Abstract
Although many strain typing methods exist for pathogenic Escherichia coli, most have drawbacks in terms of resolving power, interpretability, or scalability. For this reason, multilocus sequence typing (MLST) is an appealing alternative. However, its applicability to different pathogens in specific epidemiologic contexts is not well understood. Here, we applied a previously established MLST method based on housekeeping genes to a well-characterized collection of uropathogenic E. coli isolates to compare the discriminatory ability of this procedure with that of enterobacterial repeat intergenic consensus (ERIC2) PCR, serogrouping, and pulsed-field gel electrophoresis (PFGE). Among 45 E. coli isolates studied, 17 different multilocus sequence types (ST) were identified. One MLST group (designated ST69 complex) was comprised of 22 isolates, all belonging to uropathogenic and bacteremic E. coli strains previously defined as clonal group A (CgA) by ERIC2 PCR. The ST69 strains contained five different serogroups and 14 PFGE types. ERIC2 PCR CgA strains belonging to different MLST groups were also identified. Interestingly, one cow E. coli isolate, previously shown by PFGE to be closely related to a human uropathogenic CgA strain, was found to cluster with the ST69 strains. All of the other animal and environmental CgA isolates had different MLST profiles. The discriminatory power of this MLST method based on housekeeping genes appears to be higher than that of ERIC2 PCR but lower than that of PFGE for epidemiologic study of uropathogenic E. coli.
The morbidity and financial burden associated with urinary tract infections (UTIs) is substantial: almost half of all women experience at least one UTI in their lifetime, and overall expenditures for UTI treatment in women in the United States was approximately $2.47 billion in 2000 (3, 5). The primary etiologic agent of UTI is Escherichia coli, accounting for as much as 90% of all UTIs seen among ambulatory care patients (23). Antimicrobial resistance among uropathogenic E. coli continues to rise in the United States, contributing to greater difficulty in the management of UTI (6, 7).
The genotypic characterization of pathogens has become an important objective in epidemiologic investigations of infectious agents. Genotyping tools are amenable to validation when the diseases in question occur as recognizable outbreaks, such as those caused by E. coli O157:H7 or Salmonella spp. The problem arises in characterizing organisms that cause diseases that are not usually thought of as causing outbreaks, such as E. coli associated with community-acquired UTI. Recent studies that applied genotyping methods to differentiate uropathogenic E. coli suggest that community-acquired UTIs could occur as outbreaks.
Manges et al. reported in 2001 that a single clonal group of E. coli, designated clonal group A (CgA), based on a characteristic enterobacterial repeat intergenic consensus (ERIC2) PCR fingerprint pattern, was responsible for nearly half of trimethoprim-sulfamethoxazole-resistant E. coli isolates from 47 women with UTIs diagnosed at a California university health clinic between October 1999 and January 2000 (13). Human UTI isolates belonging to CgA were also found to share a random amplified polymorphic DNA PCR fingerprint, virulence factor profile, similar antimicrobial resistance phenotype, O antigen groups (O11, O17, O73, and O77), and indistinguishable or closely related pulsed-field gel electrophoresis (PFGE) patterns (9), suggesting that they caused an outbreak in this California community. Most recently, a gene-specific PCR was devised to identify CgA based on a single-nucleotide polymorphism (SNP) (C288T) identified within the fumC housekeeping gene (11).
Certain genotyping methods (PCR-based methods and PFGE) rely on comparison of banding patterns generated by gel electrophoresis. Such techniques are condition dependent and therefore can suffer from interlaboratory variability (20). Furthermore, visual inspection remains integral to their interpretation, introducing subjectivity and greater potential for error (22). While PFGE does offer superior reproducibility and discriminating power and remains the method most commonly used to identify food-borne outbreaks of E. coli O157:H7 at reference laboratories, it is labor intensive and technically demanding (8, 13, 21). Although gene-specific PCR is highly reproducible, a recent study evaluating 45 isolates exhibiting an ERIC2 PCR CgA pattern found that only 16 isolates contained the C288T SNP (4). Thus, a more reliable, relevant, and consistent typing method for investigating the clonal distribution of uropathogenic E. coli strains, such as CgA isolates, is needed for epidemiologic investigations. The availability of a large, well-characterized, and population-based collection of uropathogenic E. coli isolates provided us with an opportunity to evaluate another genotyping method.
