Abstract
Serotyping and other phenotypic methods are often used to characterize the capsular polysaccharide of group B streptococci (GBS). We describe a capsular genotyping method that utilizes PCR of capsular polysaccharide synthesis genes (cps) and restriction enzyme digestion. This method facilitates the detection of DNA polymorphism in cps genes and correlates well with serotyping.
Group B streptococci (GBS; Streptococcus agalactiae), although generally carried asymptomatically, can cause invasive disease in newborns, pregnant women, and immunocompromised adults. A crucial factor in GBS virulence is the production of an antigenically variable polysaccharide capsule, also used for strain typing. Certain serotypes of the nine known types (Ia, Ib, and II to VIII) are more prevalent in invasive disease, e.g., serotypes Ia, II, and III, and, since the early 1990s, serotype V (2, 4, 13, 16, 29).
Because of the role of capsule in GBS virulence, several phenotypic methods have been devised for serotyping, including the Lancefield capillary precipitin method (19), the predominant serotyping scheme, latex agglutination (22, 25), coagglutination (15), double immunodiffusion (17), and enzyme immunoassays (1). These, however, generally have limited accuracy and applicability, are expensive, and result in numerous (∼2 to 18%) nontypeable (NT) isolates (3, 4, 12). Genotypic methods, including PCR and sequencing of serotype-specific gene fragments within the cps genes (8, 18), DNA dot blot hybridization (5), and PCR-based restriction fragment length polymorphism (RFLP) analyses (24), utilize genetic polymorphisms in the capsular polysaccharide synthesis (cps) gene cluster to classify GBS strains into the corresponding serotypes. Genotypic methods complement phenotypic approaches and avoid problems of unreliable capsule expression, NT phenotypes, and new antigenic variants. Here, we evaluate a new PCR-based method that utilizes RFLP of the cps cluster to detect DNA polymorphisms in the cps cluster.
The cps cluster comprises genes cpsA-O, cpsR, cpsS, and cpsY (6, 8, 28), most of which are conserved across serotypes (8). cpsG-K, however, are highly variable in serotypes Ia, Ib, and II to VII, whereas cpsE to -K are variable in serotype VIII (see Fig. 2a in Cieslewicz et al. [8]). In addition, not all serotypes contain all cps genes. We devised a set of PCR primers (cpsG-F97, 5′-GTGTTCATTCAAACGGGTTACTCA-3′; cpsL-R200, 5′-GAACTTAAAGAACCGCTCGTCTG-3′) in the conserved cpsG and cpsL flanking the variable region of the cps cluster that yield amplicons ranging in size from 4,577 bp (serotype VI cps cluster) to 6,248 bp (serotype V cluster) (Table 1). For serotype VIII, we developed another forward primer targeting cpsR (cpsR8-F40, 5′-CCGGCAAGATATGGTGGAT-3′), which when used with the cpsL-R200 primer, produces a 4,551-bp PCR product (Table 1).
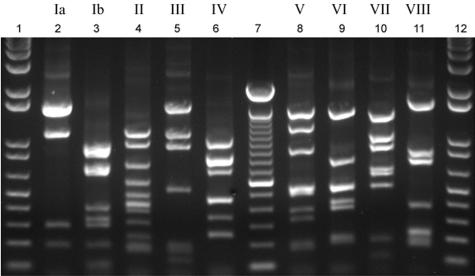
FIG. 2.
PCR-based RFLP capsular typing method illustrating the observed banding patterns specific for each of the nine group B streptococcal serotypes. Lanes 2 to 6, serotypes Ia, Ib, and II to IV, respectively; lanes 8 to 11, serotypes V to VIII; lanes 1 and 12, 1-kb plus ladder; lane 7, 100-bp ladder.
TABLE 1.
