Abstract
The Mycobacterium tuberculosis genome contains four phospholipase C (PLC)-encoding genes, designated plcA, plcB, plcC, and plcD, respectively. Each of the four genes contributes to the overall PLC activity of M. tuberculosis. PLC is hypothesized to contribute to M. tuberculosis virulence. Infection of M. tuberculosis strains carrying a truncated plcD gene is associated with the occurrence of extrathoracic tuberculosis. However, whether the other three plc genes are also associated with extrathoracic tuberculosis remains to be assessed. We investigated the insertion- and deletion-associated genetic diversity in all four plc genes among 682 epidemiologically and clinically well-characterized M. tuberculosis clinical isolates using PCR, DNA sequencing, and Southern hybridization. Two hundred sixty-six (39%) of the 682 isolates had an interruption in at least one of the four plc genes, most often associated with an IS6110 insertion. The plcD gene interruption was the most common: it was observed in 233 (34%) of the isolates, compared to 4.7%, 4.1%, and 5.9% for plcA, plcB, and plcC gene interruption, respectively. The association between the plc gene genotypes and disease presentation was adjusted for clustering using generalized estimating equations for both bivariate and multivariate analyses. After controlling for the genotypes of the plcABC genes and the host-related risk factors, interruption in the plcD gene remained significantly associated with extrathoracic tuberculosis (odds ratio, 3.27; 95% confidence interval, 1.32 to 8.14). The data suggest that the plcD gene might play a more important role in the pathogenesis of thoracic TB than it does in the pathogenesis of extrathoracic TB.
Phospholipase C (PLC) is involved in the pathogenesis of several bacterial infections (11, 16, 19, 20). For Mycobacterium leprae, Mycobacterium microti, and Mycobacterium avium, high phospholipase activity was observed among bacilli harvested from host tissues (28, 29), suggesting that phospholipase might be involved in the virulence of mycobacteria. However, the precise role of PLC in tuberculosis (TB) pathogenesis is unknown. Genome sequencing of Mycobacterium tuberculosis laboratory strain H37Rv (5) and clinical strain CDC1551 (http://www.tigr.org) revealed four genes encoding PLC enzymes. Three of these genes, plcA, plcB, and plcC, are organized in tandem (locus plcABC). The fourth gene, plcD, is located in a separate region. A recent functional study of M. tuberculosis demonstrated that all four genes encode functional PLC in M. tuberculosis, and each gene contributes to overall PLC activity (13). Furthermore, in mice, when the virulence of a plcABCD quadruple mutant was compared to a plcABC triple mutant, both showed the same level of attenuation, suggesting that either plcD does not contribute to virulence or it acts in association with other phospholipase-encoding genes.
Several previous studies using small, selected samples of M. tuberculosis clinical isolates have found insertions or deletions in the plcA, plcB, plcC, and plcD genes (9, 18, 21, 23-25). Recently, we reported the association between insertion-and deletion-associated mutations in the plcD genotype and extrathoracic TB (31); however, the potential association of mutations in the other three plc genes remains to be investigated. In order to gain a better understanding of the relative contribution of the four plc genes to the pathogenesis of human TB, we investigated the genetic polymorphisms of all four plc genes and compared the distribution of their genotypes among study subjects with different clinical presentations using a case control study design.
MATERIALS AND METHODS
Study sample and bacterial isolates.
We included 682 culture-confirmed M. tuberculosis isolates from TB patients diagnosed in Arkansas between 1 January 1996 and 31 December 2000. During the study period, a total of 973 TB cases were diagnosed in Arkansas; 719 cases were culture confirmed. Of these 719 cases, 682 (95%) isolates were available for the current study. This set included 355 unique and 327 clustered isolates belonging to 92 clusters defined by a combination of IS6110 restriction fragment length polymorphism (RFLP) and pTBN12 fingerprinting (4, 22).
Patient data, including demographics, social behaviors, and clinical characteristics, were obtained from the surveillance records of Arkansas Department of Health. Of the 682 study patients, 604 had TB disease confined within the thorax, 46 had TB outside of the thorax, and 32 had concurrent thoracic and extrathoracic involvement. Among the 32 patients that had both thoracic and extrathoracic TB, five isolates were cultured from the thoracic sites, and the remaining 27 isolates were from the various extrathoracic sites. Of the 636 patients who had thoracic involvement, 584 had a chest radiograph report available for review, and 586 had a report of a stained sputum smear. Based on chest radiograph, 216 (36.99%) had cavitation in the lungs, and the remaining 368 (63.01%) did not. Of the 586 cases with sputum smear results, 251 (42.83%) were smear positive.
