Abstract
Yersinia pseudotuberculosis is a gram-negative bacterium that infects a wide range of animals, including humans, and is transmitted by the fecal-oral route. This species is found globally and is responsible for human outbreaks, mainly in cold countries. The aim of this study was to evaluate the potential of ribotyping for the molecular typing of worldwide isolates. For this purpose, 80 strains of Y. pseudotuberculosis belonging to the six classical serotypes and nine subserotypes and isolated from various countries and different hosts were analyzed. Combination of the EcoRI and EcoRV ribopatterns allowed the delineation of 27 ribotypes. In most instances, ribotypes were associated with specific subserotypes and allowed their subdivision. No association between the ribotype and the geographical origin of the strains was observed, arguing for a global spread of this organism. Similarly, no marked association between the ribotype and the type of host was noted, confirming the circulation of this pathogen in the environment, different animal species, and human hosts. Y. pseudotuberculosis exhibited ribopatterns very close to those of Y. pestis, although not completely identical. Altogether, the present study demonstrates that ribotyping may be a useful tool for molecular typing of global isolates of Y. pseudotuberculosis but that it has some limitations due to the small number of hybridizing bands that generate the diversity of the profiles.
Yersinia pseudotuberculosis is a gram-negative bacterium that belongs to the genus Yersinia and to the Enterobacteriaceae family. Various environmental sources and a wide range of animals represent the reservoir of this organism, which is transmitted by the fecal-oral route (7, 21). Humans infect themselves after the consumption of contaminated greeneries or water or through the handling of infected animals. The ingested bacteria migrate to the ileum and reach the mesenteric lymph nodes, where they multiply. The main clinical manifestations are a pain in the right abdominal quadrant (mimicking appendicitis), fever, and sometimes diarrhea. Dissemination to deeper tissues and to the bloodstream sometimes occurs. The importance of Y. pseudotuberculosis as a causative agent of human infections is lower than that of Y. enterocolitica in most countries worldwide. However, this species remains a major cause of enteric infections and may be responsible for small human outbreaks in Japan (23), Russia (23), Scandinavia (15), and elsewhere.
According to the classical serotyping scheme (22), Y. pseudotuberculosis can be subdivided into six serotypes (O:1 to O:6) that can be further differentiated into subtypes (24). Nine additional serotypes (O:7 to O:15) have been identified (24), but they are restricted to some geographical areas, mainly in Asia. Molecular techniques represent valuable alternatives for subtyping Y. pseudotuberculosis. These include analysis of the genomic restriction profiles (14, 16-18), multilocus enzyme electrophoresis (6, 9, 10), IS fingerprinting (3, 18), and restriction pattern analysis of the Yersinia virulence plasmid (8). The three formers have been applied to a small number of Y. pseudotuberculosis strains (≤30), often isolated from a given geographical area during the same outbreak. The latter was used on a large number of isolates but exhibited a low discriminatory power and is not applicable to strains spontaneously cured of the pYV plasmid.
Ribotyping has been successfully used for distinguishing subgroups of Y. pestis, a species of low phenotypic and genetic diversity (11, 12). Previous studies performed on a small number of isolates suggested that ribotyping might also be an efficient typing tool for Y. pseudotuberculosis (18, 20). The aim of the present study was to evaluate the discriminatory power of ribotyping to subtype the species Y. pseudotuberculosis. For this purpose, 80 strains of Y. pseudotuberculosis isolated between 1960 and 2001 from 24 countries and from various sources (Table 1) were selected in the strain collection of the Yersinia National Reference Laboratory and WHO Collaborating Center at the Institut Pasteur. These strains belonged to the six classical serotypes, and their subserotype was determined by using the recently described genoserotyping method (4). Their genomic DNA was extracted, digested with EcoRI or EcoRV, electrophoresed, and transferred to nylon membranes as previously described (11). For ribotyping, ribosomal 16S+23S rRNA from Escherichia coli (Boehringer) was labeled with horseradish peroxidase by using the ECL Gene Detection System (Amersham).
TABLE 1.