Multilocus sequence typing (MLST) uses nucleotide sequences of internal fragments of selected genes as the unit of comparison and, therefore, does not suffer from the drawbacks of gel-based fingerprinting methods. Sequence data are easily comparable and transferable between laboratories and are highly reproducible (2, 12). Furthermore, the digital format of MLST data has facilitated the establishment of global, web-accessible databases for a variety of organisms and is rapidly contributing to our understanding of the clonal distribution of infectious disease agents. The most commonly used MLST schemes index the neutral genetic variation in housekeeping genes, which are believed to evolve slowly because they are under stabilizing, and not directional, selective pressure. The SNPs observed across several different MLST loci represent neutral genetic variation and presumably exhibit minimal autocorrelation, which could confound epidemiologic conclusions. MLST is thus a powerful tool for global and long-term surveillance.
In this study, we evaluated an MLST protocol standardized for E. coli maintained at the Max-Planck Institut fuer Infektionsbiologie website (http://web.mpiib-berlin.mpg.de) using a well-characterized population-based collection of CgA and other E. coli strains and compared its discriminatory power with that based on PFGE, ERIC2, and serogroup analyses.
MATERIALS AND METHODS
Bacterial isolates.
A total of 45 E. coli isolates were studied: 29 were previously identified as CgA (23 human and 6 animal and environmental) and 16 were identified as human non-CgA strains, defined by ERIC2 PCR (13, 19). Isolates were selected to include groups of strains from various geographic and host sources to investigate potential groupings by these factors. Dates of collection of isolates ranged from 1988 to 2003.
Isolates included strains from the following sources: (i) UTI isolates from California (n = 15) women with symptoms of UTI who were seen at a university health service (they were consecutively enrolled into a study between 11 October 1999 and 31 January 2000) and UTI-causing, trimethoprim-sulfamethoxazole-resistant E. coli isolates (n = 6) from Minnesota (obtained from students who were seen at university health services with uncomplicated cystitis and were enrolled in a study between June 1998 and August 1999); (ii) animal and environmental isolates (n = 6) provided by the Gastroenteric Disease Center at Pennsylvania State University (University Park, PA); (iii) human bacteremia isolates (n = 12) from San Francisco General Hospital provided by Francoise Perdreau-Remington; and (iv) other geographic comparison isolates recovered from humans (n = 6) provided by James R. Johnson of the Minneapolis VA Medical Center, University of Minnesota.
Serotyping.
Serotyping was performed on E. coli isolates at the E. coli Reference Center in University Park, Pennsylvania.
Molecular strain typing analyses.
All E. coli isolates were initially typed by the ERIC2 PCR fingerprinting assay, as described elsewhere (10, 13). The CgA ERIC2 PCR electrophoretic pattern was defined by 4 predominant bands of approximately 1,145, 1,029, 908, and 720 bp; isolates exhibiting this pattern were considered to be members of CgA. A pyelonephritogenic isolate, CFT073 (provided by Harry Mobley, University of Maryland, Baltimore), was used as a reference strain for each ERIC2 PCR run.
PFGE.
The standardized protocol for subtyping E. coli O157:H7 by PFGE, as established by the Centers for Disease Control and Prevention (Atlanta, GA), was used (1). XbaI-digested DNA was electrophoresed in the CHEF DR-II apparatus (Bio-Rad, Hercules, CA). Images of PFGE and ERIC2 PCR electrophoretic patterns were imported and analyzed with GelCompar II, version 2.0 (Applied Maths, Kortrijk, Belgium), with the pattern of the prototype CgA strain (SEQ102, deposited in the American Type Culture Collection as ATCC BAA-457) used as a reference, as previously described (19). To minimize gel-to-gel banding pattern variation, we used the autosearch band calling function of GelCompar with the following parameters: minimum profiling, 5%; gray zone, 0%; minimum area, 1%; shoulder sensitivity, 1. A distance matrix was calculated by GelCompar's Dice algorithm with a band position tolerance of 1%. A dendrogram was generated from the distance matrix by the neighbor-joining method.