Genomic DNA of the patients' isolates was extracted from Lowenstein-Jensen slant cultures using standard procedures (12). The study protocols and procedures for the protection of human subjects were approved by the Health Sciences Institutional Review Boards of the University of Michigan and the University of Arkansas for Medical Sciences.
PCR assays of the plc genes.
DNA polymorphisms in the four plc gene regions were investigated first by PCR assays using primers shown in Fig. 1. The plcD gene region was investigated with three PCR assays designated plcD-PCR1, plcD-PCR2, and plcD-PCR3, respectively. The plcA, plcB, and plcC gene regions were examined by PCR assays designated plcA-PCR, plcB-PCR, and plcC-PCR, respectively. The BD Advantage 2 PCR kit (BD-Biosciences Clontech, Palo Alto, CA) was used in all the PCRs except for plcD-PCR1 and plcD-PCR2. In plcD-PCR1, the BD Advantage GC-2 kit was used, while in plcD-PCR2, the MasterAmp Extra-Long PCR kit (Epicentre Biotechnologies, Madison, WI) was used to obtain a large PCR product for the positive reference strain (19.8 kb) and the wild-type isolates. The 16S rRNA gene PCR was performed to confirm the quantity and quality of the DNA templates (3). The plcA-PCR, plcB-PCR, plcC-PCR, and plcD-PCR1 used the same thermocycling parameters, i.e., 1 cycle at 94°C for 1 min followed by 26 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 2.5 min and a final cycle at 72°C for 10 min. The thermocycling parameters for plcD-PCR2 were as follows: 1 cycle at 94°C for 3 min followed by 31 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 14.5 min and a final cycle at 72°C for 10 min. For plcD-PCR3, the thermocycling parameters were as follows: 1 cycle at 94°C for 1 min followed by 28 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min and 10 s and a final cycle at 72°C for 10 min.
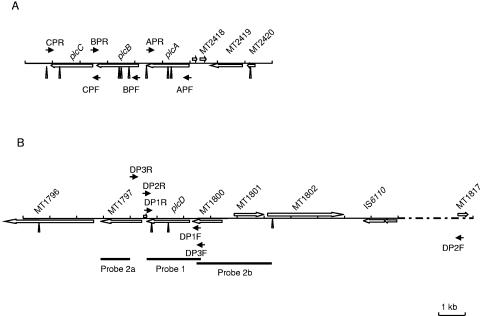
FIG. 1.
Schematic representation of the plcABC and plcD loci and the locations of the PCR primers (small arrows), PvuII restriction sites (triangles), and Southern hybridization probes (small black bars). PvuII restriction sites were obtained from the sequence of M. tuberculosis CDC1551 in GenBank. (A) plcCBA gene map of CDC1551 (GenBank accession no. NC_002755). (B) plcD gene map of CDC1551 (accession no. NC_002755). Genes between IS6110 and MT1817 are omitted due to space limit. Primers and sequences are as follows: CPF, 5′-AAGCCGAAATACACGAGGGAGAGC-3′; CPR, 5′-GGGCGGCAAAGGCGGACCAAGAG-3′; BPF, 5′-CGGCAGGCAGGCGGAATCAGAACA-3′; BPR, 5′-TCCGGCGAATGCACCTTGGCTCAC-3′; APF, 5′-GCAGGAAGGCAGGGCAAGTG-3′; APR, 5′-TCGAACGCCGGGAGATTACC-3′; DP1F, 5′-CGCGCGCCGCCGCGCCGAAATA-3′; DP1R, 5′-TCGCCCGGACAGGTCAACAAGGTG-3′; DP2F, 5′-CGCTGGTACACCTGGGGAATCTGGTGACGTAGA-3′; DP2R, 5′-GTCCACTGTCGCCCGGACAGGTCAACAAGGTGT-3′; DP3F, 5′-ACACGAGACATTTGGGCTAG-3′; DP3R, 5′-CAATGCGGATATCAGTGGAC-3′.
