Abstract
Candida nivariensis is a recently described pathogenic yeast closely related to Candida glabrata. We developed a specific set of oligonucleotide primers based on the internal transcribed spacer regions of the rRNA gene for the rapid identification of C. nivariensis. PCR with these primers amplified a 206-bp amplicon from C. nivariensis.
Invasive fungal infections are a major medical problem, particularly among immunocompromised hosts (9). The management of invasive fungal infections has been hampered by the inability to diagnose the infection at an early stage of disease. However, diagnosis of these fungal infections remains difficult, since the only clinical sign of infection may be a prolonged fever that is refractory to antibacterial treatment. In recent years, efforts have been made to develop molecular biology-based methods for rapid diagnosis, which is crucial for the treatment and recovery of patients suffering from systemic candidiasis (8).
Yeasts are usually identified through a combination of morphological features, ability to ferment selected sugars, and performance of assimilation reactions on a relatively large number of carbon and nitrogen compounds (6). Molecular studies have shown that it is not uncommon for different strains of a species to vary somewhat in their fermentation and assimilation profiles, which can lead to misidentifications (10, 4). Molecular approaches are more promising than phenotypic methods for the rapid detection and identification of pathogenic organisms (2, 3, 7, 11, 12). The recently described species Candida nivariensis differs somewhat from other known species in relation to physiological reactions (1).
A total of 35 yeast isolates, including the three available isolates of C. nivariensis, other relevant pathogenic yeasts, and four reference strains, were included in this study. The three isolates of C. nivariensis were identified as was described previously (1).
Extraction of nuclear DNA of the isolates was performed as previously described (5). Two oligonucleotides (NIV-F [AGCTCATCCTGGTTAGTTTCG] and NIV-R [CCCTCTTCGTTTGTGTTTGT]) were designed after comparison of different yeast rRNA sequences from the GenBank database. Nucleotide-nucleotide BLAST (blastn) comparisons showed that the only sequences that showed 100% identities with both primers were the internal transcribed spacer (ITS) sequence of one unidentified isolate deposited in the database (accession number AY787833.1) and the ITS sequences from the three isolates of C. nivariensis (1). The set was synthesized by Roche Diagnostics.
PCRs were carried out in 50-μl reaction volumes containing about 0.05 ng of extracted DNA added to the PCR mixture consisting of 1× reaction buffer [16 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8)], containing 0.2 mM of each of the deoxynucleoside triphosphates (Promega Corp., Madison, Wis.), 1.5 mM MgCl2, 10 pmol of each primer, and 1.25 U of Taq polymerase (Bioline). DNA amplification was performed in a GeneAmp PCR system 9700 thermocycler (PE Applied Biosystems. Foster City, Calif.) using the following thermal cycling profile: one cycle at 94°C for 5 min, followed by 30 cycles at 94°C for 30 s, at 64°C for 30 s, and at 72°C for 45 s, with a final extension step at 72°C for 10 min. After thermal cycling, 5 μl of each amplified product was separated by electrophoresis on a 1% agarose gel, stained with ethidium bromide, and visualized with UV light.
PCR products were purified using a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Then the PCR products were sequenced directly on an ABI PRISM 310 genetic analyzer using a Big Dye terminator cycle sequencing ready reaction kit (Applied Biosystems Japan Co. Ltd., Tokyo, Japan) as recommended by the kit manufacturers.
Simultaneous detection of NIV and NL amplicons by PCR.
We tested the suitability of our PCR protocol for the individual amplification of each DNA fragment, NIV (206 bp) and NL (650 bp) (5). For multiplex PCRs, a 5-μl aliquot of the DNA suspension was added to 45 μl of the PCR mixture described above, except that 20 pmol of each NL primer and 10 pmol of each NIV primer were used. The NL primers were used as an internal control to identify all species of fungus, while NIV primers were used to specifically detect C. nivariensis. In order to reduce the formation of nonspecific extension products, the protocol included a hot-start DNA amplification which was carried out using the following thermal cycling profile: one cycle at 94°C for 5 min, followed by 10 cycles at 94°C for 30 s, at 63°C for 20 s, and at 72°C for 15 s, and another 25 cycles at 94°C for 30 s, at 53.7°C for 20 s, and at 72°C for 20 s, culminating with a final extension step of 3 min.