Characteristics of the 80 strains of Y. pseudotuberculosis used for ribotyping
| Strain | Serotype | Country | Origin | EcoRI pattern | EcoRV pattern | Ribotype | Strain | Serotype | Country | Origin | EcoRI pattern | EcoRV pattern | Ribotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IP32879 | 1a | Switzerland | Bird | EI.16 | EV.5 | R.1 | IP33054 | 2a | Spain | Human | EI.1 | EV.9 | R.14 |
| IP32953 | 1b | France | Human | EI.2 | EV.16 | R.2 | IP32589 | 2a | New Zealand | Human | EI.1 | EV.9 | R.14 |
| IP31833 | 4b | England | Sheep | EI.3 | EV.3 | R.3 | IP33088 | 2a | France | Human | EI.13 | EV.15 | R.15 |
| IP32790 | 1a | Italy | Pig | EI.3 | EV.7 | R.4 | IP30215 | 2b | Denmark | Guinea pig | EI.17 | EV.11 | R.16 |
| IP32745 | 1a | Italy | Human | EI.3 | EV.7 | R.4 | IP32929 | 2b | France | Hare | EI.17 | EV.4 | R.17 |
| IP32590 | 1a | Switzerland | Human | EI.3 | EV.7 | R.4 | IP33098 | 2b | France | Hare | EI.17 | EV.4 | R.17 |
| IP31553 | 6 | Japan | Guinea-pig | EI.6 | EV.2 | R.5 | IP32934 | 2b | France | Monkey | EI.17 | EV.4 | R.17 |
| IP31554 | 6 | Japan | Guinea pig | EI.6 | EV.2 | R.5 | IP32821 | 5a | France | Human | EI.17 | EV.4 | R.17 |
| IP30284 | 1a | Italy | Pigeon | EI.5 | EV.16 | R.6 | IP32952 | 5a | France | Human | EI.17 | EV.4 | R.17 |
| IP33005 | 1a | Germany | Monkey | EI.5 | EV.16 | R.6 | IP32463 | 5a | Switzerland | Guinea pig | EI.17 | EV.4 | R.17 |
| IP32651 | 1b | Yugoslavia | Hare | EI.5 | EV.16 | R.6 | IP33061 | 5a | Germany | Monkey | EI.17 | EV.4 | R.17 |
| IP31411 | 4b | Denmark | Hare | EI.3 | EV.4 | R.7 | IP32699 | 5a | France | Wild species | EI.17 | EV.4 | R.17 |
| IP30437 | 1b | Canada | Beaver | EI.9 | EV.16 | R.8 | IP32887 | 3 | Argentina | Bovine | EI.14 | EV.12 | R.18 |
| IP31878 | 1a | Tunisia | Rodent | EI.9 | EV.16 | R.8 | IP32544 | 3 | South Africa | Pig | EI.14 | EV.12 | R.18 |
| IP32665 | 1a | Yugoslavia | Hare | E1.9 | EV.16 | R.8 | IP32564 | 3 | Belgium | Human | EI.14 | EV.12 | R.18 |
| IP33291 | 1a | France | Hare | EI.9 | EV.16 | R.8 | IP32938 | 3 | Argentina | Bovine | EI.14 | EV.12 | R.18 |
| IP32709 | 1b | England | Bird | EI.9 | EV.16 | R.8 | IP32802 | 3 | Italy | Pig | EI.14 | EV.12 | R.18 |
| IP32323 | 2a | Norway | Water | EI.9 | EV.16 | R.8 | IP32992 | 3 | Australia | Bovine | EI.14 | EV.12 | R.18 |
| IP32954 | 1a | France | Human | EI.9 | EV.3 | R.9 | IP33051 | 3 | France | Caprine | EI.14 | EV.12 | R.18 |
| IP30642 | 1a | Tunisia | Mouse | EI.9 | EV.3 | R.9 | IP33097 | 3 | Argentina | Deer | EI.14 | EV.12 | R.18 |
| IP31524 | 1a | Czechoslovakia | Human | EI.9 | EV.3 | R.9 | IP33105 | 3 | Argentina | Bovine | EI.14 | EV.12 | R.18 |
| IP33161 | 1b | Ukraine | Rodent | EI.9 | EV.3 | R.9 | IP33108 | 3 | Bulgaria | Human | EI.14 | EV.12 | R.18 |
| IP33162 | 1b | Ukraine | Human | EI.9 | EV.3 | R.9 | IP32950 | 1b | France | Human | EI.15 | EV.4 | R.19 |
| IP32721 | 2a | Italy | Hare | EI.1 | EV.5 | R.10 | IP33109 | 1b | France | Human | EI.15 | EV.4 | R.19 |
| IP32637 | 1b | France | Unknown | EI.8 | EV.8 | R.11 | IP32817 | 5b | Japan | Hare | EI.15 | EV.4 | R.19 |
| IP32949 | 1b | France | Human | EI.8 | EV.8 | R.11 | IP32816 | 5b | Japan | Hare | EI.15 | EV.4 | R.19 |
| IP32533 | 1b | New Zealand | Deer | EI.8 | EV.8 | R.11 | IP32921 | 2b | France | Hare | EI.10 | EV.11 | R.20 |
| IP33038 | 1b | Australia | Marsupial | EI.8 | EV.8 | R.11 | IP32881 | 2b | Switzerland | Monkey | EI.10 | EV.11 | R.20 |
| IP32670 | 1b | England | Pig | EI.8 | EV.8 | R.11 | IP32614 | 1a | Yugoslavia | Hare | EI.11 | EV.13 | R.21 |
