Abstract
A heterologous cluster of glycosyltransferase genes was identified in the three Moraxella catarrhalis LOS serotype strains. Multiple PCR primers designed to this region amplified products that differentiate between the serotypes more rapidly and efficiently than previously described serological analyses. This assay will be valuable for clinical and research-based studies.
Moraxella catarrhalis, a gram-negative diplococcus, is considered a significant cause of acute otitis media in children and lower respiratory infections in adults with chronic obstructive pulmonary disease (COPD) (12, 21, 29). A number of putative virulence factors have been described for M. catarrhalis (8, 11, 12, 17, 20), including a surface-exposed lipooligosaccharide (LOS) (6, 18, 23, 32). Structural and serological studies with M. catarrhalis have described only three different LOS serotypes (termed A, B, and C), which vary in length and content of the oligosaccharide branches (3-5, 13, 15, 28). One serological study by Vaneechoutte et al. grouped clinical isolates into serotypes A (60%), B (30%), and C (5%), with 5% of the strains unidentified (28). That has been the only study to investigate the prevalence of specific M. catarrhalis LOS serotypes in the population. The difficulties with serological determinations of M. catarrhalis LOS expression are the limited quantities of antibodies, the absolute requirement for purified sample, and the potential for cross-reactivity between serotypes A and C (13, 24, 25).
Recently, a cluster of three glycosyltransferase (lgt) genes were identified and characterized in a strain of M. catarrhalis 7169 expressing serotype B LOS (6). Primers 406 and 408 (Table 1) designed to this region were subsequently used in PCRs with chromosomal DNA from M. catarrhalis 25238, the previously defined LOS serotype A strain, and M. catarrhalis RS10, the previously defined LOS serotype C strain (Table 2) (3, 4). PCR was performed in 50-μl reaction mixtures containing PCR SuperMix (Invitrogen, Carlsbad, CA), 20 pmol/μl of each primer, and 1 μl chromosomal DNA prepared as previously described (26). Amplifications were performed in a GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol for 25 cycles with an annealing temperature of 53.1°C and extension time of 4 min. Primers 406 and 408 (Table 1) produced an amplicon of 4.3 kb in serotype A and C strains, which was 1 kb larger than the product amplified in the serotype B strain, 7169 (data not shown).
TABLE 1.
Nucleotide sequence of oligonucleotide primers used for PCR-based LOS typing and sequencing in this study
| Primer | Sequencea | Brief description |
|---|---|---|
| Pr 406 | CAAAAGAAGACAAACAAGCAGC | Primer (sense) designed for sequencing the entire lgt cluster and flanking DNA in all LOS serotype strains; multiplex PCR primer used to distinguish between different LOS serotypes (6) |
| Pr 408 | CATCAAAAACCCCCCTACC | Multiplex PCR primer (antisense) used to distinguish between different LOS serotypes |
| Pr 649 | ATCCTGCTCCAACTGACTTTC | Multiplex PCR primer (sense) used to distinguish between different LOS serotypes (primer only binds in LOS serotype A strains) |
| Pr 704 | GCCACCAAACTATTCACGC | Primer (antisense) designed for sequencing the entire lgt cluster and flanking DNA (downstream of lgt3) in all LOS serotype strains |
All primers are listed in the 5′ to 3′ direction.
TABLE 2.