Expected DNA fragment sizes of a 4,551- to 6,248-bp region of the group B streptococcal cps gene cluster after PCR and digestion with DdeI
| Capsule serotype | GenBank accession no. | Reference(s) | Predicted cps amplicon length (bp) | Expected no. of cut sites | Expected restriction fragment lengths (bp)a |
|---|---|---|---|---|---|
| Ia | AB028896 | 28 | 4,814 | 7 | 14, 21, 63, 192, 287, 1,100, 1,566, 1,571 |
| Ib | AB050723 | 27 | 4,717 | 12 | 14, 21, 63, 98, 100, 192, 288, 324, 390, 682, 720, 885, 940 |
| II | AY375362 | 8 | 4,645 | 11 | 14, 21, 54, 63, 122, 165, 192, 380, 521, 609, 936, 1,568 |
| III | AF163833, NC_004368 | 14,23 | 4,702 | 9 | 14, 21, 63, 122, 165, 192, 521, 936, 1,100, 1,568 |
| IV | AF355776 | 7 | 5,304 | 14 | 11, 14, 21, 43, 63, 82, 241, 245, 337, 458, 460, 693, 806, 839, 1,027 |
| V | AF349539, NC_004116 | 7,>26 | 6,248 | 14 | 11, 12, 14, 21, 63, 167, 192, 325, 368, 484, 511, 530, 883, 1,191, 1,476 |
| VI | AF337958 | 7 | 4,577 | 11 | 14, 21, 51, 63, 192, 211, 372, 409, 505, 521, 764, 1,454 |
| VII | AY376403 | 8 | 5,364 | 10 | 11, 14, 21, 114, 192, 49 8, 599, 642, 882, 1,011, 1,380 |
| VIII | AY375363 | 8 | 4,551 | 9 | 41, 163, 170, 192, 214, 224, 366, 728, 823, 1,630 |
DdeI restriction fragments of cps PCR product. Underlining denotes fragments that can be resolved on a 1.5% agarose gel.
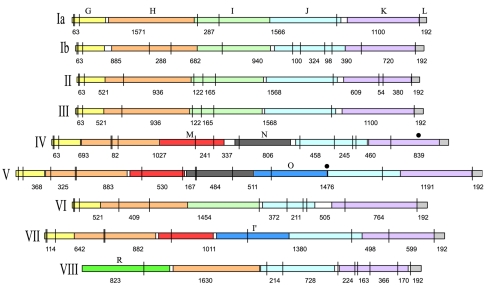
Because of the sequence variability across serotypes within the region amplified, in silico digestion of the cps amplicons yields extensive RFLP among serotypes (Fig. 1). Based on the published cps sequences of nine GBS strains, digestion with DdeI of the predicted cps amplicons should generate 8 to 15 restriction fragments (Table 1).
FIG. 1.
Diagram representing the serotype-specific variable region within the group B streptococcal capsular polysaccharide synthesis (cps) gene cluster that is amplified by PCR. Homologous cps genes are illustrated by colors. The vertical lines and numbers represent the DdeI restriction sites and the expected restriction fragment sizes, respectively. Solid circles mark fragments affected in allelic variants.
We examined 141 GBS strains representing nine serotypes and NT strains as determined previously by the Lancefield method (19) at the National Center for Streptococcus (Edmonton, Alberta, Canada). A total of 45 strains, collected in Alberta between 1998 and 2000, were isolated from patients with invasive GBS disease (10), and 36 strains were from colonized pregnant women (9). An additional 60 strains were obtained from the American Type Culture Collection (NEM316) (14), the University of Michigan (n = 26) (5, 20, 21), Czech National Collection of Type Cultures (n = 2), Norrlands Universitetssjuk-Ius (n = 20) (24), and Channing Laboratory (n = 11) (26). Approval to obtain isolates from some pregnant women and newborns in the present study was granted by the Conjoint Medical Ethics Board of the University of Calgary. Informed consent was obtained from all study participants.
DNA was isolated by growing strains overnight in Todd-Hewitt broth incubated at 37°C in CO2 (Mo Bio Laboratories, Inc., Solana Beach, CA). The three primers (10 μM each) were pooled, and DNA fragments were amplified via PCR using 1.5 U of LA Taq (Takara Bio, Inc., Kyoto, Japan). The thermal cycle, initiated by a 1-min soak at 94°C, was run for 30 cycles (94°C for 30 s, 58°C for 30 s, and 68°C for 6 min), followed by a 10-min soak at 72°C. The amplicons were digested with DdeI (New England Biolabs, Inc., Beverly, MA) overnight at 37°C, followed by electrophoresis on a 1.5% agarose gel and ethidium bromide staining.