| IP33285 | 1b | France | Human | EI.8 | EV.8 | R.11 | IP31829 | 3 | England | Ovine fetus | EI.7 | EV.6 | R.22 |
| IP32777 | 1b | France | Human | EI.8 | EV.8 | R.11 | IP30151 | 4a | Sweden | Otter | EI.12 | EV.1 | R.23 |
| IP32524 | 1b | Holland | Human | EI.8 | EV.8 | R.11 | IP32951 | 2a | France | Human | EI.16 | EV.9 | R.24 |
| IP32939 | 1a | Romania | Soil | EI.9 | EV.7 | R.12 | IP32666 | 3 | Spain | Human | EI.18 | EV.4 | R.25 |
| IP30911 | 2b | Holland | Hare | EI.10 | EV.10 | R.13 | IP32889 | 3 | Spain | Unknown | EI.18 | EV.4 | R.25 |
| IP32581 | 2a | Belgium | Human | EI.1 | EV.9 | R.14 | IP32984 | 3 | Spain | Human | EI.18 | EV.4 | R.25 |
| IP33293 | 2a | France | Human | EI.1 | EV.9 | R.14 | IP31830 | 4b | England | Human | EI.18 | EV.4 | R.25 |
| IP32584 | 2a | Spain | Pig | EI.1 | EV.9 | R.14 | IP32687 | 4b | France | Wild species | EI.4 | EV.14 | R.26 |
| IP32585 | 2a | France | Antelope | EI.1 | EV.9 | R.14 | IP33156 | 1b | Russia | Human | EI.1 | EV.17 | R.27 |
| IP33012 | 2a | Germany | Monkey | EI.1 | EV.9 | R.14 | IP33157 | 1b | Russia | Human | EI.1 | EV.17 | R.27 |
| IP33023 | 2a | Switzerland | Monkey | EI.1 | EV.9 | R.14 | IP33158 | 1b | Russia | Human | EI.1 | EV.17 | R.27 |
Agglutination with the O:1 or O:2 antiserum but PCR profile corresponding to none of the 21 described genoserotypes.
Agglutination with the O:4 antiserum but genoserotype O:8.
EcoRI ribopatterns.
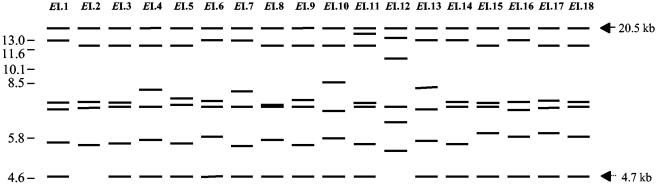
Analysis of the EcoRI patterns allowed delineation of 18 different profiles (EI.1 to EI.18) among the 80 Y. pseudotuberculosis strains studied (Fig. 1). Since the genome of Y. pseudotuberculosis contains seven rRNA operons, seven fragments were expected after hybridization of the EcoRI-digested genomic DNA with the rRNA probe. However, with the exception of one strain (IP32614, EcoRI profile EI.11), all hybridization patterns exhibited only six bands (Fig. 1). Analysis of the chromosome sequence of Y. pseudotuberculosis strain IP32953 (NCBI accession number NC 006155) (5) indicated that one EcoRI site is situated in the 5′ portion of each 16S rRNA gene and the second site is located outside the rRNA operon, at variable distances in the downstream flanking chromosomal regions. Based on the sequence data, the expected sizes for the EcoRI fragments of strain IP32953 were 4.46, 5.69, 7.29, 7.55, 12.61, 20.47, and 20.59 kb. These sizes were in accordance with those observed for the hybridizing fragments. The presence of only six hybridizing bands in most strains could thus be explained by the superposition of two large size EcoRI fragments of approximately 20.5 kb. This 20.5-kb band was found in all profiles, and a 4.7-kb fragment was conserved in all but two profiles (EI.2 and EI.12), representing only one strain each. The diversity observed was generated by the four other hybridizing fragments (Fig. 1). No dominant EcoRI pattern was observed, but five patterns—EI.9 (12 strains), EI.1 (12 strains), EI.14 (10 strains), EI.17 (9 strains), and EI.8 (8 strains)—represented 64% of the isolates (Table 1). Six profiles were limited to one strain each. No strict association between serotypes and EcoRI patterns was noted. Several patterns could be identified within a given serotype; on the other hand, a given EcoRI pattern could be found in strains of various serotypes. Some EcoRI ribopatterns were nonetheless restricted to a specific serotype (EI.5 to serotype 1, EI.8 to serotype 1b, EI.10 to serotype 2b, EI.14 to serotype 3, and EI.6 to serotype 6) (Table 1).
