Abstract
Here we present a system for adenovirus detection and genotyping based on PCR amplification and phylogenetic analysis of a conserved hexon gene fragment. The system was validated using 157 sequences (86 previously typed and 71 clinical samples) and correctly identified species and serotype in 100% and 84% of sequences, respectively. Known associations between specific serotypes and clinical syndromes are verified. Possible new associations are described to allow further independent testing.
Human adenoviruses (HAdVs) cause a wide range of clinical syndromes and are being increasingly recognized in cases of severe or fatal pneumonia, hemorrhagic cystitis, hepatitis, or disseminated disease in pediatric bone marrow transplant recipients (12). HAdVs are classified into six species, A to F, comprising 51 serotypes (5). Serotype identification is critical for epidemiological surveillance, detection of new strains, assessment of treatment efficacy, and understanding HAdV pathogenesis (16).
Molecular typing methods have been established to circumvent practical problems associated with traditional serum neutralization studies (1, 10, 13, 14). Molecular methods also have disadvantages; restriction fragment length polymorphism analysis of adenoviral DNA may fail if mutations are present within the restriction site, and multiplex PCR assays are currently not able to discriminate between serotypes. PCR amplification of the hypervariable portion of the hexon gene followed by DNA sequencing has recently been proposed as a typing method; however, this method was unable to discriminate between species B and E and was validated with only 10 clinical samples (19).
We have previously detected HAdV infection in clinical samples using generic HAdV primers in singleplex (4) and multiplex assays; these assays have been extensively validated and used routinely for clinical diagnosis (6, 8). Here, we report that DNA sequencing and phylogenetic analysis of this moderately conserved region (amino acids 540 to 662) of the hexon gene (9) are sufficient to allow HAdV speciation and, in most cases, serotype identification. We have confirmed and also noted new associations between specific serotypes and clinical presentations.
HAdV infection was detected by two distinct multiplex PCRs (8) in 46 clinical specimens and 25 HAdV culture isolates sent for diagnostic evaluation at Centro Nacional de Microbiologia, ISCIII, Spain. Clinical materials, patient characteristics, and alternate methods to detect HAdV infection, such as cell culture, direct immunofluorescence assay, and latex agglutination, are listed in Table 1.
TABLE 1.
Clinical materials, patient characteristics, and methods of detectiona
| Sampleb | Syndrome | Patient sample | Patient age | Epidemiology | Cell culture | Antigen detectionc
|
|
|---|---|---|---|---|---|---|---|
| IFA | LA | ||||||
| SO3360_01 | Bronchiolitis | NPA | 1.6 yr | Sporadic | cont | − | ND |
| SO3790_03 | Bronchiolitis | NPA | <5 yr | Sporadic | + | + | ND |
| SO4010_04 | Bronchiolitis | NPA | 1.1 yr | Sporadic | − | − | ND |
| SO4363_04 | Bronchiolitis | NPA | 10 mo | Sporadic | ND | − | ND |
| SO4366_04 | Bronchiolitis | NPA | 11 mo | Sporadic | ND | − | ND |
| SO4390_04 | Bronchiolitis | NPA | <5 yr | Sporadic | cont | + | ND |
| SO4405_04 | Bronchiolitis | NPA | <5 yr | Sporadic | + | + | ND |
| SO4408_04 | Bronchiolitis | NPA | <5 yr | Sporadic | ND | − | ND |
| SO4425_04 | Bronchiolitis | NPA | 1.5 yr | Sporadic | + | + | ND |
| SO4427_04 | Bronchiolitis | NPA | <5 yr | Sporadic | + | + | ND |