The cps region was amplified in 139 (98.5%) strains representing nine serotypes, and DdeI digestions revealed 12 distinct restriction fragment patterns (Fig. 2). With the exception of serotype II strains, the observed fragment sizes estimated using PRO-SCORE (11) matched the predicted sizes. RFLP of the cps region was concordant with the serotype in 138 (97.9%) of 141 strains (Table 2); therefore, we designated the distinct restriction patterns with numbers and letters corresponding to the serotype (e.g., cps1a for serotype Ia).
TABLE 2.
Capsule genotypes based on RFLP of the cps gene cluster
| Capsule | No. of strains | Capsule genotype (no. of strainsa) |
|---|---|---|
| Ia | 8 | cps1a (8) |
| Ib | 7 | cps1b (7) |
| II | 13 | cps2 (13) |
| III | 38 | cps3 (36), cps1a (1), cps5.1 (1) |
| IV | 16 | cps4.1 (9), cps4.2 (6), cps6 (1) |
| V | 11 | cps5.1 (11) |
| VI | 9 | cps6 (9) |
| VII | 8 | cps7.1 (6), cps7.2 (2) |
| VIII | 10 | cps8 (10) |
| NT | 21b | cps1a (2), cps1b (4), cps5.1 (9), cps5.2 (2), cps6 (1), cps8 (1) |
A complete strain list with capsule genotypes is provided in the supplemental material.
The capsular genotype could not be determined for two NT strains.
Two strains originally designated as serotype III matched the cps1a and cps5 genotypes. Blinded re-serotyping using hyperimmune rabbit antisera to the GBS polysaccharide types (21) was performed by Carol Baker. Re-serotyping confirmed the cps5 genotype in one strain, suggesting an error in the original serotype. The cps1a strain was re-serotyped as NT, a result that may be a consequence of overexpression of cpsH (6).
All 13 serotype II strains produced the identical cps2 pattern, which differed substantially from the predicted DdeI digestion of the serotype II cps sequence in GenBank (AY375362 [8]). The basis for this discrepancy is problematic; however, we suspect that the GenBank strain does not represent the primary serotype II GBS clone in circulation.
Among the 21 NT strains examined, 19 could be assigned a cps genotype after PCR amplification and DdeI digestion (Table 2). In two strains, however, no amplicons of the cps region were produced. Further evaluation of these strains with additional primer sets, including primers in cpsE and neuB paired with the reversed cpsG and cpsL primers and previously described (24) cps primers (lorfXfo and loEFrev), yielded no specific PCR products. These observations, along with reports of rare GBS strains that cannot be typed by other methods (5, 24), suggest that there are clones in the GBS population that have diverged in the conserved regions of cps locus.
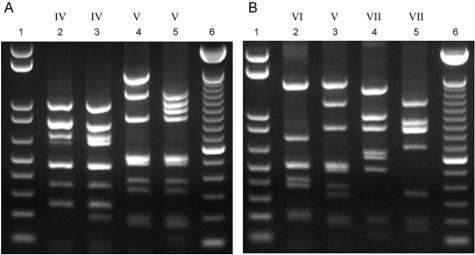
One advantage of the RFLP-based method is its ability to identify cps genotypic variants: we recovered cps variants for serotypes IV, V, and VII but not for serotype III as described previously (24) (Fig. 3). Presumably, the polymorphism underlying the type III variant described by Sellin et al. (24) lies outside the region examined by our method. For serotypes IV and V, the observed restriction patterns were similar to the predicted patterns (Fig. 3). The cps4.2 (serotype IV, allele 2) variant can be explained by an additional restriction site in the 839-bp fragment of cpsK (Fig. 1), whereas cps5.2 (serotype V, allele 2) appears to have an ∼700-bp DNA insertion with additional restriction sites between cpsO and cpsJ (Fig. 1). The serotype VII variant pattern (cps7.2), originally identified by Sellin et al. (24), differs substantially from the predicted pattern (Fig. 3); the structure of this cps variant warrants further analysis.
