (14, 29). Future application of MLST approaches (27) should be able to further clarify the contributions of horizontal gene transfer to evolution of serotype diversity among human and bovine S. agalactiae isolates.
Human host-associated subtypes may occasionally be transmitted to bovine hosts.
Interestingly, we identified two bovine isolates as part of our study that were characterized by a combination of serotype and ribotype characteristics, which were otherwise strongly associated with human S. agalactiae isolates. One of these isolates also carried tetM, a tetracycline resistance gene we had found here to be usually associated with human isolates. Isolates from additional cows on the same farm where this isolate was obtained showed identical subtype and antibiotic resistance gene patterns, indicating that isolation of this human host-associated subtype was unlikely to reflect a single contamination event but rather reflects endemic presence in this herd. We propose that these findings indicate an initial human-to-animal transfer of this specific human host-associated S. agalactiae subtype, followed by spread in this herd. This would be consistent with a previous study that also has suggested that human-to-animal transmission of S. agalactiae is possible (25).
Emergence of erythromycin and tetracycline resistance appears to largely occur independently among human and animal isolates.
While the prevalence of isolates resistant to different antibiotics was generally similar among bovine and human isolates characterized here, tetracycline and erythromycin resistance was more common among human than bovine isolates. High levels of tetracycline resistance among human S. agalactiae isolates have been reported previously (15, 49). Recognized tetracycline resistance mechanisms involve protection of the ribosome as an antibiotic target by ribosomal protection proteins or reduction of the intracellular antibiotic concentration by efflux proteins (12); previously identified tetracycline resistance genes in streptococci include tetK, tetL, tetM, tetO, tetQ, and tetT (12). Interestingly, while both tetM and tetO encode ribosomal protection proteins, we found that tetM was predominantly found among human S. agalactiae isolates, while tetO was the predominant tetracycline-resistant determinant among bovine isolates. These findings are consistent with a recent study of S. agalactiae isolates collected in Brazil, which also found a predominance of tetM and tetO among human and bovine isolates, respectively (18), possibly indicating wide host-specific distribution of tet genes and/or tetracycline-resistant strains.
The prevalence of erythromycin resistance observed here was also consistent with previously reported prevalences for macrolide resistance among S. agalactiae isolates (15, 22, 49). Resistance to macrolides in Streptococcus can be caused mainly by two mechanisms, target site modification and active drug efflux (20). Similar to our observations on the prevalence of tetracycline resistance genes, we also found that different erythromycin resistance genes were associated with human and bovine isolates: ermB, ermTR, and mefA were found among human isolates, and only ermB was found among bovine isolates. These finding were also consistent with a recent study of S. agalactiae isolates from Brazil, which found a predominance of ermB among bovine isolates (18). While ermB was associated in our study with resistance to macrolides and lincosamides, the ermTR- and mefA-carrying isolates were resistant to macrolides but sensitive to lincosamides. This is consistent with reports that target site modification, mediated by rRNA adenine methylases encoded by erm genes, can produce cross-resistance to macrolides, lincosamides, and streptogramin B antibiotics (20). On the other hand, active drug efflux mediated by mef-encoded transmembrane pumps has been reported to yield resistance only to 14- and 15-member macrolides (erythromycin, azithromycin, and clarithromycin); 16-member macrolides (e.g., josamycin) are not affected and neither are lincosamides or streptogramin B-type compounds (macrolide phenotype) (13, 20). In addition, many S. agalactiae isolates carrying ermTR have been shown to display an inducible macrolides, lincosamides, and streptogramin B resistance, which can only be detected by a double-disk diffusion test, possibly explaining why ermTR-positive isolates tested here were clindamycin sensitive by single-disk testing (15). Importantly, ermB-positive bovine isolates represented a different ribotype than ermB-positive human isolates, indicating a different clonal origin of human and bovine ermB-positive isolates, possibly through horizontal gene transfer either between S. agalactiae or another closely related organism, e.g., Streptococcus pyogenes, which was previously shown to carry ermB (16).
Spread of antibiotic resistance determinants in S. agalactiae appears to include both clonal spread of antibiotic-resistant strains and horizontal gene transfer.