| SO4429_04 | Bronchiolitis | NPA | <5 yr | Sporadic | − | − | ND |
| G1066_01 | Influenza-like | TS | 33 yr | Sporadic | − | − | ND |
| G1108_01 | Influenza-like | TS | 26 yr | Sporadic | − | − | ND |
| G1253_02 | Influenza-like | TS | 2 yr | Sporadic | − | − | ND |
| G1507_04 | Influenza-like | TS | 33 yr | Sporadic | − | ND | ND |
| G1508_04 | Influenza-like | TS | 25 yr | Sporadic | − | − | ND |
| G1634_04 | Influenza-like | TS | 29 yr | Sporadic | − | − | ND |
| R1625_04 | Pneumonia | NW | 20 d | Sporadic | ND | ND | ND |
| 1184X_93 | Pneumonia | BAL | <5 yr | Sporadic | + | ND | ND |
| R1629_04 | HIV+, pneumonia | BAL | 30 yr | Sporadic | − | ND | ND |
| 591Fi_01 | LRI | BAL | Sporadic | − | ND | ND | |
| 992Fi_01 | LRI | BAL | Sporadic | − | ND | ND | |
| 593Fi_01 | LRI | BAL | Sporadic | − | ND | ND | |
| 130C_99 | LRI | NW | 19 mo | Sporadic | ND | ND | ND |
| R1612_04 | BMTx, hemorrhagic cystitis | NW | 14 yr | Sporadic | − | ND | ND |
| 226l_03 | BMTx, hemorrhagic cystitis | Urine | 40 yr | Sporadic | ND | ND | ND |
| 1938l_89 | BMTx, hemorrhagic cystitis | Urine | 30 yr | Sporadic | + | ND | ND |
| 1370l_03 | BMTx, hemorrhagic cystitis | Urine | 15 yr | Sporadic | ND | ND | ND |
| 411I_04 | LITx, fatal hepatitis | Liver | 5 yr | Sporadic | ND | ND | ND |
| 1803F_03 | Exanthema, fever | TS | 3 yr | Sporadic | ND | ND | ND |
| 2549F_03 | Exanthema, fever | TS | 4 yr | Sporadic | ND | ND | ND |
| 1804F_03 | Exanthema, fever | TS | 6 yr | Sporadic | ND | ND | ND |
| 1292F_01 | Exanthema, not fever | TS | 2 yr | Sporadic | + | ND | ND |
| 824O_02 | Keratoconjunctivitis | Eye swab | 56 yr | Sporadic | ND | ND | ND |
| 928O_04 | Keratoconjunctivitis | Eye swab | 30 yr | Out-Jaén | ND | ND | ND |
| 926O_04 | Keratoconjunctivitis | Eye swab | 55 yr | Out-Jaén | ND | ND | ND |
| 925O_04 | Keratoconjunctivitis | Eye swab | 61 yr | Out-Jaén | ND | ND | ND |
| D0005_01 | Diarrhea | Feces | <14 yr | Sporadic | − | ND | + |
| D0007_01 | Diarrhea | Feces | <14 yr | Sporadic | − | ND | + |
| D0001_01 | Diarrhea | Feces | <14 yr | Sporadic | − | ND | + |
| D0011_01 | Diarrhea | Feces | <14 yr | Sporadic | − | ND | + |
| D0002_01 | Diarrhea | Feces | <14 yr | Sporadic | − | ND | + |
| D0004_01 | Diarrhea | Feces | <14 yr | Sporadic | − | ND | + |
| D0015_01 | Diarrhea | Feces | <14 yr | Sporadic | − | ND | + |
| D0016_01 | Diarrhea | Feces | <14 yr | Sporadic | − | ND | + |
| D0017_01 | Diarrhea | Feces | <14 yr | Sporadic | − | ND | + |
| R1650_04 | BMTx, diarrhea | Feces | <5 yr | Sporadic | Hep-2 | + | ND |
| R1641_04 | BMTx, diarrhea | Feces | <5 yr | Sporadic | Hep-2 | + | ND |
| C1640_03 | BMTx, diarrhea | Feces | 6 yr | Sporadic | HEF | + | ND |
| R1647_04 | BMTx, diarrhea | Feces | <5 yr | Sporadic | Hep-2 | + | ND |
| C5335_01 | BMTx, pneumonia | BAL + biopsy | 49 yr | Sporadic | HEF | + | ND |
| 594_89 | HIV+, diarrhea | Feces | >40 yr | Sporadic | Hep-2 | + | ND |
| 4030_96 | Influenza-like | Feces | 3 yr | Sporadic | Hep-2 | + | ND |
| C1662_03 | Bronchiolitis | NPA | 5 mo | Sporadic | HEF | + | ND |
| C1491_02 | Bronchiolitis | NPA | 1 mo | Sporadic | HEF | + | ND |
| C1519_02 | Bronchiolitis | NPA | 15 mo | Sporadic | HEF | + | ND |
| C1201_00 | Bronchiolitis | NPA | 8 mo | Sporadic | HEF | + | ND |
| G3093_03 | LRI | Feces | 30 yr | Sporadic | Hep-2 | + | ND |
| C1167_00 | LRI | NPA | 3 mo | Sporadic | HEF | + | ND |
| 859_96 | Keratoconjunctivitis | Eye swab | >40 yr | Out-Tenerife | Hep-2 | + | ND |
| 860_96 | Keratoconjunctivitis | Eye swab | >40 yr | Out-Tenerife | Hep-2 | + | ND |