FIG. 3.
Restriction fragment banding patterns of cps allelic variants. (A) Lane 2, cps4.1; lane 3, cps4.2; lane 4, cps5.1; lane 5, cps5.2. (B) Lane 2, cps6; lane 3, cps5.1; lane 4, cps7.1; lane 5, cps7.2. In both panels, lanes 1 and 6 show the 1-kb plus ladder and 100-bp ladder, respectively. Capsule serotypes are indicated above in Roman numerals.
The amplification and RFLP analysis of the cps gene cluster accurately reveals the genetic variation underlying the capsular variation of GBS strains. Genotyping the cps locus as described is inexpensive and easy to perform, and can be used to classify NT isolates and identify cps allelic variants. In addition, it can identify novel, emergent, capsular types based on restriction pattern differences.
Supplementary Material
Acknowledgments
We thank Clare Fraser of The Institute of Genomic Research for authorizing the use of 11 GBS strains used in the phylogenetic analysis of the serotype V strain 2603 V/R; Michael Cieslewicz and Lawrence Madoff of the Channing Laboratory at Harvard University for supplying original cultures of TIGR strains; Patricia Tallman of the University of Michigan, Helena Zemlickova of The Czech National Collection of Type Cultures, and Mats Sellin of Norrlands Universitetssjuk-Ius for supplying GBS strains; and Carol Baker at the Streptococcal Immunology Laboratory for re-serotyping discrepant isolates. We also thank Cody Springman for technical assistance.
This study was supported by Public Health Service grant AI066081 from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Arakere, G., A. E. Flores, P. Ferrieri, and C. E. Frasch.1999. Inhibition enzyme-linked immunosorbent assay for serotyping of group B streptococcal isolates. J. Clin. Microbiol. 37:2564-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, C. J., and M. S. Edwards.2003. Group B streptococcal conjugate vaccines. Arch. Dis. Child. 88:375-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bliss, S. J., S. D. Manning, P. Tallman, C. J. Baker, M. D. Pearlman, C. F. Marrs, and B. Foxman.2002. Group B Streptococcus colonization in male and nonpregnant female university students: a cross-sectional prevalence study. Clin. Infect. Dis. 34:184-190. [DOI] [PubMed] [Google Scholar]
- 4.Blumberg, H. M., D. S. Stephens, M. Modansky, M. Erwin, J. Elliot, R. R. Facklam, A. Schuchat, W. Baughman, and M. M. Farley.1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 173:365-373. [DOI] [PubMed] [Google Scholar]
- 5.Borchardt, S. M., B. Foxman, D. O. Chaffin, C. E. Rubens, P. A. Tallman, S. D. Manning, C. J. Baker, and C. F. Marrs.2004. Comparison of DNA dot blot hybridization and Lancefield capillary precipitin methods for group B streptococcal capsular typing. J. Clin. Microbiol. 42:146-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaffin, D. O., S. B. Beres, H. H. Yim, and C. E. Rubens.2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182:4466-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaffin, D. O., K. McKinnon, and C. E. Rubens.2002. CpsK of Streptococcus agalactiae exhibits alpha2,3-sialyltransferase activity in Haemophilus ducreyi. Mol. Microbiol. 45:109-122. [DOI] [PubMed] [Google Scholar]
- 8.Cieslewicz, M. J., D. Chaffin, G. Glusman, D. Kasper, A. Madan, S. Rodrigues, J. Fahey, M. R. Wessels, and C. E. Rubens.2005. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 73:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, H. D., C. Adair, A. McGeer, D. Ma, S. Robertson, M. Mucenski, L. Kowalsky, G. Tyrell, and C. J. Baker.2001. Antibodies to capsular polysaccharides of group B Streptococcus in pregnant Canadian women: relationship to colonization status and infection in the neonate. J. Infect. Dis. 184:285-291. [DOI] [PubMed] [Google Scholar]
- 10.Davies, H. D., S. Raj, C. Adair, J. Robinson, and A. McGeer.2001. Population-based active surveillance for neonatal group B streptococcal infections in Alberta, Canada: implications for vaccine formulation. Pediatr. Infect. Dis. J. 20:879-884. [DOI] [PubMed] [Google Scholar]
- 11.DNA ProScan.1999. PRO-SCORE, 3.36 ed. DNA ProScan, Inc., Nashville, Tenn.