For the plcA, plcB, and plcC genes, M. tuberculosis clinical strain CDC1551 was used as a positive control in the PCR assays, and the BCG Australian strain was used as a negative control (1). For the plcD gene, genomic DNA from clinical strain CDC1551 was used as a positive control, and H37Rv was used as a negative control, because the plcD gene region is truncated in strain H37Rv (5) and is intact in strain CDC1551 (http://www.tigr.org).
Southern blot and hybridization.
For isolates that had consistently negative PCR results, Southern blot hybridization was conducted to confirm the potential partial or complete absence of the plc genes. The restriction endonuclease PvuII was used to digest the chromosomal DNA. Because the plcA, plcB, plcC, and plcD genes share about 70% sequence homology, the purified plcD-PCR1 product of CDC1551, named probe 1, was used as a probe to detect the presence or absence of all four plc genes. For plcD detection, two additional probes, named probes 2a and 2b, complement to the flanking regions of the plcD gene, were used after probe 1 to confirm the presence or absence of the 3.9-kb and 4.6-kb restriction fragments of plcD (Fig. 1). One microgram of DNA of each isolate was digested using PvuII and subjected to electrophoresis in 1% (wt/vol) agarose gel in 1× Tris-borate-EDTA (pH 8.0) buffer. The DNA was blotted onto nylon membranes (Hybond-N+; Amersham Biosciences, Piscataway, NJ) by using a Vacuum Blotter (model 785; Bio-Rad Laboratories, Hercules, CA) and hybridized with probes that were covalently labeled with chemiluminescence and then detected using the Gene Images AlkPhos Direct Labeling and Detection system (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instructions, except that the stringency temperature was changed from the recommended 55°C to 60°C.
DNA sequencing.
For the isolates, the PCR products of which showed an altered size compared with that of the positive control, automated DNA sequencing was performed at the Sequencing Core at the University of Michigan to confirm the insertion and deletion events in the genes examined using the PCR products purified with a QIAquick PCR purification kit (QIAGEN, Valencia, CA). Primers used for the PCR assays were used for DNA sequencing. Sequence comparisons were performed using Edit Seq 5.02 and MegAlign 5.01 software (DNAStar Inc., Madison, WI).
Definitions.
For this study, a mutant type was defined only when an isolate had a large sequence variation consisting of an IS6110 insertion and/or deletion in one of the plc gene regions; isolates that generated a PCR product of the same size as that of the positive control strain were classified as the wild type. Isolates that had a partial plcD gene deletion and deletion of adjacent genes were classified separately as plcD and adjacent gene mutant group and were treated as a separate group in the data analyses. Since plcA, plcB, and plcC are not cotranscribed (8, 13) as a polycistron even though they are organized as a cluster in the genome of M. tuberculosis, the plcA, plcB, and plcC mutant types were defined solely based on the interruption of the individual genes.
To study the association between the insertion- and deletion-associated interruption in the plc genes and clinical TB phenotypes, we classified the study patients into thoracic TB and extrathoracic TB groups. Patients whose disease sites were confined to lungs, pleura, and intrathoracic lymph nodes were classified into the thoracic TB group; patients who had extrathoracic involvement with or without concurrent diseases within the thorax were considered to have extrathoracic involvement. For those who had thoracic involvement, we grouped them into cavitary and noncavitary TB groups and sputum smear-positive and -negative groups.
Statistical analysis.
We assessed the associations between the four plc gene genotypes and the clinical characteristics of the disease, including extrathoracic TB involvement, cavitation in the lungs, and positive sputum smear. The genotypes of the plcD gene and plcABC genes were treated as two individual variables in the analysis. The functional effect of a mutation in the plcA, plcB, or plcC gene was considered to be the same as that previously shown by Raynaud et al. (13). Considering the likelihood that mutations observed among cases in a cluster defined by IS6110 RFLP and the pTBN12 pattern (4, 22) would not be independent, we used generalized estimating equations (GEE) to control for intracluster dependence (10, 32) in both bivariate and multivariate analyses. The magnitude of the associations was estimated using the odds ratio (OR) and 95% confidence intervals (95% CI). Multivariate analyses using the logistic regression GEE model were conducted to control for potential confounding by the genotypes of plcABC and host-related factors. All the statistical analyses were done using SAS version 8.0 (SAS Institute, Cary, NC).