MLST. (i) Allele templates selected.
Housekeeping genes for typing were selected from the E. coli database at the MLST website maintained at the Max-Planck Institut fuer Infektionsbiologie (http://web.mpiib-berlin.mpg.de) (Table 1). The genes were shown to be unlinked on an E. coli K-12 genome map. Product lengths varied from 583 to 932 bp (Table 1). Allele templates, included on the website, were based on the genome sequence of E. coli strain MG1655.
TABLE 1.
Forward and reverse sequences of primersc
| Gene (function) | Primer sequence (5′-3′)a | Annealing temp (°C) | Amplicon size (bp)b |
|---|---|---|---|
| adk (adenylate kinase) | ATTCTGCTTGGCGCTCCGGG (F) | 52 | 583 |
| CCGTCAACTTTCGCGTATTT (R) | |||
| fumC (fumarate hydratase) | TCACAGGTCGCCAGCGCTTC (F) | 52 | 806 |
| GTACGCAGCGAAAAAGATTC (R) | |||
| icd (isocitrate/isopropylmalate dehydrogenase) | ATGGAAAGTAAAGTAGTTGTTCCGGCACA (F) | 52 | 878 |
| GGACGCAGCAGGATCTGTT (R) | |||
| purA (adenylosuccinate dehydrogenase) | CGCGCTGATGAAAGAGATGA (F) | 54 | 816 |
| CATACGGTAAGCCACGCAGA (R) | |||
| gyrB (DNA gyrase) | TCGGCGACACGGATGACGGC (F) | 58 | 911 |
| ATCAGGCCTTCACGCGCATC (R) | |||
| recA (ATP/GTP binding motif) | CGCATTCGCTTTACCCTGACC (F) | 58 | 780 |
| TCGTCGAAATCTACGGACCGGA (R) | |||
| mdh (malate dehydrogenase) | ATGAAAGTCGCAGTCCTCGGCGCTGCTGGCGG (F) | 58 | 932 |
| TTAACGAACTCCTGCCCCAGAGCGATATCTTTCTT (R) |
F, forward; R, reverse.
The sequenced DNA strands were shorter than the original PCR amplicon.
Primers are maintained at http://web.mpiib-berlin.mpg.de and http://www.mlst.net.
(ii) DNA isolation.
E. coli isolates were obtained from a collection of strains stored in 15% glycerol at −80°C. Isolates were incubated at 37°C on LB agar plates overnight. Single colonies were picked and inoculated into 2 ml LB media and further incubated in a shaking incubator for 12 to 15 h at 37°C. A 1-ml suspension of bacteria was centrifuged, and DNA was extracted from the bacterial pellet with the QIAGEN DNeasy tissue kit (QIAGEN, Valencia, CA).
PCR.
Amplifications were carried out in a total volume of 50 μl with 2 μl of template DNA, 4 μl of each 10 mM primer (Sigma, Genosys, The Woodlands, TX), 5 μl 10× buffer, 15 mM MgCl2 (Applied Biosystems, Foster City, CA), 5 μl 2 mM deoxynucleoside triphosphate mix (Invitrogen, Carlsbad, CA), and 0.5 U AmpliTaq Gold (Applied Biosystems). Reaction conditions used were: 2 min of denaturation at 95°C; 30 cycles of 1 min of denaturation at 95°C, 1 min of annealing at specific temperature (Table 1), 2 min of extension at 72°C; and a final additional 5 min at 72°C in an Applied Biosystems GeneAmp PCR System 2400 thermocycler.
Sequencing.