FIG. 1.
Schematic representation of the EcoRI hybridization profiles of the genomic DNA of 80 strains of Y. pseudotuberculosis obtained after hybridization with an E. coli 16S+23S rRNA probe. A solid arrow points to a band conserved in all strains, and a dotted arrow points to a band present in all except two strains. Tick marks with numbers on the left indicate the sizes (in kilobases) of the molecular mass standards (Xenorhabdus sp. strain 278).
EcoRV ribopatterns.
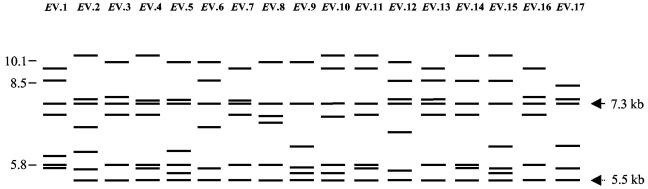
Seventeen profiles (EV.1 to EV.17) were obtained after digestion of the DNA of the 80 Y. pseudotuberculosis strains with EcoRV (Fig. 2). Seven hybridizing bands were seen in most profiles, except for seven profiles which displayed six fragments, likely because of the superposition of two fragments of approximately the same size. This was confirmed in strain IP32953 whose genome sequence predicted seven fragments of 8.7, 7.5, 7.3, 6.9, 5.8, 5.8, and 5.5 kb, respectively, two of which were of the same size (5.8 kb). The 7.3-kb band was found in all profiles, and a 5.5-kb fragment was conserved in all but one profiles (EV.1) composed of a single strain (Fig. 2). The most frequent EcoRV patterns were EV.4 (17 strains), EV.12 (10 strains), EV.16 (10 strains), EV.9 (9 strains), and EV.8 (8 strains). They represented 67% of the isolates (Table 1). Six profiles were found in one strain each. As for EcoRI, no strict association between serotypes and EcoRV patterns was observed, although some EcoRV ribopatterns were restricted to a specific serotype (EV.7 to serotype 1a, EV.8 and EV.17 to serotype 1b, EV.9 to serotype 2a, and EV.12 to serotype 3) (Table 1).
FIG. 2.
Schematic representation of the EcoRV hybridization profiles of the genomic DNA of 80 strains of Y. pseudotuberculosis obtained after hybridization with an E. coli 16S+23S rRNA probe. A plain arrow points to a band conserved in all strains, and a dotted arrow points to a band present in all except one strain. Tick marks with numbers on the left indicate the sizes (in kilobases) of the molecular mass standards (Xenorhabdus sp. strain 278).
Genomic analysis of the EcoRI and EcoRV ribopatterns.
A change in the size of an hybridizing fragment may be due either to a point mutation (or a short deletion or insertion), which creates or abolishes a restriction site, or to a large chromosome rearrangement that modifies the regions flanking the rRNA locus. Since a unique EcoRI and EcoRV site is located within each rRNA locus, at its 5′ extremity, the variability in the fragment size is essentially generated by the polymorphism of the 3′ flanking region. Interestingly, the 20.5-kb EcoRI and 7.3-kb EcoRV hybridizing fragments, which were systematically present in all strains studied (Fig. 1 and 2), corresponded to the same region carrying the rRNA locus located at positions 150833 to 155951 on the Y. pseudotuberculosis IP32953 chromosome (5). The absence of size polymorphism for this band suggests that the region adjacent to the 3′ extremity of this rRNA locus is less prone to mutations or rearrangements than the chromosomal regions flanking the other rRNA loci. An EcoRI (4.7 kb) and an EcoRV (5.5 kb) fragment were also found in all except one or two strains (Fig. 1 and 2). Again, these EcoRI and EcoRV bands corresponded to the same rRNA locus, located at positions 320391 to 325633 on the Y. pseudotuberculosis chromosome. The conservation of this band is most likely due to the fact that the EcoRI and EcoRV sites flanking the 3′ end of this rRNA locus are located very close to its extremity, the occurrence of point mutations or rearrangements being statistically less probable for short regions of DNA. The unique combined EcoRI and EcoRV profiles found in five strains (IP33088, IP32687, IP30151, IP32614, and IP31829) is more likely attributable to a large chromosomal rearrangement involving the two restriction sites than to the simultaneous modifications of these two sites.