RESULTS
Polymorphisms of plc genes.
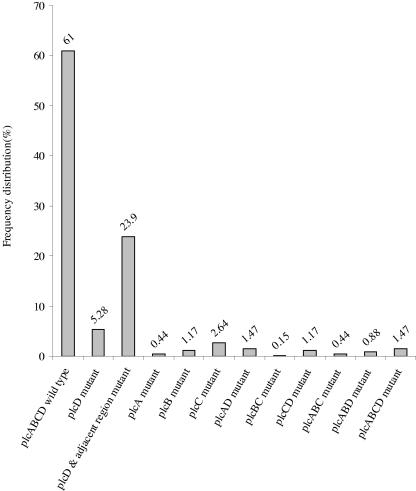
We observed an interruption in at least one of the four plc genes in 266 (39%) out of 682 M. tuberculosis isolates investigated. Among the 266 isolates with a mutation, 228 (33.43%) had a mutation in only one of the four plc genes, 19 (2.78%) had a mutation in two of the plc genes, 9 (1.32%) had a mutation in three plc genes, and 10 (1.47%) had mutations in all four plc genes. Most of the mutations occurred in the plcD gene region (233/266 [88%]). Among the 32, 28, and 40 isolates that had a mutation in plcA, plcB, and plcC, respectively, 26 (81.25%), 16 (57.14%), and 18 (45%) isolates also had a mutation in plcD. Of the 233 plcD mutants, 10 (4.29%) also had a mutation in the plcA gene, 16 (6.87%) had a mutation in the plcB gene, and 18 (7.72%) had a mutation in the plcC gene. The profiles of plc gene genotypes, in terms of the frequency distribution of different combinations of mutations, are shown in Fig. 2.
FIG. 2.
Frequency distribution of combined plc gene genotypes among 682 isolates. x axis, profiles of plcABCD genotypes; y axis, frequency distribution.
IS6110 insertion.
When PCR products showed a different size in comparison with the positive control, the PCR products were sequenced; 13, 9, 27, and 144 products were sequenced for plcA, plcB, plcC, and plcD, respectively. Twelve of the 13 isolates sequenced for plcA had an IS6110 insertion within the gene in the same orientation as that of plcA transcription; the remaining isolate had an IS6110 in the reverse direction. Of the nine isolates sequenced for plcB, seven had an IS6110 insertion within the gene in the direction of gene transcription, while the other two isolates had an IS6110 insertion within the gene in the reverse direction. Of the 27 isolates sequenced for the plcC mutation, 16 had an IS6110 insertion within the gene in the direction of plcC gene transcription, 10 had an IS6110 insertion within the gene in the reverse direction, and 1 isolate had a 115-bp deletion in plcC without an IS6110 insertion. Of the 144 isolates sequenced for plcD, 17 had an IS6110 insertion in the direction of plcD gene transcription, 14 had an IS6110 insertion in the reverse direction, 7 isolates had an IS6110 insertion in the reverse direction followed by a partial plcD deletion, and 106 isolates had an IS6110 insertion and a deletion of partial plcD and adjacent genes. Of these 106 isolates, 70 had an IS6110 insertion within the gene in the same direction as the orientation of gene transcription, and 36 had an IS6110 insertion within the gene in the reverse direction.
The numbers of IS6110 insertion sites found in the plcA, plcB, plcC, and plcD genes were 6, 5, 16, and 27, respectively. The most common sites of IS6110 insertion in plcA, plcB, plcC, and plcD are shown in Table 1. When IS6110 was inserted without a partial deletion of plc genes and the adjacent region, a 3- to 4-bp sequence duplication at the site of the insertion was observed; otherwise, this 3- to 4-bp duplication was not seen.
TABLE 1.