- 12.Edwards, M. S., M. A. Rench, D. L. Palazzi, and C. J. Baker.2005. Group B streptococcal colonization and serotype-specific immunity in healthy elderly persons. Clin. Infect. Dis. 40:352-357. [DOI] [PubMed] [Google Scholar]
- 13.Farley, M. M.2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 14.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst.2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 15.Hakansson, S., L. G. Burman, J. Henrichsen, and S. E. Holm.1992. Novel coagglutination method for serotyping group B streptococci. J. Clin. Microbiol. 30:3268-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, A. Schuchat, et al.1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, D. R., and P. Ferrieri.1984. Group B streptococcal Ibc protein antigen: distribution of two determinants in wild-type strains of common serotypes. J. Clin. Microbiol. 19:506-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert.2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lancefield, R.1934. Serological differentiation of specific types of bovine hemolytic streptococci (group B). J. Exp. Med. 59:441-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning, S. D., B. Foxman, C. L. Pierson, P. Tallman, C. J. Baker, and M. D. Pearlman.2003. Correlates of antibiotic-resistant group B Streptococcus isolated from pregnant women. Obstet. Gynecol. 101:74-79. [DOI] [PubMed] [Google Scholar]
- 21.Manning, S. D., P. Tallman, C. J. Baker, B. Gillespie, C. F. Marrs, and B. Foxman.2002. Determinants of co-colonization with group B Streptococcus among heterosexual college couples. Epidemiology 13:533-539. [DOI] [PubMed] [Google Scholar]
- 22.Park, C. J., N. M. Vandel, D. K. Ruprai, E. A. Martin, K. M. Gates, and D. Coker.2001. Detection of group B streptococcal colonization in pregnant women using direct latex agglutination testing of selective broth. J. Clin. Microbiol. 39:408-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubens, C. E., L. M. Heggen, R. F. Haft, and M. R. Wessels.1993. Identification of cpsD, a gene essential for type III capsule expression in group B streptococci. Mol. Microbiol. 8:843-855. [DOI] [PubMed] [Google Scholar]
- 24.Sellin, M., C. Olofsson, S. Hakansson, and M. Norgren.2000. Genotyping of the capsule gene cluster (cps) in nontypeable group B streptococci reveals two major cps allelic variants of serotypes III and VII. J. Clin. Microbiol. 38:3420-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slotved, H. C., J. Elliott, T. Thompson, and H. B. Konradsen.2003. Latex assay for serotyping of group B Streptococcus isolates. J. Clin. Microbiol. 41:4445-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser.2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe, M., K. Miyake, K. Yanae, Y. Kataoka, S. Koizumi, T. Endo, A. Ozaki, and S. Iijima.2002. Molecular characterization of a novel beta1,3-galactosyltransferase for capsular polysaccharide synthesis by Streptococcus agalactiae type Ib. J. Biochem. 131:183-191. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto, S., K. Miyake, Y. Koike, M. Watanabe, Y. Machida, M. Ohta, and S. Iijima.1999. Molecular characterization of type-specific capsular polysaccharide biosynthesis genes of Streptococcus agalactiae type Ia. J. Bacteriol. 181:5176-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaleznik, D. F., M. A. Rench, S. Hillier, M. A. Krohn, R. Platt, M. L. Lee, A. E. Flores, P. Ferrieri, and C. J. Baker.2000. Invasive disease due to group B Streptococcus in pregnant women and neonates from diverse population groups. Clin. Infect. Dis. 30:276-281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.