PCR products were purified for sequencing with the QIAGEN QIAquick PCR purification kit. Both the forward and reverse strands were sequenced with the PCR primer set, with the exception of mdh, which was sequenced by the forward primer 5′-TCTGGTGAAGATGCGACTCC-3′ and reverse primer 5′-CCCAGGGCGATATCTTTCTT-3′. Sequencing was performed at the University of California at Berkeley Sequencing Facility, which is equipped with an Applied Biosystems 48 capillary 3730 and two Applied Biosystems 16 capillary 3100 genetic analyzer systems. A Perkin Elmer (Wellesley, MA) 9600, an Eppendorf (Hamburg, Germany) Mastercycler, and three MJ Research (Bio-Rad, Hercules, CA) PTC-200s were used for cycle sequencing. The facility runs a 25-cycle sequencing reaction with the following program: 96°C for 10 s plus 50°C for 5 s plus 60°C for 4 min.
Sequence analysis.
Raw sequences were reviewed by visual inspection with Chromas, version 1.45 (32-bit) (Technelysium Pty. Ltd., Tewantin Qld 4565, Australia). DNA sequences were aligned by the neighbor-joining method with 1,000 bootstrap iterations in ClustalW (Hinxton, England). Forward and reverse sequences were aligned for each strain for comparison and editing purposes. Alignments were edited to equally sized fragments and aligned against allele templates based on E. coli strain MG1655, provided at http://www.mlst.net. Sequence editing was conducted with BioEdit (Carlsbad, CA), version 7.0.1. Sequences for the seven genes of each strain were concatenated to produce an alignment sequence of 3,423 bp. A dendrogram of SNP groupings based on these alignments was then constructed by the Phylip programs (Seattle, WA) DNADIST and NEIGHBOR, and the final dendrogram was visualized with the software TreeView (Win32) (Glasgow, Scotland), version 1.6.6.
ST designation.
Gene sequences were submitted to the curator of the MLST E. coli submission page maintained at http://web.mpiib-berlin.mpg.de. Sequence types (STs) and sequence complexes were designated by the curator of the MLST database for sequence submissions that contained tracings found to be acceptable by the curator (Table 2). ST complexes were defined as STs that are linked by distances of one to two allelic differences.
TABLE 2.
STs and ST complexes
| Isolate | Source (location) | Serotype(s) | CgA | ST | ST complex |
|---|---|---|---|---|---|
| 102 | UTI (California) | 011 | Yes | ST69 | ST69 |
| 160 | UTI (California) | 011 | Yes | ST69 | ST69 |
| 403 | UTI (California) | 011 | Yes | ST69 | ST69 |
| 431 | UTI (California) | 011 | Yes | ST69 | ST69 |
| 477 | UTI (California) | 011 | Yes | ST69 | ST69 |
| 220 | UTI (California) | 077 | Yes | ST69 | ST69 |
| 283 | UTI (California) | 077 | Yes | ST69 | ST69 |
| 44 | UTI (California) | 077 | Yes | ST69 | ST69 |
| A1707 | UTI (Maryland) | 017, 077 | Yes | ST407 | ST69 |
| A556 | UTI (Munich, Germany) | 017, 077 | Yes | ST69 | ST69 |
| A925 | UTI (Barcelona, Spain) | 077 | Yes | ST69 | ST69 |
| PY3 | UTI (Los Angeles, Calif.) | 017, 077 | Yes | ST69 | ST69 |
| B | UTI (Minnesota) | NTa | Yes | ST69 | ST69 |
| G | UTI (Minnesota) | 017 | Yes | ****c | **** |
| Y | UTI (Minnesota) | 077 | Yes | ST69 | ST69 |
| AN559 | Cow | 017 | Yes | ST408 | ST69 |
| AN298 | Cow | 011 | Yes | ST101 | ST101 |
| AN45 | Pig | 011 | Yes | ST101 | ST101 |