Using the newly designed primers, we were able to amplify a 206-bp fragment, as expected, from the three strains of C. nivariensis. In contrast, we failed to amplify the genomes from the list of unrelated microorganisms listed in Table 1.
TABLE 1.
Strains examined in this study and PCR results using NIV primers
| Species | Straina | NIV productb |
|---|---|---|
| C. nivariensis | HC 4292-20T | + |
| C. nivariensis | HC 7609-30 | + |
| C. nivariensis | HC 5937-63 | + |
| C. albicans | ATCC 90028T | − |
| C. albicans | HC 6597-20 | − |
| C. glabrata | ATCC 90030T | − |
| C. glabrata | HC 9460-30 | − |
| C. parasilopsis | ATCC 22019T | − |
| C. parasilopsis | HC 0832-30 | − |
| C. krusei | ATCC 6258T | − |
| C. krusei | HC 6224-20 | − |
| C. tropicalis | HC 5531-30 | − |
| C. tropicalis | HC 3131-20 | − |
| C. dubliniensis | M-2/04/CC | − |
| C. dubliniensis | HC 5233-63 | − |
| C. guilliermondii | HC 5729-65 | − |
| C. norvegensis | HC 9912-97 | − |
| C. famata | HC 3771-38 | − |
| C. lusitaniae | HC 7524-38 | − |
| C. globosa | HC 438-88 | − |
| C. famata | HC 3771-88 | − |
| Saccharomyces cerevisiae | HC 5056-88 | − |
| Kloeckera japonica | HC 0618-48 | − |
| Rhodotorula glutinis | HC 7360-38 | − |
| Geotricum capitatum | HC 8488-20 | − |
| Trichosporon asashii | HC 7142-65 | − |
| Trichosporon mucoide | HC 1359-38 | − |
| Cryptococcus humicola | HC 7750-38 | − |
| Cryptococcus neoformans | HC 7613-95 | − |
| Cryptococcus laurentii | HC 5737-38 | − |
| Candida sp. | HC 6607-36 | − |
| Candida sp. | HC 0454-88 | − |
| Candida sp. | HC 8228-88 | − |
| Candida sp. | HC 6396-30 | − |
| Candida sp. | HC 5539-38 | − |
T, type strain; ATCC, American Type Culture Collection; HC, Hospital Universitario Nuestra Señora de Candelaria Cuture Collection, Tenerife, Canary Islands, Spain.
+, PCR product obtained; −, PCR product not obtained.
The ITS sequences of the three strains of C. nivariensis revealed that they do not have intraspecies variation, although further studies including new strains whenever detected will display a more reliable variation measure. However, the interspecies variation of C. nivariensis with other Candida species is remarkable. Indeed, the assay based on our newly designed primer set was optimized to yield the expected band for C. nivariensis but not for any of the other species examined.
The species-specific primers for C. nivariensis presented here provide a molecular diagnostic method that can be used, in conjunction with current clinical tools, for the diagnosis of C. nivariensis infections with greater confidence and accuracy.
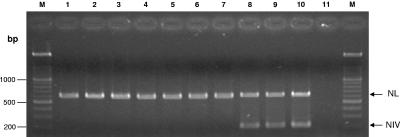
Once the specific PCR for C. nivariensis was optimized, we approached the development of a multiplex PCR assay for detection of all fungal species and specific identification of C. nivariensis. In this respect, we performed a double amplification of the D1/D2 (large subunit rRNA gene) and NIV fragments. Figure 1 shows an agarose gel illustrating typical results obtained with the optimized multiplex PCR assay.