| 519_93 | Keratoconjunctivitis | Eye swab | >40 yr | Out-Toledo | Hep-2 | + | ND |
| 856_96 | Keratoconjunctivitis | Eye swab | >40 yr | Out-Tenerife | Hep-2 | + | ND |
| 615_96 | Keratoconjunctivitis | Eye swab | >40 yr | Out-Pamplona | Hep-2 | + | ND |
| 636_96 | Keratoconjunctivitis | Eye swab | >40 yr | Out-Pamplona | Hep-2 | + | ND |
| 647_96 | Keratoconjunctivitis | Eye swab | >40 yr | Out-Pamplona | Hep-2 | + | ND |
| 841_94 | Keratoconjunctivitis | Eye swab | >40 yr | Sporadic | Hep-2 | + | ND |
| 43024_02 | Keratoconjunctivitis | TS | >40 yr | Out-Madrid | HEF | + | ND |
| C4292_02 | Keratoconjunctivitis | Eye swab | >40 yr | Out-Madrid | HEF | + | ND |
| C1629_02 | Newborn control | NPA | 0 d | Sporadic | HEF | + | ND |
| G1T4_03 | Environmental water | A549 | + | ND | |||
Abbreviations: BMTx, bone marrow transplant; LITx, liver and intestinal transplant; LRI, low respiratory infection; NPA, nasopharyngeal aspirates; TS, throat swab; BAL, bronchoalveolar lavage; NW, nasal wash; LA, latex agglutination; cont, contamination of cell cultures; ND, not done; Out-, outbreak; HEF, human embryonic fibroblast cell line.
Clinical sample identification no._year or isolate_year (boldface).
Cells were collected and stained by standard methods. The monoclonal antibodies used for detection of the fusion protein (F0/F1) of all strains of parainfluenza virus type 1, hemagglutinin of all strains of parainfluenza viruses types 2 and 3, nucleoproteins of influenza A and B, hemagglutinins H1 and H3 of influenza A, and fusion protein and nuclear protein of respiratory syncytial virus strains and enteroviruses were obtained from Chemicon. The immunofluorescence assay was performed with fluorescein isothiocyanate-conjugated goat antimouse immunoglobulin G (Sigma). All specimens for culture were collected in 3 ml of virus transport medium (MEM, Gibco-BRL, Life Technologies, Paisley, Scotland; penicillin, 200 U/ml; and streptomycin, 200 mg/ml; BioWhittaker, MA; mycostatin, 200 U/ml, Sigma; bovine serum albumin, 0.25%, Merck, Darmstadt, Germany). Stool samples were tested for HAdV (HAdV-40 and -41) by latex agglutination (Adenolex, Orion, Helsinki, Finland).
For comparison, 47 prototype HAdV strains, each representing a distinct serotype, were obtained from the American Type Culture Collection (Manassas, VA) or from an existing collection in our institute (see Table S1 in the supplemental material).
Nucleic acids were extracted (7) and the singleplex PCR assay performed as described previously (4). Briefly, 5 μl of the nucleic acid extraction was added to 45 μl of reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 500 μM (each) deoxynucleoside triphosphates, 4 mM MgCl2, 2.5 units of Taq polymerase (Amplitaq; Perkin-Elmer Cetus, Norwalk, Conn.), and 20 pmol of the degenerate primers ADHEX1F (5′CAACACCTAYGASTACATGAA3′) and ADHEX1R (5′KATGGGGTARAGCATGTT3′). Temperature and time profiles were as follows: 94°C for 1 min, 50°C for 1 min, and 68°C for 1 min for 30 cycles and a final incubation at 68°C for 5 min. Amplification products (475 bp) from this PCR were visualized by agarose gel electrophoresis and sequenced directly. For clinical samples where direct sequencing was not possible due to low DNA yield, two independent nested reactions were performed with 20 pmol of the degenerate primers ADHEX2F (5′CCCITTYAACCACCACCG3′) and ADHEX1R or 20 pmol of ADHEX1F and ADHEX2R (5′ACATCCTTBCKGAAGTTCCA3′). Amplified products were purified and sequenced in both directions using an automated ABI PRISM 377 model sequencer.