Ribotype.
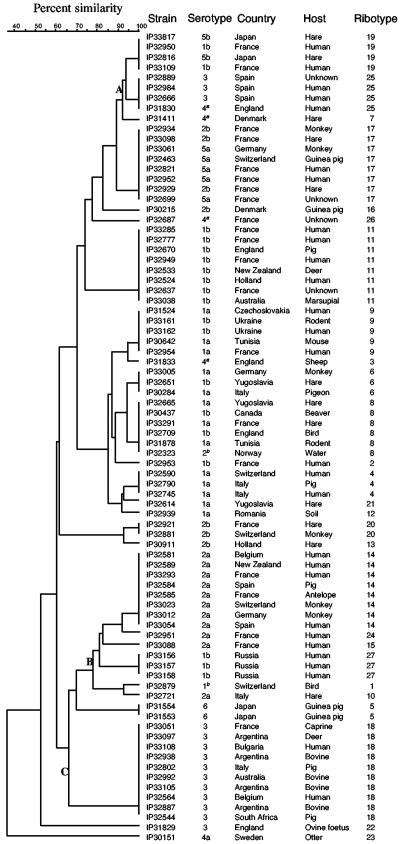
The ribotypes were defined as the combination of the EcoRI and EcoRV patterns (11). The EcoRI and EcoRV profiles were frequently but not systematically associated, leading to the delineation of 27 ribotypes (R.1 to R.27) among the 80 Y. pseudotuberculosis strains analyzed (Table 2). The dominant ribotype was R.18 (10 strains), followed by R.11, R.14, and R.17 (8 strains each). Fourteen strains had a unique ribotype. Cluster analysis of the combined EcoRI and EcoRV ribopatterns was done by the unweighted pair group method with average linkages (UPGMA), using the Dice coefficient to analyze the similarities of the banding patterns. The dendrogram derived from this analysis shows that, with the exception of R.22 and R.23 which formed outgroups, all other ribotypes were related (mean similarity of 70%) and no major clusters were delineated (Fig. 3). Analysis of the association between ribotype and serotype indicated that in six cases (R.6, R.8, R.9, R.17, R.19, and R.25) the same ribotype was found among strains belonging to two different serotypes or subserotypes (Table 2). However, the 21 other ribotypes were linked to a single serotype, and the UPGMA dendrogram showed that different ribotypes associated with the same serotype were frequently clustered (Fig. 3). Ribotyping thus allowed the subtyping of the strains within a given serotype (Table 2). For instance, the 13 strains of serotype 1a could be subdivided into six ribotypes, the 19 strains of serotype 1b into seven ribotypes, the 14 strains of serotype three into four ribotypes, and the 11 strains of serotype 2a into four ribotypes. The discrimination index (D) of ribotyping, based on the Simpson's index of diversity (13), was 0.94, while that of serotyping was lower (D = 0.72), thus indicating that the discriminatory power of ribotyping is superior to that of serotyping. This may be explained by the fact that (i) the chances of neutral point mutations or chromosomal rearrangements occurring in regions flanking the rRNA loci are higher than within the O-Ag gene cluster and (ii) the size of the chromosomal regions flanking the rRNA loci is much larger than that of the O-Ag repeats locus, thus allowing a statistically higher number of mutational events. No association between the ribotype and the geographical origin of the strains was noted (Fig. 3). Strains from the same continent or country were dispersed in the various clusters. On the other hand, isolates from different continents were found in the same cluster and sometimes had the same ribotype. For instance, the dominant ribotype R.18 was found in Africa, America, Oceania, and Europe. This argues for a global circulation of Y. pseudotuberculosis strains, although homoplasies cannot be ruled out. One exception was the three strains from Russia, which had a unique and specific ribotype (R.27). No strong association between the type of host and the ribotype of the strains was observed (Fig. 3). This fits with the known epidemiological features of Y. pseudotuberculosis, which is found in the environment and in a wide variety of animals that form the reservoir for human infections. However, some branches of the dendrogram (A and B, Fig. 3) contained predominantly strains isolated from humans, whereas branch C was composed almost exclusively of strains of animal origins. Although a much larger number of strains would be needed to draw solid conclusions, these data nonetheless suggest that some ribotypes may be associated with strains having a higher pathogenic potential for humans.