Numbers and most common locations of IS6110 insertion sites found in four plc genes of M. tuberculosis clinical strains collected from Arkansas between 1 January 1996 and 31 December 2000
| Gene (no.) | No. of IS6110 insertion loci | Sequence of the most common insertion sitea | Position of the most common insertion siteb | Frequency (%)c |
|---|---|---|---|---|
| plcD (n = 144) | 27 | GGTCACACTG↓CGCCGCCAAG | 1978100 | 29/122 (23.77) |
| plcA (n = 13) | 6 | CAGCTTCGGC↓AGCGCTCCCA | 2626532 | 4/11 (36.36) |
| plcB (n = 9) | 5 | ACCGTCGCGT↓CCCGCCAAGC | 2624861 | 5/9 (55.56) |
| plcC (n = 27) | 16 | GGACCGGGTG↓GTCCAGATTG | 2623172 | 3/21 (14.28) |
A 3- to 4-bp duplication at the site of the IS6110 insertion is shown with underlining. This duplication happened when IS6110 was inserted without a partial deletion of the plc genes and adjacent regions.
The genome sequence of CDC1551 was used as a reference.
The number of nominators and denominators was counted by strain, which was defined by RFLP and pTBN12 genotyping.
Association of plc gene polymorphism with clinical presentation.
The associations between the plc gene polymorphisms and three clinical characteristics of the disease, including extrathoracic TB involvement, cavitation in the lungs, and positive sputum smear, were explored using GEE. In bivariate analysis using GEE, we did not find any statistically significant association between any of the plc gene genotype profiles and cavitation in the lungs or a positive sputum smear. There was, however, a statistically significant association between plcD gene mutation and extrathoracic TB involvement (Table 2). After controlling for mutations in the plcABC genes in a GEE logistic regression model, the association between plcD gene mutation and extrathoracic TB involvement remained almost the same (OR of 2.87 and 95% CI of 1.33 and 6.16 after adjustment versus OR of 2.86 and 95% CI of 1.32 and 6.19 prior to adjustment). When the three previously identified host-related risk factors for extrathoracic TB (human immunodeficiency virus [HIV] seropositive, black ethnicity, and female) (30) along with the genotypes of plcABC genes were included in the GEE logistic regression model, interruption in the plcD gene remained significantly associated with extrathoracic TB (Table 3).
TABLE 2.
Bivariate analysis of the association between plc gene genotypes and clinical characteristics using GEE
| plc gene genotype | Clinical characteristic
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Extrathoracic TB involvement (n = 682)
|
Cavitation in the lungs (n = 584)
|
Sputum smear positive (n = 586)
|
|||||||
| No. with TTBa | No. with ETTBb | OR (95% CI) | No | Yes | OR (95% CI) | No. negative | No. positive | OR (95% CI) | |
| plcD genotypes | |||||||||
| plcD wild type | 405 | 44 | 1.00 (referent) | 243 | 144 | 1.00 (referent) | 228 | 162 | 1.00 (referent) |
| plcD mutant | 29 | 9 | 2.86 (1.32-6.19) | 21 | 11 | 0.88 (0.41-1.89) | 22 | 10 | 0.64 (0.30-1.39) |
| plcD and adjacent gene, mutant group | 170 | 25 | 1.35 (0.82-2.23) | 104 | 61 | 0.99 (0.68-1.44) | 85 | 79 | 1.31 (0.91-1.89) |
| plcABC genotypes | |||||||||
| plcABC wild type | 546 | 69 | 1.00 (referent) | 326 | 197 | 1.00 (referent) | 298 | 227 | 1.00 (referent) |
| One of plcABC mutant | 40 | 7 | 1.38 (0.58-3.30) | 28 | 13 | 0.77 (0.40-1.48) | 24 | 17 | 0.93 (0.50-1.74) |
| Two of plcABC mutant | 6 | 1 | 1.32 (0.15-1.68) | 5 | 2 | 0.66 (0.16-2.67) | 5 | 2 | 0.52 (0.13-2.13) |
| plcABC all mutant | 12 | 1 | 0.66 (0.08-5.30) | 9 | 4 | 0.74 (0.27-2.00) | 8 | 5 | 0.82 (0.25-2.66) |
TTB, thoracic tuberculosis.
ETTB, extrathoracic tuberculosis.
TABLE 3.