The consensus sequence was compared and aligned against other sequences from samples or the DNA database using the program CLUSTAL X (version 1.83). The relationships between individual viruses were established using neighbor-joining, unweighted-pair group method using average linkages, and nucleotide substitution methods (Tamura-Nei, Kimura-2p, and Jukes-Cantor). Phylogenetic trees were reconstructed through the neighbor-joining method (MEGA package, version 3) by 1,000 bootstrap resamplings. Pairwise comparisons were also made by global alignment using the Needleman Wunsch algorithm (17), implemented by a program from EMBOSS (18).
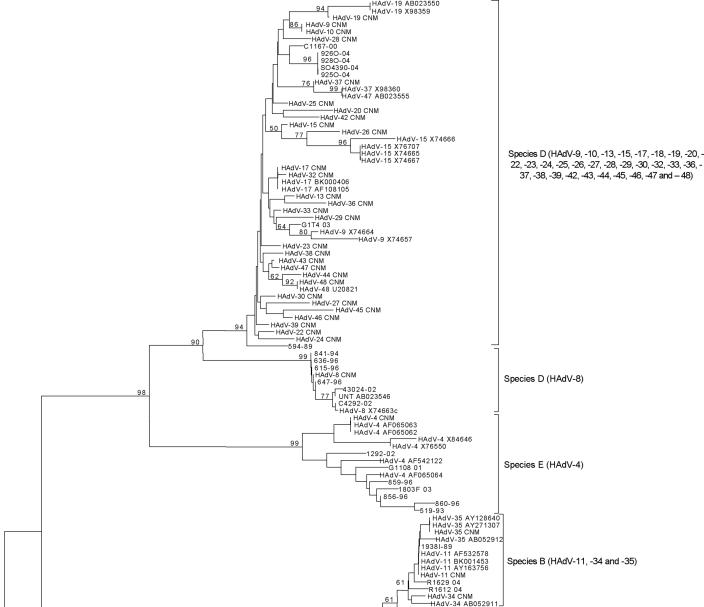
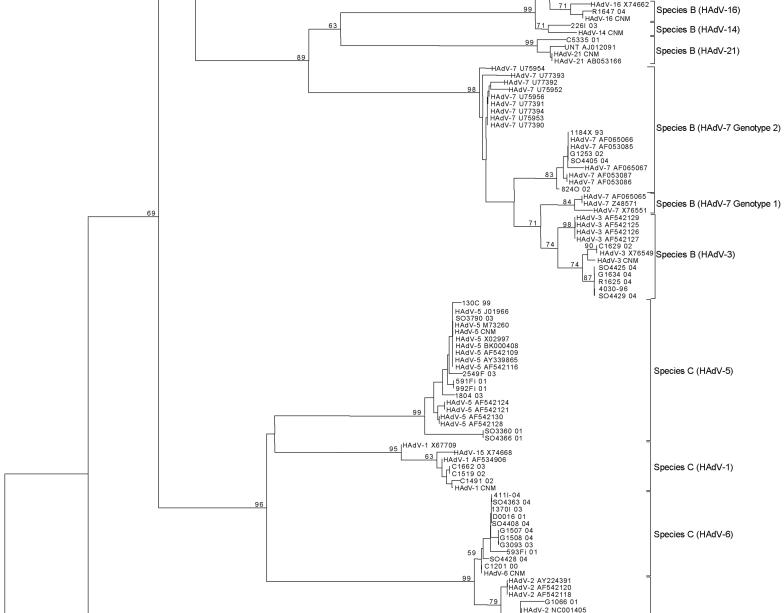
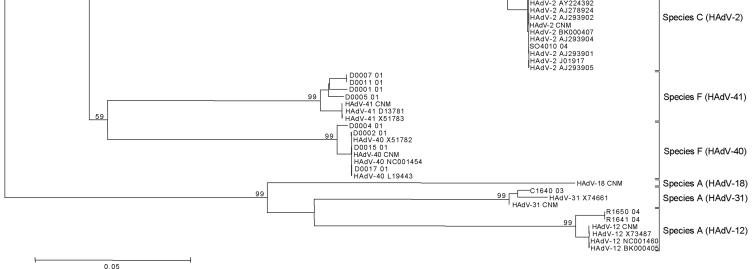
The phylogenetic tree showed six different clusters representing species A to F at the nucleotide (Fig. 1) and amino acid levels, with bootstrap values ranging from 59 to 99. Results obtained when the sequences from 46 clinical samples and the 25 HAdV isolates were compared with sequences from the reference strains are presented in Table 2. All clinical samples were speciated, and 42 (91%) of 46 were serotyped. All 25 isolates were speciated, and 22 (88%) of 25 were serotyped. Phylogenetic analysis revealed subclusterings, except in species E, D, and B2; serotype HAdV-7 could be separated into two lineages (14). Serotypes of species D were not clearly discriminated because of high homology. Serotypes 11, 34, and 35 of subgroup B2 were indistinguishable. HAdV-4 is the only member of species E. The Needleman Wunsch pairwise algorithm produced results identical to those obtained via phylogenetic analyses (Table 2) (see Table S2 in the supplemental material). Although both analyses permitted accurate HAdV classification, pairwise similarity analysis has the advantage of speed and simplicity.