TABLE 2.
Scheme used to define the ribotype of the 80 strains of Y. pseudotuberculosis studied
| EcoRI profile | EcoRV profile | Ribotype | No. of strains | Serotype |
|---|---|---|---|---|
| EI.16 | EV.5 | R.1 | 1 | 1a |
| EI.2 | EV.16 | R.2 | 1 | 1b |
| EI.3 | EV.3 | R.3 | 1 | 4b |
| EI.3 | EV.7 | R.4 | 3 | 1a |
| EI.6 | EV.2 | R.5 | 2 | 6 |
| EI.5 | EV.16 | R.6 | 3 | 1a, 1b |
| EI.3 | EV.4 | R.7 | 1 | 4b |
| EI.9 | EV.16 | R.8 | 6 | 1a, 1b, 2a |
| EI.9 | EV.3 | R.9 | 5 | 1a, 1b |
| EI.1 | EV.5 | R.10 | 1 | 2a |
| EI.8 | EV.8 | R.11 | 8 | 1b |
| EI.9 | EV.7 | R.12 | 1 | 1a |
| EI.10 | EV.10 | R.13 | 1 | 2b |
| EI.1 | EV.9 | R.14 | 8 | 2a |
| EI.13 | EV.15 | R.15 | 1 | 2a |
| EI.17 | EV.11 | R.16 | 1 | 2b |
| EI.17 | EV.4 | R.17 | 8 | 2b, 5a |
| EI.14 | EV.12 | R.18 | 10 | 3 |
| EI.15 | EV.4 | R.19 | 4 | 1b, 5b |
| EI.10 | EV.11 | R.20 | 2 | 2b |
| EI.11 | EV.13 | R.21 | 1 | 1a |
| EI.7 | EV.6 | R.22 | 1 | 3 |
| EI.12 | EV.1 | R.23 | 1 | 4b |
| EI.16 | EV.9 | R.24 | 1 | 2a |
| EI.18 | EV.4 | R.25 | 4 | 3, 4b |
| EI.4 | EV.14 | R.26 | 1 | 4b |
| EI.1 | EV.7 | R.27 | 3 | 1b |
Agglutination with the O:1 or O:2 antiserum but PCR profile corresponding to none of the 21 described genoserotypes.
Agglutination with the O:4 antiserum but genoserotype O:8.
FIG. 3.
Dendrogram derived from the UPGMA clustering analysis of the Y. pseudotuberculosis ribotypes. Analysis of the combined EcoRI and EcoRV ribopatterns was done with the BioNumerics software package version 4.0 (Applied Maths, Kortrijk, Belgium). A position tolerance of 1% for EcoRI and 1.8% for EcoRV was chosen to allow 100% matching of the banding patterns obtained with duplicate samples. The Dice coefficient was used to calculate similarities. Superscripts: a, agglutination with the O:4 antiserum but genoserotype O:8; b, agglutination with the O:1 or O:2 antiserum but PCR profile corresponding to none of the 21 described genoserotypes.
Comparison of the ribotypes of Y. pestis and Y. pseudotuberculosis.