Our data showed that the tetracycline resistance genes tetM and tetO, as well as erythromycin resistance gene ermB, were broadly distributed among different serotypes and ribotypes, suggesting that the presence of these resistance genes among human- and bovine-associated S. agalactiae populations may represent a consequence of multiple horizontal gene transfer events. Since ermB-carrying human and bovine isolates represented different ribotypes, horizontal gene transfer of ermB between human and bovine isolates or from another source (e.g., S. pyogenes) (16) may have occurred. The observed association between the presence of ermB and either the presence of tetM (in human isolates) or the presence of tetO (in bovine isolates) may also further support the importance of horizontal gene transfer in antibiotic resistance gene evolution in S. agalactiae, since tetM and ermB have previously been identified on the same conjugative transposon in S. pneumoniae (43). In addition, our data, which show an association between serotypes, ribotypes, and the presence of specific erythromycin resistance genes (e.g., ermTR and mefA) in S. agalactiae, also suggest that at least some erythromycin-resistant strains may represent examples of the clonal spread of resistant strains. Interestingly, while most human-associated erythromycin-resistant isolates appear to represent examples of clonal spread, bovine-associated erythromycin-resistant isolates appear to be more likely to have evolved by multiple independent horizontal gene transfer events (since ermB among bovine isolates has been found in three different ribotypes and serotypes). It may be noteworthy that a variety of other studies have also revealed a potential association between serotype V human S. agalactiae isolates and erythromycin resistance (19, 33, 49), which may further support that clonal spread of erythromycin-resistant serotype V strains, the only serotype that carried ermTR in our study, may contribute to the increased prevalence of erythromycin-resistant S. agalactiae in human hosts. Interestingly, it has also been shown that clonal expansion of erythromycin-resistant strains is an important contributor the spread of erythromycin-resistant S. pyogenes among human hosts in France and Taiwan (4, 54). Since ribotyping and serotyping do not allow for determination of evolutionary relationships between isolates, future studies using MLST will be necessary to confirm the occurrence of horizontal gene transfer of resistance genes and to define the relative contributions of horizontal gene transfer and clonal expansion to the spread of tetracycline and erythromycin resistance among S. agalactiae isolates from human and bovine hosts.
Acknowledgments
This publication was developed under the auspices of the Cornell University Center for Biotechnology, a New York State Center for Advanced Technology supported by New York State and West Agro, Inc., located in Hamilton, N.Y. (in a grant to Y.H.S.), and was also partially supported by New York State dairy farmers through the New York State Dairy Promotion Order.
We thank Martin Wiedmann for his helpful comments.
REFERENCES
- 1.Aarestrup, F. M. 1999. Association between the consumption of antimicrobial agents in animal husbandry and the occurrence of resistant bacteria among food animals. Int. J. Antimicrob. Agents 12:279-285. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, J. I., D. J. Diekema, S. K. Hunter, P. R. Rhomberg, M. A. Pfaller, R. N. Jones, and G. V. Doern. 2000. Group B streptococci causing neonatal bloodstream infection: antimicrobial susceptibility and serotyping results from SENTRY centers in the Western Hemisphere. Am. J. Obstet. Gynecol. 183:859-862. [DOI] [PubMed] [Google Scholar]
- 3.Baker, C. J., and F. F. Barrett. 1974. Group B streptococcal infections in infants. The importance of the various serotypes. JAMA 230:1158-1160. [PubMed] [Google Scholar]
- 4.Bingen, E., F. Fitoussi, C. Doit, R. Cohen, A. Tanna, R. George, C. Loukil, N. Brahimi, I. Le Thomas, and D. Deforche. 2000. Resistance to macrolides in Streptococcus pyogenes in France in pediatric patients. Antimicrob. Agents Chemother. 44:1453-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisharat, N., D. W. Crook, J. Leigh, R. M. Harding, P. N. Ward, T. J. Coffey, M. C. Maiden, T. Peto, and N. Jones. 2004. Hyperinvasive neonatal group B streptococcus has arisen from a bovine ancestor. J. Clin. Microbiol. 42:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumberg, H. M., D. S. Stephens, M. Modansky, M. Erwin, J. Elliot, R. R. Facklam, A. Schuchat, W. Baughman, and M. M. Farley. 1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 173:365-373. [DOI] [PubMed] [Google Scholar]
