Abstract
Hemagglutinin sequences of 146 human influenza A/H3N2 strains identified in respiratory specimens from Asia and Europe during the 2001-2003 influenza seasons were analyzed by DNA sequencing. Our results suggest that four amino acid substitutions, L25I, H75Q, H155T, and Q156H, led to the antigenic conversion of the previously predominant A/Panama/2007/99-like strains to the more recent A/Fujian/411/2002-like strains.
Influenza virus genomes are well known to undergo antigenic drifts that enable escape from preexisting immunity and potentially cause epidemics in humans (10). Monitoring antigenic variations in circulating influenza viruses is crucial for anticipating epidemics and for vaccine design.
The surface glycoprotein hemagglutinin (HA) is the major surface antigen of influenza viruses, against which neutralizing antibodies are elicited during virus infection and vaccination (12). HA is synthesized as a single polypeptide that is subsequently cleaved into two polypeptides, HA1 and HA2. The HA1 polypeptide mutates more frequently than HA2 and plays a crucial role in natural selection (13, 14). Five antigenic sites (A to E) on the three-dimensional structure of the HA protein of A/Aichi/2/68 (H3N2) have been proposed to be the antibody-binding sites of the protein (13, 14). About one-third of the HA1 amino acids lie on or near these five antigenic sites, although the importance of these amino acid positions is not clear (6). Recently, several models have been proposed to predict the antigenic evolution of the A/H3N2 viruses (6, 8, 9, 11).
A/Panama/2007/99-like viruses have been circulating worldwide since 1999, which predates the emergence of A/Fujian/411/2002-like viruses. The appearance of A/Fujian/411/2002-like viruses prompted a change in the selection of vaccine components for the Southern Hemisphere in 2003 and for the Southern and Northern Hemispheres in 2004. In this report, we utilized a high-throughput sequencing method to determine the nucleotide sequence of the HA1 gene segments of influenza virus in nasal swabs collected from infected children aged 6 months and older during the 2001-2003 influenza seasons. The specimens were divided into the following five groups, based on the date of collection (Table 1): groups 1, 3, and 5 were from Asian countries, including Bangladesh, Singapore, Malaysia, and South Korea, and groups 2 and 4 were from European countries, including the United Kingdom, Scotland, Finland, The Netherlands, Poland, and Italy. Viral RNA was extracted from influenza viruses propagated once in MDCK cells as previously described (4). HA1 fragments corresponding to nucleotides 37 to 1154 were amplified by reverse transcription-PCR using oligonucleotides 5′-CTATCATTGCTTTGAGCTAC (primer 1) and 5′-ATCTGCTGCTTGTCCTGTGC (primer 2). The PCR products were purified and subjected to sequencing on a genetic analyzer (ABI PRISM 3100; Applied Biosystems, Foster City, CA) with the above-mentioned oligonucleotides and two additional oligonucleotides, 5′-ATGCCAAACAATGACAAATT (primer 3) and 5′-TGTTTGGCATAGTCACGTTC (primer 4), except for group 3 specimens, for which only primers 1 and 4 were used for sequencing. Multiple alignments of the sequences were performed by the ClustalV method, and the phylogenetic tree was constructed by the neighbor-joining method using MegAlign version 5.03 (DNASTAR) as previously described (4).
TABLE 1.
Changes in the deduced amino acid sequences of the HA1 protein of influenza A/H3N2 virusesa
| Group | Region | Period (mo/yr) | No. of specimens | No. of amino acid changes detectedb | Changes in amino acid residuesb |
|---|---|---|---|---|---|
| 1 | Asia | 11/2001 to 03/2002 | 16 | 9 | S21P, R50G, E83K, A131T, L183H, S186G, V202I, W222R, G225D |
| 2 | Europe | 11/2001 to 03/2002 | 17 | 8 | Same as group 1 but without A131T |
| 3 | Asia | 04/2002 to 08/2002 | 58 | 12 | Same as group 1 plus L25I, H75Q, and H155T |
| 4 | Europe | 11/2002 to 05/2003 | 28 | 13 | Same as group 1 plus L25I, H75Q, H155T, and Q156H |
| 5 | Asia | 11/2002 to 05/2003 | 27 | 13 | Same as group 1 plus L25I, H75Q, H155T, and Q156H |
Compared with the HA1 sequence of A/Panama/2007/99.
Only changes corresponding to those in A/Fujian/411/2002 are included.