FIG. 1.
Nucleotide sequences were aligned by using Clustal W. Phylogenetic analyses were performed using the Kimura two-parameter model as a model of nucleotide substitution and using the neighbor-joining method to reconstruct phylogenetic trees (MEGA version 2.1 software package). The statistical significance of the phylogenies constructed was estimated by bootstrap analysis with 1,000 pseudoreplicate data sets.
TABLE 2.
Sequence analysis of clinical samples and HAdV isolates
| Name | Speciesa | Typing by phylogeny
|
Typing by pairwise alignment
|
||
|---|---|---|---|---|---|
| Bootstrap value | Serotypeb | NW homology score (%)c | Serotype | ||
| Clinical samples | |||||
| G1066_01 | C | 79 | HAdV-2 | 98.5 | HAdV-2 |
| SO4010_04 | C | 79 | HAdV-2 | 100.0 | HAdV-2 |
| G1634_04 | B | 74 | HAdV-3 | 99.0 | HAdV-3 |
| R1625_04 | B | 74 | HAdV-3 | 99.0 | HAdV-3 |
| SO4425_04 | B | 74 | HAdV-3 | 99.0 | HAdV-3 |
| SO4429_04 | B | 74 | HAdV-3 | 99.0 | HAdV-3 |
| G1108_01 | E | 99 | HAdV-4 | 93.1 | HAdV-4 |
| 1292F_01 | E | 99 | HAdV-4 | 95.6 | HAdV-4 |
| 1803F_03 | E | 99 | HAdV-4 | 93.1 | HAdV-4 |
| SO3360_01 | C | 99 | HAdV-5 | 96.1 | HAdV-5 |
| SO3790_03 | C | 99 | HAdV-5 | 100.0 | HAdV-5 |
| SO4366_04 | C | 99 | HAdV-5 | 96.1 | HAdV-5 |
| 591F_01 | C | 99 | HAdV-5 | 99.5 | HAdV-5 |
| 992F_01 | C | 99 | HAdV-5 | 99.5 | HAdV-5 |
| 130C_99 | C | 99 | HAdV-5 | 99.5 | HAdV-5 |
| 2549F_03 | C | 99 | HAdV-5 | 99.0 | HAdV-5 |
| 1804F_03 | C | 99 | HAdV-5 | 98.5 | HAdV-5 |
| G1507_04 | C | 59 | HAdV-6 | 99.0 | HAdV-6 |
| SO4408_04 | C | 59 | HAdV-6 | 99.5 | HAdV-6 |
| SO4427_04 | C | 59 | HAdV-6 | 99.5 | HAdV-6 |
| G1508_04 | C | 59 | HAdV-6 | 99.0 | HAdV-6 |
| 593F_01 | C | 59 | HAdV-6 | 98.5 | HAdV-6 |
| 1370l_03 | C | 59 | HAdV-6 | 99.5 | HAdV-6 |
| 411l_04 | C | 59 | HAdV-6 | 99.5 | HAdV-6 |
| D0016_01 | C | 59 | HAdV-6 | 99.5 | HAdV-6 |
| SO4363_04 | C | 59 | HAdV-6 | 99.1 | HAdV-6 |
| SO4405_04 | B | 98 | HAdV-7 genotype 2 | 99.5 | HAdV-7 genotype 2 |
| G1253_02 | B | 98 | HAdV-7 genotype 2 | 99.5 | HAdV-7 genotype 2 |
| 1184X_93 | B | 98 | HAdV-7 genotype 2 | 99.5 | HAdV-7 genotype 2 |
| 824O_02 | B | 98 | HAdV-7 genotype 2 | 99.0 | HAdV-7 genotype 2 |
| R1629_04 | B | 61 | HAdV-11, 34 or 35 | 98.2* | HADV-11, 34 or 35 |