Multivariate analysis of the association between plcD interruption and extrathoracic TB after adjusting for the genotypes of plcABC genes and the three host-related risk factors using GEEa
| Variable in the model | Adjusted OR | 95% CI |
|---|---|---|
| plcD genotypes | ||
| plcD wild type | 1.00 | |
| plcD mutant | 3.27 | 1.32-8.14 |
| plcD and adjacent gene mutant group | 1.25 | 0.73-2.14 |
| plcABC genotypes | ||
| plcABC wild type | 1.00 | |
| One of plcABC mutant | 0.93 | 0.34-2.53 |
| Two of plcABC mutant | 1.15 | 0.08-16.01 |
| plcABC all mutant | 0.77 | 0.08-7.27 |
| HIV serological test | ||
| HIV seronegative | 1.00 | |
| HIV seropositive | 5.22 | 1.78-15.23 |
| Unknown | 1.05 | 0.61-1.80 |
| Race/ethnicity | ||
| Non-Hispanic white | 1.00 | |
| Non-Hispanic black | 2.39 | 1.31-4.38 |
| Other | 2.16 | 0.92-5.08 |
| Gender | ||
| Male | 1.00 | |
| Female | 2.46 | 1.52-4.00 |
Adjusted OR is adjusted for mutations in other plc genes and the three host-related factors using logistic regression GEE (n = 682).
DISCUSSION
To gain a better understanding of the potential contribution of the plc genes of M. tuberculosis to human TB, we extended our previous population-based study of plc gene genetic diversity and its clinical association from focusing only on the plcD gene (31) to focusing on all four plc genes and increasing the number of isolates/cases from 496 to 682. The present study confirmed our previous finding of a significant association between the plcD gene mutation and the occurrence of extrathoracic TB after controlling for host-related risk factors and the genotypes of the plcABC genes. It has provided a more complete assessment of the relative contribution of all four M. tuberculosis plc genes to the pathogenesis of human TB. The data suggest that the plcD gene might play a more important role in the pathogenesis of thoracic TB than it does in the pathogenesis of extrathoracic TB; in contrast, the other three plc genes may have an equally important role in the pathogenesis of both thoracic and extrathoracic TB.
M. tuberculosis clinical strains with major polymorphisms in all four plc genes and impaired plc gene expression have been reported previously by others (25). Raynaud et al. recently demonstrated that plcABC triple mutants and plcABCD quadruple mutants were attenuated for growth in the lungs and spleens of mice, and the expression of the different plc genes was important for virulence at different time points of the infection (13). In our study, 10 isolates from 10 patients with only thoracic TB had a plcABCD quadruple mutant. These 10 isolates were defined as eight different strains by IS6110 and pTBN12 fingerprinting. Among these 10 patients, 3 had cavities in the lung, and 5 were sputum smear positive. All 10 patients were HIV seronegative. Thus, it is unclear whether or not the PLC function is necessary for the bacilli to cause thoracic TB in all hosts. This also suggests that PLC may not be the only enzyme that is needed by M. tuberculosis to establish pathology in the lung.
PLC expressed within host cells might serve several functions related to virulence. First, these enzymes might provide the bacterium with nutrients by releasing fatty acids from host phospholipids, which might be a major energy source for M. tuberculosis in chronically infected lung tissue (15, 27). A second possible role for PLC would be to degrade the phagosome/endosome membrane to change its permeability or to interrupt its function (28, 29). Finally, PLC can hydrolyze membrane phosphatidylinositol of macrophages to release free arachidonic acid. Arachidonic acid metabolites can mediate inflammation by stimulating macrophage aggregation and granulocytic chemotaxis, thus activating the host immune responses (11, 20). Given these possible functions, we hypothesize that an alteration of the plcD gene will decrease PLC activity and impair the ability of tubercle bacilli to degrade the phagosome/endosome membrane, causing persistence of M. tuberculosis within the macrophage and allowing the microbe to travel to distant organs via hematogenous spread. An alternative hypothesis is that interruption of the plcD gene reduces the release of arachidonic acid from the macrophage membrane, thereby reducing the migration of monocyte-derived macrophages and T and B cells to the initial lung infection, thus increasing the chance of the bacillary spread to other organs. To explore the role of plc genes in the pathogenesis of extrathoracic TB, studies could be conducted by infecting animals with M. tuberculosis strains having different profiles of plc gene genotypes to compare the risk and extent of extrathoracic TB among animals infected with various genotypes.