| AN300 | Cow | 011 | Yes | ST101 | ST101 |
| AN431 | Pig | 077 | Yes | ST394 | ST69 |
| AN437 | Water | 077 | Yes | ST394 | ST69 |
| H42937 | Blood | 015 | Yes | ST393 | ST31 |
| M32569 | Blood | 015 | Yes | ST393 | ST31 |
| F21065 | Blood | 073 | Yes | ST69 | ST69 |
| H3708 | Blood | 017 | Yes | ST69 | ST69 |
| S48427 | Blood | 077 | Yes | ST69 | ST69 |
| W55291 | Blood | 077 | Yes | ST69 | ST69 |
| X19714 | Blood | 086 | Yes | ST69 | ST69 |
| X47726 | Blood | 011 | Yes | ST69 | ST69 |
| SEQ 111 | UTI (California) | †b | No | **** | **** |
| SEQ 020 | UTI (California) | † | No | **** | **** |
| SEQ 329 | UTI (California) | † | No | ST355 | ST73 |
| SEQ 338 | UTI (California) | † | No | ST88 | ST29 |
| SEQ 240 | UTI (California) | † | No | ST127 | None |
| SEQ 336 | UTI (California) | † | No | **** | **** |
| 1702 | UTI (California) | † | No | ST402 | ST405 |
| A219 | UTI (Toulouse, France) | † | No | ST23 | ST23 |
| A563 | UTI (Munich, Germany) | † | No | ST10 | ST10 |
| Jo J | UTI (Minnesota) | † | No | ST73 | ST73 |
| Jo K | UTI (Minnesota) | † | No | ST73 | ST73 |
| Jo L | UTI (Minnesota) | † | No | ST73 | ST73 |
| W38035 | Blood | † | No | ST144 | None |
| F591 | Blood | † | No | ST131 | None |
| X42574 | Blood | † | No | ST372 | None |
| W18993 | Blood | † | No | ST12 | None |
NT, nontypeable.
†, serotype not available.
****, MLST designation not yet available.
RESULTS
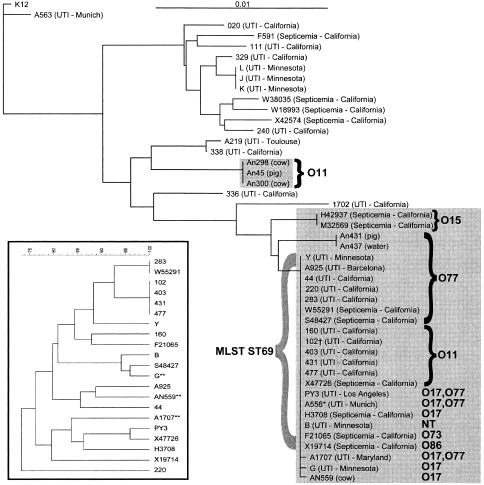
Of 45 E. coli isolates analyzed by MLST, 41 generated sequence tracings acceptable for an ST number assignment. The 41 isolates were grouped into 17 distinct STs (Table 2). Of the 17 STs, 12 were already included in the E. coli MLST website database. Of the 23 human isolates defined by ERIC2 PCR as CgA, one MLST cluster comprised of 21 human isolates, designated ST69 complex, was observed (Table 2; Fig. 1). Of these, 19 showed identical sequences in all seven genes regardless of geographic or clinical sources. One isolate from Maryland (A1707) and one isolate from Minnesota (G) differed by one single-nucleotide polymorphism each, in the fumC gene and the gyrB gene, respectively. Of the 8 human bacteremia CgA isolates, two isolates (H42937 and M32569) fell outside cluster ST69 and shared exact sequence identity. The non-CgA bacteremia isolates (n = 4) each belonged to a distinct MLST type (Fig. 1). Five of the six animal/environmental CgA isolates (AN300, AN298, AN45, AN431, and AN437) fell outside the ST69 grouping. The 21 human strains in the ST69 complex were composed of five serogroups (O11, O77, O17, O73, and O86). The non-ST69 complex CgA isolates that belonged to serogroups O77, O11, and O15 fell into three clusters in which serogroup and sequence type were shared among the isolates in that branch.
FIG. 1.
Neighbor-joining tree constructed from the concatenated sequences of the 7 MLST genes described in the text. The 29 previously identified CgA isolates are shown with shaded backgrounds. Serogroups are indicated for the CgA isolates. The inset shows a dendrogram based on Dice distance coefficient measurements of PFGE banding patterns among the 21 isolates belonging to the ST69 complex. *, no PFGE data available; **, ST69 complex members, not ST69 (see Table 2); †, prototype human uropathogenic CgA strain ATCC BAA-457.