- 7.Bohnsack, J. F., A. A. Whiting, G. Martinez, N. Jones, E. E. Adderson, S. Detrick, A. J. Blaschke-Bonkowsky, N. Bisharat, and M. Gottschalk. 2004. Serotype III Streptococcus agalactiae from bovine milk and human neonatal infections. Emerg. Infect. Dis. 10:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley, A. 2002. Bovine mastitis: an evolving disease. Vet. J. 164:116-128. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, J. R., S. L. Hillier, M. A. Krohn, P. Ferrieri, D. F. Zaleznik, and C. J. Baker. 2000. Group B streptococcal colonization and serotype-specific immunity in pregnant women at delivery. Obstet. Gynecol. 96:498-503. [DOI] [PubMed] [Google Scholar]
- 10.Cascone, C., M. Santagati, S. Noviello, F. Iannelli, S. Esposito, G. Pozzi, and S. Stefani. 2002. Macrolide-resistance genes in clinical isolates of Streptococcus pyogenes. Microb. Drug Resist. 8:129-132. [DOI] [PubMed] [Google Scholar]
- 11.Chatellier, S., H. Huet, S. Kenzi, A. Rosenau, P. Geslin, and R. Quentin. 1996. Genetic diversity of rRNA operons of unrelated Streptococcus agalactiae strains isolated from cerebrospinal fluid of neonates suffering from meningitis. J. Clin. Microbiol. 34:2741-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 14.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryniewicz, J. C. Paton, and B. G. Spratt. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73-83. [DOI] [PubMed] [Google Scholar]
- 15.de Azavedo, J. C., M. McGavin, C. Duncan, D. E. Low, and A. McGeer. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desjardins, M., K. L. Delgaty, K. Ramotar, C. Seetaram, and B. Toye. 2004. Prevalence and mechanisms of erythromycin resistance in group A and group B streptococcus: implications for reporting susceptibility results. J. Clin. Microbiol. 42:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duarte, R. S., O. P. Miranda, B. C. Bellei, M. A. Brito, and L. M. Teixeira. 2004. Phenotypic and molecular characteristics of Streptococcus agalactiae isolates recovered from milk of dairy cows in Brazil. J. Clin. Microbiol. 42:4214-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duarte, R. S., B. C. Bellei, O. P. Miranda, M. A. Brito, and L. M. Teixeira. 2005. Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob. Agents Chemother. 49:97-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez, M., M. E. Hickman, and C. J. Baker. 1998. Antimicrobial susceptibilities of group B streptococci isolated between 1992 and 1996 from patients with bacteremia or meningitis. Antimicrob. Agents Chemother. 42:1517-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fluit, A. C., M. R. Visser, and F. J. Schmitz. 2001. Molecular detection of antimicrobial resistance. Clin. Microbiol. Rev. 14:836-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furrer, B., U. Candrian, C. Hoefelein, and J. Luethy. 1991. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J. Appl. Bacteriol. 70:372-379. [DOI] [PubMed] [Google Scholar]
- 22.Guerin-Faublee, V., F. Tardy, C. Bouveron, and G. Carret. 2002. Antimicrobial susceptibility of Streptococcus species isolated from clinical mastitis in dairy cows. Int. J. Antimicrob. Agents 19:219-226. [DOI] [PubMed] [Google Scholar]
- 23.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, and A. Schuchat. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. Maryland Emerging Infections Program. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 24.Hauge, M., C. Jespersgaard, K. Poulsen, and M. Kilian. 1996. Population structure of Streptococcus agalactiae reveals an association between specific evolutionary lineages and putative virulence factors but not disease. Infect. Immun. 64:919-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen, N. E. 1982. Experimental bovine group-B streptococcal mastitis induced by strains of human and bovine origin. Nord. Vet. Med. 34:441-450. [PubMed] [Google Scholar]
- 26.Jensen, N. E., and F. M. Aarestrup. 1996. Epidemiological aspects of group B streptococci of bovine and human origin. Epidemiol. Infect. 117:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keefe, G. P. 1997. Streptococcus agalactiae mastitis: a review. Can. Vet. J. 38:429-437. [PMC free article] [PubMed] [Google Scholar]
- 29.King, S. J., J. A. Leigh, P. J. Heath, I. Luque, C. Tarradas, C. G. Dowson, and A. M. Whatmore. 2002. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40:3671-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lachenauer, C. S., D. L. Kasper, J. Shimada, Y. Ichiman, H. Ohtsuka, M. Kaku, L. C. Paoletti, P. Ferrieri, and L. C. Madoff. 1999. Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 179:1030-1033. [DOI] [PubMed] [Google Scholar]