Previous investigations of the genetic diversity of M. tuberculosis plc genes used either convenience samples or selected samples (7, 18, 21, 25). In our population-based sample, we observed that the plcD mutation accounted for 88% (233/266) of the mutations found in the plc genes; this is consistent with the previous findings of Talarico and colleagues, who used a convenience sample collected from Turkey (18). In the current study, almost all the deletion and insertion events in the plc genes were IS6110 related. The existence of numerous preferential insertion sites of IS6110 has been demonstrated previously (2, 6, 14, 26), and phospholipase C genes of M. tuberculosis have been reported to be preferential loci for IS6110 transposition (23). Our results suggest that within the plc genes, some sites for IS6110 insertion are much more common than others.
There is no statistically significant association between simultaneous mutation in plcD and adjacent genes and extrathoracic TB involvement. Some of the isolates in our study were classified as having simultaneous mutations in the plcD gene and adjacent genes by Southern hybridization, but the mutations in the adjacent genes were not accurately characterized. The potential effect of mutations of the adjacent genes on the plcD gene mutation that favors development of extrathoracic TB remains to be assessed after the genetic profiles of the genes adjacent to plcD are better characterized in future studies.
One limitation of this study is that the method we used is restricted in its capacity to identify small sequence variations, such as single-nucleotide polymorphisms, and such small variations might also have an influence on gene expression or gene function. Thus, it is possible that some isolates with point mutations or small deletions were misclassified as wild-type isolates. However, this type of nondifferential misclassification would tend to bias the result toward that of no association between the plc gene genotypes and extrathoracic TB (17). If the point mutation or small insertions or deletions in the plc genes were counted in defining the mutation in plc genes, the observed associations would have been enhanced.
Acknowledgments
We are indebted to Annadell H. Fowler, Bill Starrett, and Deborah Witonski for their valuable efforts that contributed to patient data and M. tuberculosis isolate collection during the study. We acknowledge Kashef Ijaz's contribution to the establishment of the Arkansas Department of Health's surveillance database that was used for the study and Sarah E. Talarico and Peter J Boldenow for help with the plcABC genotype data collection. We also thank Jack T. Crawford and Laura S. Cowan at the Centers for Disease Control and Prevention, Atlanta, Ga., for providing the DNA preparation of CDC1551.
Financial support was provided by the National Institutes of Health (grant number NIH-R01-AI151975).
REFERENCES
- 1.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin, W. H., Jr., K. H. Lok, R. Harris, N. Brook, L. Bond, D. Mulcahy, N. Robinson, V. Pruitt, D. P. Kirkpatrick, M. E. Kimerling, and N. E. Dunlap. 2001. Identification of a contaminating Mycobacterium tuberculosis strain with a transposition of an IS6110 insertion element resulting in an altered spoligotype. J. Clin. Microbiol. 39:1092-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boddinghaus, B., T. Rogall, T. Flohr, H. Blocker, and E. C. Bottger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaves, F., Z. Yang, H. el Hajj, M. Alonso, W. J. Burman, K. D. Eisenach, F. Dronda, J. H. Bates, and M. D. Cave. 1996. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J. Clin. Microbiol. 34:1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, S., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. Quail, M. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 6.Fang, Z. G., and K. J. Forbes. 1997. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J. Clin. Microbiol. 35:479-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 8.Johansen, K. A., R. E. Gill, and M. L. Vasil. 1996. Biochemical and molecular analysis of phospholipase C and phospholipase D activity in mycobacteria. Infect. Immun. 64:3259-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang, K. Y., and S. Zeger. 1986. Longitudinal data analysis using generalized linear models. Biometrika 73:13-22. [Google Scholar]
- 11.Meyers, D. J., and R. S. Berk. 1990. Characterization of phospholipase C from Pseudomonas aeruginosa as a potent inflammatory agent. Infect. Immun. 58:659-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray, M. G., and W. F. Thompson. 1980. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res. 8:4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raynaud, C., C. Guilhot, J. Rauzier, Y. Bordat, V. Pelicic, R. Manganelli, I. Smith, B. Gicquel, and M. Jackson. 2002. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 45:203-217. [DOI] [PubMed] [Google Scholar]
- 14.Sampson, S. L., R. M. Warren, M. Richardson, G. D. van der Spuy, and P. D. van Helden. 1999. Disruption of coding regions by IS6110 insertion in Mycobacterium tuberculosis. Tuberc. Lung Dis. 79:349-359. [DOI] [PubMed] [Google Scholar]
- 15.Segal, W. 1984. Growth dynamics of in vivo and in vitro grown mycobacterial pathogens, p. 547-573. In G. P. Kubica, and L. G. Wayne (ed.), The mycobacteria: a sourcebook. Marcel Dekker, Inc., New York, N.Y.