FIG. 1.
Agarose gel electrophoresis of multiplex PCR amplification products D1/D2 (NL) and ITS1 (NIV). Lanes: M, 100-bp DNA ladder (Roche Diagnostics, Mannheim, Germany); 1, C. albicans ATCC 90028; 2, C. glabrata ATCC 90030; 3, C. krusei ATCC 6258; 4, C. parasilopsis ATCC 22019; 5, C. lusitaniae; 6, C. dubliniensis; 7, C. tropicalis; 8 to 10, C. nivariensis; 11, control without a DNA template.
Amplification of the D1/D2 and NIV targets produced easily identifiable bands consistent with their respective molecular sizes (650 and 206 bp, respectively). The NIV fragments were always amplified in the case of C. nivariensis strains but not in the case of infections by other Candida spp. The D1/D2 fragment was detected in all yeast strains.
To understand the clinical significance and epidemiological role of C. nivariensis, it is very important to correctly identify this yeast in clinical specimens. The method reported is a very reliable assay for this purpose.
Acknowledgments
This research was supported by Project BIO 2002/00953 from the Ministerio de Educacion y Ciencia (Spain) to S.M.-A. (partially supported by FIS contract 99/3060) and PI62/02 from the Fundación Canaria de Investigación y Salud (FUNCIS). S.M.-A. is an Associated Scientist of the Centro de Investigaciones Biológicas (CIB), Consejo Superior de Investigaciones Científicas (CSIC), Madrid, Spain.
REFERENCES
- 1.Alcoba-Flórez, J., S. Méndez-Álvarez, J. Cano, J. Guarro, E. Pérez-Roth, and M. P. Arévalo. 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 43:4107-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, Y. C., J. D. Eisner, and M. M. Kattar. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2303-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurtzman, C. P., and H. J. Phaff. 1987. Molecular taxonomy, p. 63-94. In A. H. Rose and J. S. Harrison (ed.), The yeasts, vol. 1: biology of yeast. Academic Press, London, England.
- 4.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurtzman, C. P., and J. W. Fell. 1998. Summary of species characteristics, p.915-947. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, a taxonomic study, 4th ed. Elsevier Science B. V., Amsterdam, The Netherlands.
- 6.Louws, F. J., J. L. W. Rademaker, and F. J. de Bruijin. 1999. The three Ds of PCR-based genomic analysis of phytobacteria: diversity, detection, and disease diagnosis. Annu. Rev. Phytopathol. 37:81-125. [DOI] [PubMed] [Google Scholar]
- 7.Maaroufi, Y., N. Ahariz, M. Husson, and F. Crokaert. 2004. Comparison of different methods of isolation of DNA of commonly encountered Candida species and its quantitation by using a real-time PCR-based assay. J. Clin. Microbiol. 42:3159-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marr, K. A., and R. A. Bowden. 1999. Fungal infections in patients undergoing blood and marrow transplantation. Transpl. Infect. Dis. 1:237-246. [DOI] [PubMed] [Google Scholar]
- 9.Price, C. W., G. B. Fuson, and H. J. Phaff. 1978. Genome comparison in yeast systematics: delimitation of species within the genera Schwanniomyces, Saccharomyces, Debaryomyces, and Pichia. Microbiol. Rev. 42:161-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin, J. H., F. S. Nolte, and C. J. Morrison. 1997. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J. Clin. Microbiol. 35:1454-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin, J. H., F. S. Nolte, B. P. Holloway, and C. J. Morrison. 1999. Rapid identification of up to three Candida species in a single reaction tube by a 5′ exonuclease assay using fluorescent DNA probes. J. Clin. Microbiol. 37:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trama, J. P., E. Mordechai, and M. E. Adelson. 2005. Detection and identification of Candida vaginitis by real-time PCR and pyrosequencing. Mol. Cell. Probes 19:145-152. [DOI] [PubMed] [Google Scholar]