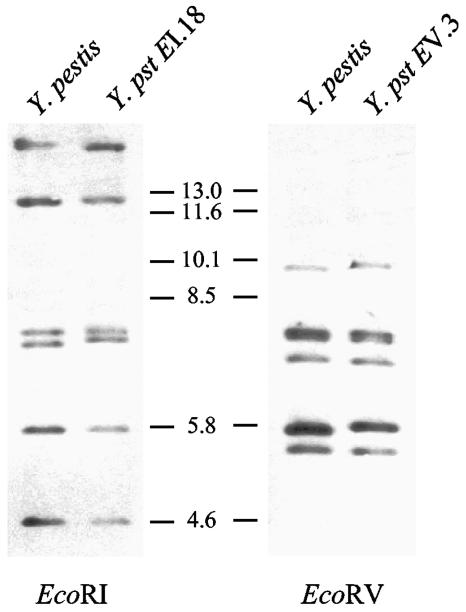
Since Y. pestis was shown to be a clone recently emerged from Y. pseudotuberculosis (2), we wondered whether the two species displayed different profiles or whether common ribotypes could be identified. Sixteen ribotypes were previously identified among 70 isolates of Y. pestis of worldwide origin (11). The most frequent one was ribotype B, which was found in strains of biotype Orientalis. However, this biotype is characterized by the loss of one rRNA operon (19). The second most common Y. pestis ribotype was O, which was present in both Medievalis and Antiqua strains (11). Ribotype O was characterized by the same EcoRI and EcoRV profiles as ribotype B, plus an additional band corresponding to the rRNA locus, lost in strains of ribotype B. Since ribotype B/O is the most common ribotype and is found in the three biotypes of Y. pestis, it may therefore be considered anterior to the split of Y. pestis into various branches (1). We thus compared the EcoRI and EcoRV patterns defining ribotype O of Y. pestis with the Y. pseudotuberculosis patterns. Overall, the Y. pseudotuberculosis patterns resembled those of Y. pestis. In particular, the conserved and highly conserved hybridizing bands in Y. pseudotuberculosis were present and also conserved among Y. pestis isolates (11). The most similar EcoRI and EcoRV profiles were EI.18(with one band slightly higher) and EV.3, respectively (Fig. 4). However, no ribotype corresponding to the association EI.18+EV.3 was identified among the 80 Y. pseudotuberculosis strains analyzed. Therefore, the ancestral ribotype O of Y. pestis was not found in any of the Y. pseudotuberculosis strains studied.
FIG. 4.
Comparison of the EcoRI and EcoRV ribopatterns of Y. pestis strain PKH-3 (biotype Medievalis and ribotype O) with the most similar ribopatterns found in Y. pseudotuberculosis (Y. pst). Numbers between the autoradiograms correspond to the sizes (in kilobases) of the molecular mass standards (Xenorhabdus sp. strain 278).
Conclusion.
The aim of the present study was to evaluate the potential of ribotyping for molecular typing of Y. pseudotuberculosis strains of worldwide origin. Twenty-seven ribotypes were identified among the 80 strains studied belonging to six serotypes and nine subserotypes, indicating that ribotyping has a much higher discriminatory potential than serotyping. The method known to have the highest power to discriminate Y. pseudotuberculosis isolates is pulsed-field gel electrophoresis (14, 16-18). This method is indeed valuable to compare strains within a given focus and determine the origin of a contamination. However, the complexity of the profiles makes the comparison of large numbers of strains of various geographical origins difficult. Ribotyping has the advantage over PFGE to generate less complex and more reproducible profiles (11), thus allowing a simpler and more reliable global comparison of strains. It also has the advantage of not requiring a sophisticated electrophoresis apparatus and therefore to be applicable in most laboratories. However, ribotyping has some limitations: (i) the polymorphism of the profiles is restricted to a small number of bands (four to five), thus limiting the discriminatory power; (ii) the varying bands may exhibit only slight differences in size, making the distinction between several ribopatterns sometimes uneasy; and (iii) it does not clearly differentiate Y. pestis from Y. pseudotuberculosis since some profiles are highly similar among the two species. Therefore, the present study demonstrates that ribotyping may be a useful tool for molecular typing of global isolates of Y. pseudotuberculosis but that this technique has some intrinsic limitations.
Acknowledgments
This study was funded in part by the Action Concertées des Instituts Pasteur et Instituts Associés.