| R1612_04 | B | 61 | HAdV-11, 34 or 35 | 97.8* | HADV-11, 34 or 35 |
| 1938l_89 | B | 61 | HAdV-11, 34 or 35 | 99.2* | HADV-11, 34 or 35 |
| 226l_03 | B | 71 | HAdV-14 | 96.6 | HAdV-14 |
| D0002_01 | F | 99 | HAdV-40 | 100.0 | HAdV-40 |
| D0004_01 | F | 99 | HAdV-40 | 98.5 | HAdV-40 |
| D0017_01 | F | 99 | HAdV-40 | 100.0 | HAdV-40 |
| D0015_01 | F | 99 | HAdV-40 | 100.0 | HAdV-40 |
| D0005_01 | F | 99 | HAdV-41 | 97.0 | HAdV-41 |
| D0007_01 | F | 99 | HAdV-41 | 98.0 | HAdV-41 |
| D0001_01 | F | 99 | HAdV-41 | 97.0 | HAdV-41 |
| D0011_01 | F | 99 | HAdV-41 | 98.0 | HAdV-41 |
| 928O_04 | D | 94 | NIB; Species D (not HAdV-8) | 95.5* | Species D (not HAdV-8) |
| 926O_04 | D | 94 | NIB; Species D (not HAdV-8) | 95.6* | Species D (not HAdV-8) |
| 925O_04 | D | 94 | NIB; Species D (not HAdV-8) | 95.6* | Species D (not HAdV-8) |
| SO4390_04 | D | 94 | NIB; Species D (not HAdV-8) | 95.0* | Species D (not HAdV-8) |
| HAdV isolates | |||||
| C1519_02 | C | 95 | HAdV-1 | 99.0 | HAdV-1 |
| C1662_03 | C | 95 | HAdV-1 | 99.0 | HAdV-1 |
| C1491_02 | C | 95 | HAdV-1 | 99.5 | HAdV-1 |
| C1629_02 | B | 74 | HAdV-3 | 99.0 | HAdV-3 |
| 4030_96 | B | 74 | HAdV-3 | 99.0 | HAdV-3 |
| 859_96 | E | 99 | HAdV-4 | 92.2 | HAdV-4 |
| 860_96 | E | 99 | HAdV-4 | 89.4 | HAdV-4 |
| 519_93 | E | 99 | HAdV-4 | 90.7 | HAdV-4 |
| 856_96 | E | 99 | HAdV-4 | 92.7 | HAdV-4 |
| C1201_00 | C | 59 | HAdV-6 | 100.0 | HAdV-6 |
| G3093_03 | C | 59 | HAdV-6 | 99.0 | HAdV-6 |
| 615_96 | D | 99 | HAdV-8 | 100.0 | HAdV-8 |
| 636_96 | D | 99 | HAdV-8 | 100.0 | HAdV-8 |
| 647_96 | D | 99 | HAdV-8 | 100.0 | HAdV-8 |
| 841_94 | D | 99 | HAdV-8 | 100.0 | HAdV-8 |
| 43024_02 | D | 99 | HAdV-8 | 98.0 | HAdV-8 |
| C4292_02 | D | 99 | HAdV-8 | 98.5 | HAdV-8 |
| R1650_04 | A | 99 | HAdV-12 | 97.5 | HAdV-12 |
| R1641_04 | A | 99 | HAdV-12 | 97.5 | HAdV-12 |
| C5335_01 | B | 99 | HAdV-21 | 96.5 | HAdV-21 |
| R1647_04 | B | 71 | HAdV-16 | 99.5 | HAdV-16 |
| C1640_03 | A | 99 | HAdV-31 | 98.5 | HAdV-31 |
| G1T4_03 | D | 94 | NIB; species D (not HAdV-8) | 97.0* | Species D (not HAdV-8) |
| 594_89 | D | 94 | NIB; species D (not HAdV-8) | 96.1* | Species D (not HAdV-8) |
| C1167_00 | D | 94 | NIB; species D (not HAdV-8) | 96.6* | Species D (not HAdV-8) |
Species identified by bootstrap.
NIB, serotype not identified by bootstrap.