- 16.Songer, J. G. 1997. Bacterial phospholipases and their role in virulence. Trends Microbiol. 5:156-161. [DOI] [PubMed] [Google Scholar]
- 17.Szklo, M., and F. J. Nieto. 2004. Epidemiology: beyond the basics. Jones and Bartlett Publishers, Sudbury, Mass.
- 18.Talarico, S., R. Durmaz, and Z. Yang. 2005. Insertion- and deletion-associated genetic diversity of Mycobacterium tuberculosis phospholipase C-encoding genes among 106 clinical isolates from Turkey. J. Clin. Microbiol. 43:533-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terada, L. S., K. A. Johansen, S. Nowbar, A. I. Vasil, and M. L. Vasil. 1999. Pseudomonas aeruginosa hemolytic phospholipase C suppresses neutrophil respiratory burst activity. Infect. Immun. 67:2371-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Titball, R. W. 1993. Bacterial phospholipases C. Microbiol. Rev. 57:347-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vera-Cabrera, L., M. A. Hernandez-Vera, O. Welsh, W. M. Johnson, and J. Castro-Garza. 2001. Phospholipase region of Mycobacterium tuberculosis is a preferential locus for IS6110 transposition. J. Clin. Microbiol. 39:3499-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vera-Cabrera, L., S. T. Howard, A. Laszlo, and W. M. Johnson. 1997. Analysis of genetic polymorphism in the phospholipase region of Mycobacterium tuberculosis. J. Clin. Microbiol. 35:1190-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viana-Niero, C., P. E. De Haas, D. Van Soolingen, and S. C. Leao. 2004. Analysis of genetic polymorphisms affecting the four phospholipase C (plc) genes in Mycobacterium tuberculosis complex clinical isolates. Microbiology 150:967-978. [DOI] [PubMed] [Google Scholar]
- 26.Warren, R. M., S. L. Sampson, M. Richardson, G. D. Van Der Spuy, C. J. Lombard, T. C. Victor, and P. D. van Helden. 2000. Mapping of IS6110 flanking regions in clinical isolates of Mycobacterium tuberculosis demonstrates genome plasticity. Mol. Microbiol. 37:1405-1416. [DOI] [PubMed] [Google Scholar]
- 27.Wheeler, P. R., K. Bulmer, and C. Ratledge. 1990. Enzymes for biosynthesis de novo and elongation of fatty acids in mycobacteria grown in host cells: is Mycobacterium leprae competent in fatty acid biosynthesis? J. Gen. Microbiol. 136:211-217. [DOI] [PubMed] [Google Scholar]
- 28.Wheeler, P. R., and C. Ratledge. 1992. Control and location of acyl-hydrolysing phospholipase activity in pathogenic mycobacteria. J. Gen. Microbiol. 138:825-830. [DOI] [PubMed] [Google Scholar]
- 29.Wheeler, P. R., and C. Ratledge. 1991. Phospholipase activity of Mycobacterium leprae harvested from experimentally infected armadillo tissue. Infect. Immun. 59:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, Z., Y. Kong, F. Wilson, B. Foxman, A. H. Fowler, C. F. Marrs, M. D. Cave, and J. H. Bates. 2004. Identification of risk factors for extrapulmonary tuberculosis. Clin. Infect. Dis. 38:199-205. [DOI] [PubMed] [Google Scholar]
- 31.Yang, Z., D. Yang, Y. Kong, L. Zhang, C. F. Marrs, B. Foxman, J. H. Bates, F. Wilson, and M. D. Cave. 2005. Clinical relevance of Mycobacterium tuberculosis plcD gene mutations. Am. J. Respir. Crit. Care Med. 171:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeger, S. L., and K. Y. Liang. 1986. Longitudinal data analysis for discrete and continuous outcomes. Biometrics 42:121-130. ′ [PubMed] [Google Scholar]