One CgA animal (cow) isolate (AN559) was found to belong to the MLST complex ST69, which included only human UTI and bacteremia CgA isolates (Fig. 1). None of the strains previously identified as non-CgA fell into this ST69 complex. Of the non-CgA strains, the three UTI isolates from Minnesota all belonged to another MLST complex (Fig. 1). These isolates were indistinguishable by ERIC2 PCR and were the only clustered non-CgA isolates.
Of 22 ST69 complex isolates, 19 had identical sequences in all 7 gene templates. These 19 ST69 strains were further separated into 14 subtypes by PFGE (Fig. 1, inset).
DISCUSSION
The evaluation of new molecular strain typing techniques with well-characterized isolates and previously established typing methods is an important component of infectious disease epidemiologic research. Although the limitations of ERIC2 PCR have been reported (14), it was demonstrated here that an MLST scheme based on housekeeping genes revealed highly similar clustering of human isolates previously defined as CgA by ERIC2 PCR (13). However, this MLST method was able to further discriminate human and animal/environmental CgA strains. Five of six animal/environmental isolates defined as CgA did not cluster with human CgA isolates, whereas one did. Thus, while ERIC2 PCR is not as discriminating as the MLST based on housekeeping genes, the observations made in this study suggest that it is still a reliable screening tool that can be used to rapidly separate a large collection of uropathogenic E. coli isolates into groups that have possible epidemiologic relationships.
Interestingly, strain AN559, isolated from a cow in 1988 that was previously shown to be 94% similar by PFGE to a human UTI-causing CgA E. coli isolate (19), was found to occur within the ST69 complex. This was the only animal strain to cluster within this group, differing by a single nucleotide in the adk gene.
Recently, CgA isolates shown to be highly similar by multiple techniques, including C288T SNP analysis, drug susceptibility testing, presence of the 1.8-kb class I integron bearing the dfrA17 gene, and virulence gene profile analysis, were each shown to be distinct by PFGE profile (4). Our investigation of the 21 CgA strains evaluated by PFGE showed that there was no difference in clustering by MLST among the strains that shared PFGE types and the strains that were unique by PFGE. Thus, PFGE still has superior discriminatory ability for uropathogenic E. coli than any of the other methods, including this MLST scheme. A similar observation was made by Noller et al., who compared PFGE and MLST for their discriminatory ability using a collection of diarrheagenic E. coli serotype O157:H7 (16). In one study, MLST was found to be more discriminatory than PFGE only if gene templates in addition to the 7 housekeeping genes were included (15).
ST69 belongs to ECOR D group. A previous study of CgA strains by virulence factor profile analysis found them to have a profile similar to that of the O15:K52:H1 clonal group that was reported to be responsible for community outbreaks of UTI and bacteremia in Europe (8, 17, 18).
Ultimately, the appropriate level of discriminatory power of any strain typing test is determined by the epidemiologic question of interest. Just as highly discriminatory typing methods can obscure important epidemiologic relationships between related strains, typing schemes with low discriminatory power can obscure important epidemiologic associations. Uropathogenic CgA E. coli strains clearly include multiple subtypes, as was shown by the present MLST technique, PFGE analysis, and analysis by France et al. (4). Here, a housekeeping gene-based MLST applied to a well-characterized CgA collection demonstrated discriminatory power that fell between ERIC2 PCR and PFGE. Therefore, for an MLST scheme to be applicable for community-based microepidemiologic studies of uropathogenic E. coli, it will have to be based on allelic templates that show a greater degree of variation.
Acknowledgments
We thank the staff of Tang Health Center, James R. Johnson, Francoise Perdreau-Remington, and Chitrita DebRoy for providing isolates; Sherry Smith for editorial input; Amy White for logistical support; Mark Achtman for advice in executing this MLST analysis; and Linda and David Tartof for general support.
This work was supported by NIH grant RO1 AI059523.