*, average score from all members of the group. Boldfacing indicates that the Needleman Wunsch (NW) score for the second-highest serotype was higher than 93%. Due to their close relationship, all cases of HAdV-2 and -6 present this situation. In all cases the highest score pointed to the correct serotype.
Using our method, we found indications of possible new associations between specific clinical syndromes and HAdV serotypes. In the past, given the complexity of adenovirus typing, association between adenovirus subtypes and disease were considered after isolation and characterization of a virus from the main affected organ (12). However, due to prolonged viral shedding after infection, as well as the relative persistence of DNA beyond which viable virus can be detected, more in-depth studies should be done nowadays to prove causation. Additionally, it would also be necessary to rule out other etiologies. In our study, other etiologies were ruled out using multiplex PCR techniques that are capable of diagnosing 14 different respiratory etiological agents (8). Typically, respiratory tract diseases are associated with species B1, C, and E, gastrointestinal diseases with species A and F, eye diseases with species D and E, and kidney and urinary tract diseases with species B2.
Acute respiratory diseases due to HAdV are attributed primarily to serotypes 3, 4, 7, 14, and 21 (species B and E) (12). We also observed serotypes 1, 2, 5, and 6 (species C) and species D. Pneumonia in children has been associated with serotypes 1 to 3 and 7, whereas pneumonia in adults is predominantly associated with serotypes 4 and 7. We also found serotypes 5 and 6 in young children and subgroup B2 in immunosuppressed patients.
Gastrointestinal manifestations of HAdV infection include diarrhea and hepatitis. In addition to serotypes 40 and 41 (2, 12), we identified cases of diarrhea associated with serotypes 6 (immunocompetent patient); 12, 16, and 31 and a member of species D (bone marrow transplants or human immunodeficiency virus-positive case). The significance of these findings is not clear, since members of species C may be excreted in feces during subclinical infection. HAdV hepatitis has been reported in child recipients of liver transplants associated with serotypes 1, 2, and 5 (species C); in this study we detected the remaining member of species C, serotype 6, in a case of fatal hepatitis.
Epidemic keratoconjunctivitis has been associated with serotypes 8, 19, and 37 (species D) and serotype 11 (species B2). Our sequences revealed not only serotype 8 (species D) but also serotypes 4 and 7 (species E and B1, respectively). Adenovirus 4 (species E) was recently discovered as the cause of either respiratory or mild ocular infections (21, 12) and nosocomial epidemic conjunctivitis in Japan (3).
Cases of acute hemorrhagic cystitis in young children have been associated with species B2 serotypes 11 and 21. In our study, this disease is associated with serotypes 16 and 14 (species B1 and B2), and an indistinguishable member of the cluster 11, 34, and 35 (species B2). Fatal infections due to species B1, serotypes 3 and 7, have been reported (11, 12, 15). The fact that subgroups B1 and B2 use different cellular receptors for viral entry underscores the importance of typing HAdV for epidemiology and pathobiology (20).
Finally, we found serotypes 4 and 5 (species E and C) in throat swabs of four patients with fever, morbilliform rash, Koplik's spots, and cough who had a history of measles, mumps, and rubella vaccination and were negative for measles and rubella. To our knowledge this is the first report of HAdV presenting as a syndrome compatible with measles infection. Case numbers are not sufficient to establish causality between specific serotypes and specific syndromes. Further independent testing is needed to verify these associations.
Using the database and classification system from this study, we have deployed a website (www.greeneidlab.columbia.edu) wherein clinical laboratories can submit hexon sequences to generate an automatic report detailing the serotype, date, and location of the most similar sequence isolate in the database. This system will allow new genotypes to be readily identified, because the classification scheme will fail to relate them to any described serotype. Epidemiological surveillance of HAdV serotypes will improve our understanding of the global burden of HAdV infection. High-throughput systems described here will facilitate HAdV surveillance and enhance understanding of HAdV pathogenesis.
Nucleotide sequence accession numbers.
The sequences determined in this work were deposited in the GenBank sequence database under accession numbers AY819809 to AY819926.
.
Supplementary Material
Acknowledgments
We acknowledge technical assistance from Manuela López-Valero in the Respiratory Viruses Laboratory, CNM, ISCIII, Majadahonda, Madrid, Spain. We thank Neil Renwick for his critical reviewing of the manuscript and helpful comments. We also thank the reviewers for their constructive criticism and helpful suggestions on the initial draft.