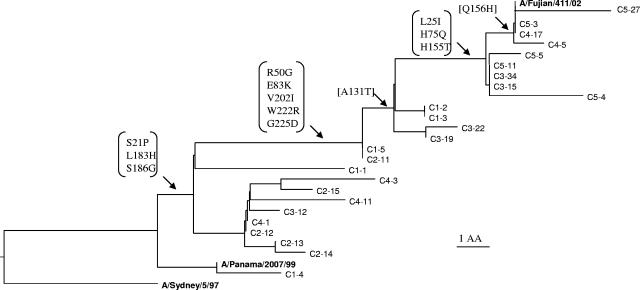
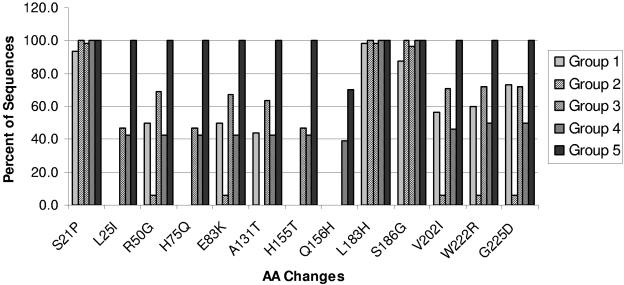
A total of 13 amino acid changes were found in the HA1 protein of the reference vaccine strain, A/Fujian/411/2002, compared to that of the A/Panama/2007/99 strain, including changes at positions 21, 25, 50, 75, 83, 131, 155, 156, 183, 186, 202, 222, and 225. These changes were allocated to the HA1 sequences for the five groups of specimens; the results are summarized in Table 1. For phylogenetic analysis, 25 HA1 sequences (five from each of the five groups) were selected from 146 specimens in an attempt to represent the varieties within each group. The sequences formed clusters (Fig. 1) that corresponded to different stages of antigenic drift. We attempted to establish a pattern by further examining the HA1 sequences of all 146 specimens with respect to these 13 amino acid changes. Figure 2 shows the frequencies of individual amino acid changes within each group. The HA1 sequences in groups 1 and 2 all exhibited amino acid changes S21P, L183H, and S186G, regardless of origin (Asia or Europe), suggesting that these three changes were inherited from earlier drifts. Six additional changes (R50G, E83K, A131T, V202I, W222R, and G225D) were detected in group 1 sequences, five of which (all but A131T) were also observed in group 2. More changes were detected in specimens in later groups. Nearly half (47%) of the sequences from group 3 exhibited 12 of the 13 changes found in the HA1 sequence of A/Fujian/411/2002, including the three amino acid substitutions L25I, H75Q, and H155T, which were also found in group 4 at a frequency of 43%. The occurrence of these three substitutions increased significantly, to 100%, in group 5. Moreover, 39% of group 4 (European) viruses and 70% of group 5 (Asian) viruses also exhibited an additional amino acid substitution, Q156H, which appears to be critical for the antigenic evolution of the viruses from A/Panama/2007/99-like to A/Fujian/411/2002-like, as reported previously (3).
FIG. 1.
Phylogenetic analyses of influenza A/H3N2 viruses based on 320 amino acid sequences of HA1. Twenty-five HA1 sequences (five from each of the five groups) were selected from 146 specimens in an attempt to represent the varieties within the same group. The changes in the deduced amino acid sequences were compared with the sequence of A/Panama/2007/99. Only the amino acid changes corresponding to the sequence of A/Fujian/411/2002 are shown in square brackets. Bold letters indicate the reference vaccine strains. C1, C2, C3, C4, and C5 correspond to groups 1, 2, 3, 4 and 5, respectively. The group numbers are followed by hyphens and then the isolate numbers. AA, amino acid.
FIG. 2.
Prevalence of genetic variations in the HA1 sequences of influenza A/H3N2 viruses. The percentages of HA1 sequences exhibiting the 13 amino acid changes reflect the differences in the A/Fujian/411/2002 strain relative to A/Panama/2007/99.
A hallmark of influenza virus is its ability to undergo rapid antigenic variation (14). New variants are usually subject to intense selective pressure from existing human immunity gained from exposure to their viral ancestors (5). In this study, we investigated the antigenic drift of the HA1 sequence of the human influenza A/H3N2 virus during the 2001-2003 influenza seasons. This study confirms a progressive antigenic drift from predominantly A/Panama/2007/99-like strains to A/Fujian/411/2002-like strains in our A/H3N2 influenza specimens during the 2001-2003 seasons. Among the 13 amino acid differences between the HA1 protein of A/Fujian/411/2002 and that of A/Panama/2007/99, many were detected before March 2002 in both Asia and Europe. Changes were more frequent and advanced in Asia since only 1 out of 17 specimens in group 2 exhibited amino acid changes other than S21P, L183H, and S186G (Fig. 2). Three amino acid changes—L25I, H75Q, and H155T—were found later in the year 2002 but prior to the emergence of the key amino acid change Q156H. These three substitutions were not found separately from each other and appeared to occur at the same time, suggesting that these three amino acid changes arose from a single genetic event. Although most of the viruses examined during this period were genetically closer to A/Fujian/411/2002 than to A/Panama/2007/99 based on their HA1 sequences, it was not until the emergence of the last amino acid change, Q156H, that the antigenic property of the viruses completely shifted to that of A/Fujian/411/2002. In addition, 39% of the European specimens collected from November 2002 to May 2003 contained A/Fujian/411/2002-like viruses, based on the HA1 sequence data. This percentage is significantly higher than that previously reported by Paget et al. (7), based mostly on classical serology assays. However, it is not clear whether the difference is due to sampling or to the sensitivity of the assay method. The sharp rise of the A/Fujian/411/2002-like viruses in the population coincides with the emergence of the key amino acid change Q156H. This particular amino acid located near antigenic site B is considered 1 of the 18 amino acids in the HA1 protein under positive selection and exhibiting a rapid rate of replacement (2). It is likely that these changes altered the antigenic property of the virus and facilitated the spread of A/Fujian/411/2002-like viruses in Asia and Europe as well as in other parts of the world such as South Africa (1).