- 32.Lin, F. Y., J. D. Clemens, P. H. Azimi, J. A. Regan, L. E. Weisman, J. B. Philips III, G. G. Rhoads, P. Clark, R. A. Brenner, and P. Ferrieri. 1998. Capsular polysaccharide types of group B streptococcal isolates from neonates with early-onset systemic infection. J. Infect. Dis. 177:790-792. [DOI] [PubMed] [Google Scholar]
- 33.Lin, F. Y., P. H. Azimi, L. E. Weisman, J. B. Philips III, J. Regan, P. Clark, G. G. Rhoads, J. Clemens, J. Troendle, E. Pratt, R. A. Brenner, and V. Gill. 2000. Antibiotic susceptibility profiles for group B streptococci isolated from neonates, 1995-1998. Clin. Infect. Dis. 31:76-79. [DOI] [PubMed] [Google Scholar]
- 34.Martinez, G., J. Harel, R. Higgins, S. Lacouture, D. Daignault, and M. Gottschalk. 2000. Characterization of Streptococcus agalactiae isolates of bovine and human origin by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morales, W. J., S. S. Dickey, P. Bornick, and D. V. Lim. 1999. Change in antibiotic resistance of group B streptococcus: impact on intrapartum management. Am. J. Obstet. Gynecol. 181:310-314. [DOI] [PubMed] [Google Scholar]
- 36.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. USA 86:4731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Committee for Clinical Laboratory Standards. 1997. Performance standards for antimicrobial disk susceptibility tests, 6th ed. Approved standard M2-A6, vol. 17. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 38.Ng, L. K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 39.Reinert, R. R., S. Simic, A. Al-Lahham, S. Reinert, M. Lemperle, and R. Lutticken. 2001. Antimicrobial resistance of Streptococcus pneumoniae recovered from outpatients with respiratory tract infections in Germany from 1998 to 1999: results of a national surveillance study. J. Clin. Microbiol. 39:1187-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrag, S. J., S. Zywicki, M. M. Farley, A. L. Reingold, L. H. Harrison, L. B. Lefkowitz, J. L. Hadler, R. Danila, P. R. Cieslak, and A. Schuchat. 2000. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. N. Engl. J. Med. 342:15-20. [DOI] [PubMed] [Google Scholar]
- 41.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuchat, A. 1999. Group B streptococcus. Lancet 353:51-56. [DOI] [PubMed] [Google Scholar]
- 43.Seral, C., F. J. Castillo, M. C. Rubio-Calvo, A. Fenoll, C. Garcia, and R. Gómez-Lus. 2001. Distribution of resistance genes tet(M), aph3′-III, catpC194 and the integrase gene of Tn1545 in clinical Streptococcus pneumoniae harbouring erm(B) and mef(A) genes in Spain. J. Antimicrob. Chemother. 47:863-866. [DOI] [PubMed] [Google Scholar]
- 44.Sukhnanand, S., B. Dogan, M. O. Ayodele, R. N. Zadoks, M. P. Craver, N. B. Dumas, Y. H. Schukken, K. J. Boor, and M. Wiedmann. 2005. Molecular subtyping and characterization of bovine and human Streptococcus agalactiae isolates. J. Clin. Microbiol. 43:1177-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trijbels-Smeulders, M. A., L. A. Kollee, A. H. Adriaanse, J. L. Kimpen, and L. J. Gerards. 2004. Neonatal group B streptococcal infection: incidence and strategies for prevention in Europe. Pediatr. Infect. Dis. J. 23:172-173. [DOI] [PubMed] [Google Scholar]
- 48.Tyrrell, G. J., L. D. Senzilet, J. S. Spika, D. A. Kertesz, M. Alagaratnam, M. Lovgren, and J. A. Talbot. 2000. Invasive disease due to group B streptococcal infection in adults: results from a Canadian, population-based, active laboratory surveillance study—1996. Sentinel Health Unit Surveillance System Site Coordinators. J. Infect. Dis. 182:168-173. [DOI] [PubMed] [Google Scholar]
- 49.Uh, Y., I. H. Jang, G. Y. Hwang, K. J. Yoon, and W. Song. 2001. Emerging erythromycin resistance among group B streptococci in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 20:52-54. [DOI] [PubMed] [Google Scholar]
- 50.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkinson, H. W., L. G. Thacker, and R. R. Facklam. 1973. Nonhemolytic group B streptococci of human, bovine, and ichthyic origin. Infect. Immun. 7:496-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, D. J., R. N. Gonzalez, and H. H. Das. 1997. Bovine mastitis pathogens in New York and Pennsylvania: prevalence and effects on somatic cell count and milk production. J. Dairy Sci. 80:2592-2598. [DOI] [PubMed] [Google Scholar]
- 53.Witte, W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996-997. [DOI] [PubMed] [Google Scholar]
- 54.Yan, J.-J., H.-M. Wu, A.-H. Huang, H.-M. Fu, C.-T. Lee, and J.-J. Wu. 2000. Prevalence of polyclonal mefA-containing isolates among erythromycin-resistant group A streptococci in southern Taiwan. J. Clin. Microbiol. 38:2475-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]