This work was supported by grants from the National Institutes of Health (AI51292, AI55466, AI056118, and U54AI057158-Northeast Biodefense Center), the Ellison Medical Foundation, and ISCIII (MPY 1251/00 and MPY 1018/04).
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Allard, A., B. Albinsson, and G. Wadell.2001. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 39:498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allard, A., A. Kajon, and G. Wadell.1994. Simple procedure for discrimination and typing of enteric adenoviruses after detection by polymerase chain reaction. J. Med. Virol. 44:250-257. [DOI] [PubMed] [Google Scholar]
- 3.Ariga, T., Y. Shimada, K. Ohgami, Y. Tagawa, H. Ishiko, K. Aoki, and S. Ohno.2004. New genome type of adenovirus serotype 4 caused nosocomial infections associated with epidemic conjunctivitis in Japan. J. Clin. Microbiol. 42:3644-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avellon, A., P. Perez, J. C. Aguilar, R. Lejarazu, and J. E. Echevarria.2001. Rapid and sensitive diagnosis of human adenovirus infections by a generic polymerase chain reaction. J. Virol. Methods 92:113-120. [DOI] [PubMed] [Google Scholar]
- 5.Benko, M., B. Harrach, and W. C. Russell.2000. Adenoviridae, p. 227-237. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 6.Briese, T., G. Palacios, M. Kokoris, O. Jabado, Z. Liu, N. Renwick, V. Kapoor, I. Casas, F. Pozo, R. Limberger, P. Perez-Brena, J. Ju, and W. I. Lipkin.2005. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg. Infect. Dis. 11:310-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casas, I., L. Powell, P. E. Klapper, and G. M. Cleator.1995. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods 53:25-36. [DOI] [PubMed] [Google Scholar]
- 8.Coiras, M. T., J. C. Aguilar, M. L. Garcia, I. Casas, and P. Perez-Brena.2004. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J. Med. Virol. 72:484-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford-Miksza, L., and D. P. Schnurr.1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elnifro, E. M., R. J. Cooper, P. E. Klapper, and A. S. Bailey.2000. PCR and restriction endonuclease analysis for rapid identification of human adenovirus subgenera. J. Clin. Microbiol. 38:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hierholzer, J. C., R. Wigand, L. J. Anderson, T. Adrian, and J. W. Gold.1988. Adenoviruses from patients with AIDS: a plethora of serotypes and a description of five new serotypes of subgenus D (types 43-47). J. Infect. Dis. 158:804-813. [DOI] [PubMed] [Google Scholar]
- 12.Horwitz, M. S.2001. Adenoviruses, p. 2301-2326. In B. N. Fields, D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 13.Kidd, A. H., M. Jonsson, D. Garwicz, A. E. Kajon, A. G. Wermenbol, M. W. Verweij, and J. C. De Jong.1996. Rapid subgenus identification of human adenovirus isolates by a general PCR. J. Clin. Microbiol. 34:622-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Q. G., A. Henningsson, P. Juto, F. Elgh, and G. Wadell.1999. Use of restriction fragment analysis and sequencing of a serotype-specific region to type adenovirus isolates. J. Clin. Microbiol. 37:844-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mistchenko, A. S., J. F. Robaldo, F. C. Rosman, E. R. Koch, and A. E. Kajon.1998. Fatal adenovirus infection associated with new genome type. J. Med. Virol. 54:233-236. [DOI] [PubMed] [Google Scholar]
- 16.Morfin, F., S. Dupuis-Girod, S. Mundweiler, D. Falcon, D. Carrington, P. Sedlacek, M. Bierings, P. Cetkovsky, A. C. Kroes, M. J. van Tol, and D. Thouvenot.2005. In vitro susceptibility of adenovirus to antiviral drugs is species-dependent. Antivir. Ther. 10:225-229. [PubMed] [Google Scholar]
- 17.Needleman, S. B., and C. D. Wunsch.1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 18.Rice, P., I. Longden, and A. Bleasby.2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 19.Sarantis, H., G. Johnson, M. Brown, M. Petric, and R. Tellier.2004. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J. Clin. Microbiol. 42:3963-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segerman, A., N. Arnberg, A. Erikson, K. Lindman, and G. Wadell.2003. There are two different species B adenovirus receptors: sBAR, common to species B1 and B2 adenoviruses, and sB2AR, exclusively used by species B2 adenoviruses. J. Virol. 77:1157-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wadell, G.1984. Molecular epidemiology of human adenoviruses. Curr. Top. Microbiol. Immunol. 110:191-220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.