The HA1 polypeptide has been proposed to contain five antigenic sites, based on a study of its three-dimensional structure (13). Among the 13 amino acid changes in the HA1 protein of A/Fujian/411/2002 compared to that of A/Panama/2007/99, at least 7 of them were located in one of the five antigenic sites: R50G (site C), H75Q and E83K (site E), A131T (site A), and H155T, Q156H, and S186G (site B) (6). In addition, the L183H change is involved in sialic acid receptor binding (12). However, not all amino acid changes in the HA1 sequence were sustained. The majority (94%) of HA1 sequences of European specimens from group 2 exhibited the changes A106V and N144D, which are not found in A/Fujian/411/2002, and the frequency of these changes declined to 50 to 60% in later European specimens (group 4). Similarly, about 30 to 40% of HA1 sequences in Asian specimens found in groups 1 and 3 also exhibited these two changes. However, none of the specimens in group 5 exhibited A106V, and less than 10% exhibited N144D.
Nucleotide sequence accession numbers.
The nucleotide sequences of the HA1 gene can be found in GenBank under theindicated accession numbers: strain A/Panama/2007/99, ISDNCDA001; strain A/Fujian/411/2002, ISDN38157; 146 clinical specimens, DQ179382 to DQ179527.
Acknowledgments
We thank Giuseppe Palladino for providing wild-type influenza reference strains and Fenglan Li and Wafa Al-Rimawi for technical assistance.
REFERENCES
- 1.Besselaar, T. G., L. Botha, J. M. McAnerney, and B. D. Schoub. 2004. Antigenic and molecular analysis of influenza A (H3N2) virus strains isolated from a localised influenza outbreak in South Africa in 2003. J. Med. Virol. 73:71-78. [DOI] [PubMed] [Google Scholar]
- 2.Bush, R. M., C. A. Bender, K. Subbarao, N. J. Cox, and W. M. Fitch. 1999. Predicting the evolution of human influenza A. Science 286:1921-1925. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2003. Information for the Vaccines and Related Biological Products Advisory Committee, CBER, FDA. [Online.] http://www.fda.gov/ohrms/dockets/ac/03/briefing/3922B1_2.pdf.
- 4.Chi, X. S., T. V. Bolar, P. Zhao, R. Rappaport, and S. M. Cheng. 2003. Cocirculation and evolution of two lineages of influenza B viruses in Europe and Israel in the 2001-2002 season. J. Clin. Microbiol. 41:5770-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferguson, N. M., and R. M. Anderson. 2002. Predicting evolutionary change in the influenza A virus. Nat. Med. 8:562-563. [DOI] [PubMed] [Google Scholar]
- 6.Lee, M. S., and J. S. Chen. 2004. Predicting antigenic variants of influenza A/H3N2 viruses. Emerg. Infect. Dis. 10:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paget, W. J., T. J. Meerhoff, and H. Rebelo de Andrade. 2003. Heterogeneous influenza activity across Europe during the winter of 2002-2003. Euro. Surveill. 8:230-239. [DOI] [PubMed] [Google Scholar]
- 8.Plotkin, J. B., and J. Dushoff. 2003. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc. Natl. Acad. Sci. USA 100:7152-7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plotkin, J. B., J. Dushoff, and S. A. Levin. 2002. Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus. Proc. Natl. Acad. Sci. USA 99:6263-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potter, C. W. 2001. A history of influenza. J. Appl. Microbiol. 91:572-579. [DOI] [PubMed] [Google Scholar]
- 11.Smith, D. J., A. S. Lapedes, J. C. de Jong, T. M. Bestebroer, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Fouchier. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371-376. [DOI] [PubMed] [Google Scholar]
- 12.Wiley, D. C., and J. J. Skehel. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365-394. [DOI] [PubMed] [Google Scholar]
- 13.Wiley, D. C., I. A. Wilson, and J. J. Skehel. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373-378. [DOI] [PubMed] [Google Scholar]
- 14.Wilson, I. A., and N. J. Cox. 1990. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 8:737-771. [DOI] [PubMed] [Google Scholar]