REFERENCES
- 1.Bender, J. B., C. W. Hedberg, J. M. Besser, D. J. Boxrud, K. L. MacDonald, and M. T. Osterholm. 1997. Surveillance by molecular subtype for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N. Engl. J. Med. 337:388-394. [DOI] [PubMed] [Google Scholar]
- 2.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 3.Foxman, B. 2003. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis. Mon. 49:53-70. [DOI] [PubMed] [Google Scholar]
- 4.France, A. M., K. M. Kugeler, A. Freeman, C. A. Zalewski, M. Blahna, L. Zhang, C. F. Marrs, and B. Foxman. 2005. Clonal groups and the spread of resistance to trimethoprim-sulfamethoxazole in uropathogenic Escherichia coli. Clin. Infect. Dis. 40:1101-1107. [DOI] [PubMed] [Google Scholar]
- 5.Griebling, T. L. 2005. Urologic diseases in America project: trends in resource use for urinary tract infections in women. J. Urol. 173:1281-1287. [DOI] [PubMed] [Google Scholar]
- 6.Gupta, K. 2003. Addressing antibiotic resistance. Dis. Mon. 49:99-110. [DOI] [PubMed] [Google Scholar]
- 7.Gupta, K., D. F. Sahm, D. Mayfield, and W. E. Stamm. 2001. Antimicrobial resistance among uropathogens that cause community-acquired urinary tract infections in women: a nationwide analysis. Clin. Infect. Dis. 33:89-94. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, J. R., A. R. Manges, T. T. O'Bryan, and L. W. Riley. 2002. A disseminated multidrug-resistant clonal group of uropathogenic Escherichia coli in pyelonephritis. Lancet 359:2249-2251. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, J. R., A. C. Murray, M. A. Kuskowski, S. Schubert, M. F. Prere, B. Picard, R. Colodner, and R. Raz. 2005. Distribution and characteristics of Escherichia coli clonal group A. Emerg. Infect. Dis. 11:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, J. R., and T. T. O'Bryan. 2000. Improved repetitive-element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenetic groups within Escherichia coli. Clin. Diagn. Lab. Immunol. 7:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, J. R., K. Owens, A. R. Manges, and L. W. Riley. 2004. Rapid and specific detection of Escherichia coli clonal group A by gene-specific PCR. J. Clin. Microbiol. 42:2618-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007-1013. [DOI] [PubMed] [Google Scholar]
- 14.Meacham, K. J., L. Zhang, B. Foxman, R. J. Bauer, and C. F. Marrs. 2003. Evaluation of genotyping large numbers of Escherichia coli isolates by enterobacterial repetitive intergenic consensus-PCR. J. Clin. Microbiol. 41:5224-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemoy, L. L., M. Kotetishvili, J. Tigno, A. Keefer-Norris, A. D. Harris, E. N. Perencevich, J. A. Johnson, D. Torpey, A. Sulakvelidze, J. G. Morris, Jr., and O. C. Stine. 2005. Multilocus sequence typing versus pulsed-field gel electrophoresis for characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates. J. Clin. Microbiol. 43:1776-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noller, A. C., M. C. McEllistrem, O. C. Stine, J. G. Morris, Jr., D. J. Boxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips, I., S. Eykyn, A. King, W. R. Gransden, B. Rowe, J. A. Frost, and R. J. Gross. 1988. Epidemic multiresistant Escherichia coli infection in West Lambeth Health District. Lancet i:1038-1041. [DOI] [PubMed]
- 18.Prats, G., F. Navarro, B. Mirelis, D. Dalmau, N. Margall, P. Coll, A. Stell, and J. R. Johnson. 2000. Escherichia coli serotype O15:K52:H1 as a uropathogenic clone. J. Clin. Microbiol. 38:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramchandani, M., A. R. Manges, C. DebRoy, S. P. Smith, J. R. Johnson, and L. W. Riley. 2005. Possible animal origin of human-associated, multidrug-resistant, uropathogenic Escherichia coli. Clin. Infect. Dis. 40:251-257. [DOI] [PubMed] [Google Scholar]
- 20.Riley, L. W. 2004. Molecular epidemiology of infectious diseases: principles and practices. ASM Press, Washington, D.C.
- 21.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Forey, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O'Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. El Solh, R. de Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, L., and B. Foxman. 2003. Molecular epidemiology of Escherichia coli mediated urinary tract infections. Front. Biosci. 8:e235-e244. [DOI] [PubMed] [Google Scholar]