REFERENCES
- 1.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, D. Wagner, C. Allender, R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 101:17837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., K. Zurth, C. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobrov, A. G., and A. A. Filippov. 1997. Prevalence of IS285 and IS100 in Y. pestis and Y. pseudotuberculosis genomes. Mol. Genet. Mikrobiol. Virusol. 2:36-40. [PubMed] [Google Scholar]
- 4.Bogdanovich, T., E. Carniel, H. Fukushima, and M. Skurnik. 2003. Use of O-antigen gene cluster-specific PCRs for the identification and O-genotyping of Yersinia pseudotuberculosis and Yersinia pestis. J. Clin. Microbiol. 41:5103-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolina, M., and R. Peduzzi. 1993. Population genetics of human, animal, and environmental Yersinia strains. Appl. Environ. Microbiol. 59:442-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukushima, H., M. Gomyoda, and S. Kaneko. 1991. Wild animals as the source of infection with Yersinia pseudotuberculosis in Shimane Prefecture, Japan, p. 1-4. In T. Une, T. Maruyama, and M. Tsubokura (ed.), Current investigations of the microbiology of yersiniae, vol. 12. Karger, Basel, Switzerland. [PubMed] [Google Scholar]
- 8.Fukushima, H., M. Gomyoda, S. Kaneko, M. Tsubokura, N. Takeda, T. Hongo, and F. N. Shubin. 1994. Restriction endonuclease analysis of virulence plasmids for molecular epidemiology of Yersinia pseudotuberculosis infections. J. Clin. Microbiol. 32:1410-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goullet, P., and B. Picard. 1988. Characterization of Yersinia enterocolitica, Y. intermedia, Y. aldovae, Y. frederiksenii, Y. kristensenii, and Y. pseudotuberculosis by electrophoretic polymorphism of acid phosphatase, esterases, and glutamate and malate dehydrogenases. J. Gen. Microbiol. 134:317-325. [DOI] [PubMed] [Google Scholar]
- 10.Goullet, P., and B. Picard. 1984. Distinctive electrophoretic and isoelectric focusing patterns of esterases from Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Gen. Microbiol. 130:1471-1480. [DOI] [PubMed] [Google Scholar]
- 11.Guiyoule, A., F. Grimont, I. Iteman, P. A. D. Grimont, M. Lefevre, and E. Carniel. 1994. Plague pandemics investigated by ribotyping of Yersinia pestis strains. J. Clin. Microbiol. 32:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guiyoule, A., B. Rasoamanana, C. Buchrieser, P. Michel, S. Chanteau, and E. Carniel. 1997. Recent emergence of new variants of Yersinia pestis in Madagascar. J. Clin. Microbiol. 35:2826-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iteman, I., H. Najdenski, and E. Carniel. 1995. High genomic polymorphism in Yersinia pseudotuberculosis, p. 106-111. In G. Ravagnan and C. Chiesa (ed.), Yersiniosis: present and future, vol. 13. Karger, Basel, Switzerland. [PubMed] [Google Scholar]
- 15.Jalava, K., S. Hallanvuo, U. M. Nakari, P. Ruutu, E. Kela, T. Heinasmaki, A. Siitonen, and J. P. Nuorti. 2004. Multiple outbreaks of Yersinia pseudotuberculosis infections in Finland. J. Clin. Microbiol. 42:2789-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niskanen, T., M. Fredriksson-Ahomaa, and H. Korkeala. 2002. Yersinia pseudotuberculosis with limited genetic diversity is a common finding in tonsils of fattening pigs. J. Food Prot. 65:540-545. [DOI] [PubMed] [Google Scholar]
- 17.Nuorti, J. P., T. Niskanen, S. Hallanvuo, J. Mikkola, E. Kela, M. Hatakka, M. Fredriksson-Ahomaa, O. Lyytikainen, A. Siitonen, H. Korkeala, and P. Ruutu. 2004. A widespread outbreak of Yersinia pseudotuberculosis O:3 infection from iceberg lettuce. J. Infect. Dis. 189:766-774. [DOI] [PubMed] [Google Scholar]
- 18.Odaert, M., P. Berche, and M. Simonet. 1996. Molecular typing of Yersinia pseudotuberculosis by using an IS200-like element. J. Clin. Microbiol. 34:2231-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeño-Tárraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. F. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 20.Picard-Pasquier, N., B. Picard, S. Heeralal, R. Krishnamoorthy, and P. Goullet. 1990. Correlation between ribosomal DNA polymorphism and electrophoretic enzyme polymorphism in Yersinia. J. Gen. Microbiol. 136:1655-1666. [DOI] [PubMed] [Google Scholar]
- 21.Slee, K. J., and N. W. Skilbeck. 1992. Epidemiology of Yersinia pseudotuberculosis and Y. enterocolitica Infections in Sheep in Australia. J. Clin. Microbiol. 30:712-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thal, E. 1973. Observations on immunity in Yersinia pseudotuberculosis. Contrib. Microbiol. Immunol. 2:190-195. [Google Scholar]
- 23.Toyokawa, Y., Y. Ohtomo, T. Akiyama, K. Masuda, M. Kasai, S. Kaneko, and T. Maruyama. 1993. Large scale outbreak of Yersinia pseudotuberculosis serotype 5a infection at Noheji-machi in Aomori Prefecture. J. Jpn. Assoc. Infect. Dis. 67:36-44. [DOI] [PubMed] [Google Scholar]
- 24.Tsubokura, M., and S. Aleksic. 1995. A simplified antigenic scheme for serotyping of Yersinia pseudotuberculosis: phenotypic characterization of reference strains and preparation of O and H factor sera. Contrib. Microbiol. Immunol. 13:99-105. [PubMed] [Google Scholar]