M. catarrhalis strains used in this study
| Strain | Description | Source and/or reference |
|---|---|---|
| M. catarrhalis strains | ||
| 3P3B1 | Adult COPD sputum isolate | Timothy Murphy (VA Medical Center, Buffalo, N.Y.) (1) |
| 5P26B1 | Adult COPD sputum isolate | Timothy Murphy (VA Medical Center, Buffalo, N.Y.) |
| 7P94B1 | Adult COPD sputum isolate | Timothy Murphy (VA Medical Center, Buffalo, N.Y.) |
| 18P6B1 | Adult COPD sputum isolate | Timothy Murphy (VA Medical Center, Buffalo, N.Y.) |
| 12P80B1 | Adult COPD sputum isolate | Timothy Murphy (VA Medical Center, Buffalo, N.Y.) |
| 51P9B1 | Adult COPD sputum isolate | Timothy Murphy (VA Medical Center, Buffalo, N.Y.) (1, 22) |
| HF-006 | Pediatric middle-ear isolate | Howard Faden (Children's Hospital, Buffalo, N.Y.) (7) |
| HF-039 | Pediatric middle-ear isolate | Howard Faden (Children's Hospital, Buffalo, N.Y.) |
| HF-165 | Pediatric middle-ear isolate | Howard Faden (Children's Hospital, Buffalo, N.Y.) |
| HF-218 | Pediatric middle-ear isolate | Howard Faden (Children's Hospital, Buffalo, N.Y.) |
| HF-2246 | Pediatric middle-ear isolate | Howard Faden (Children's Hospital, Buffalo, N.Y.) |
| 7169 | Pediatric middle-ear isolate; LOS type B strain | Howard Faden (Children's Hospital, Buffalo, N.Y.) (6, 9, 18, 19) |
| 2951 | Nasopharyngeal isolate; LOS type A strain | Mike Apicella (Iowa City, Iowa) (32) |
| 27335 | Unknown origin isolate from Paris, France | Mark Achtman (Max-Planck Institute, Germany) |
| 035E | Middle-ear fluid isolate | Eric Hansen (Dallas, Tex.) (10) |
| ATCC 25238 | LOS serotype A control strain | American Type Culture Collection (ATCC) (4) |
| ATCC 43617 | Transtracheal aspirate | ATCC (30) |
| CCUG 3292 | LOS serotype B control strain | Culture Collection, University of Göteborg, Göteborg, Sweden (CCUG) (5) |
| CCUG 26391 | LOS serotype C strain | CCUG (14) |
| RS10 | LOS serotype C control strain | Motiur Rahman (3) |
| Non-M. catarrhalis strains | ||
| Acinetobacter baumanni strain 19606 | ATCC | |
| Moraxella bovis strain 10900 | ATCC | |
| Moraxella caviae strain 14659 | ATCC | |
| Moraxella nonliquefaciens strain 17593 | ATCC | |
| Moraxella osloensis strain 15276 | ATCC | |
| Neisseria cinerea strain 658 | Dave Dyer (Oklahoma City, Okla.) | |
| Neisseria gonorrhoeae (two strains) strain GC6 and GC10 | Dave Dyer (Oklahoma City, Okla.) | |
| Neisseria meningitidis strain 121 | Dave Dyer (Oklahoma City, Okla.) | |
| Haemophilus ducreyi (two strains) strains 35000 and CIP542 | 2 | |
| Haemophilus influenzae (two strains) strains 7502 and 2019 | Timothy Murphy (VA Medical Center, Buffalo, N.Y.) | |
| Haemophilus parainfluenzae strain 9P20 | Timothy Murphy (VA Medical Center, Buffalo, N.Y.) | |
| Klebsiella pneumoniae strain 94-339-0352 | Thomas Russo (Buffalo, N.Y.) | |
| Enterobacter aerogenes strain 94-347-0274 | Thomas Russo (Buffalo, N.Y.) | |
| Escherichia coli (two strains) strain CP9 and XL-1 Blue | Thomas Russo (Buffalo, N.Y.) (27) and Stratagene (La Jolla, Calif.) | |
| Pseudomonas aeruginosa strain 94-343-0448 | Thomas Russo (Buffalo, N.Y.) | |
| Proteus mirabilis strain 94-341-0610 | Thomas Russo (Buffalo, N.Y.) |
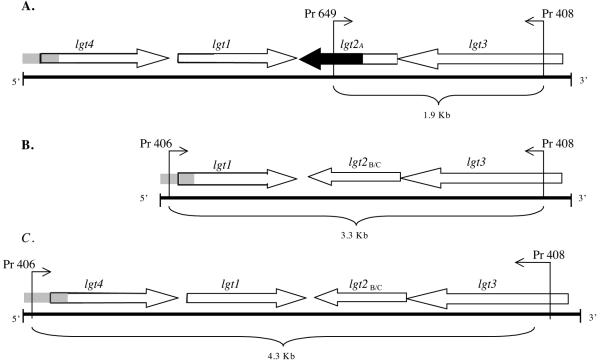
Sequence analyses (MacVector 7.2 software; Accelrys, San Diego, CA) of the entire region amplified by primer 406 and flanking primer 704 (Table 1) identified an additional open reading frame upstream of the original lgt cluster described in 7169 and in the same orientation as lgt1, as illustrated in Fig. 1A and C. A ClustalW alignment with the translated sequence of this open reading frame (Lgt4) in both strains revealed 46% identity and 60% similarity to Lgt1, an α(1-2) glucosyltransferase in M. catarrhalis 7169 (6). The 5′ end of this cluster in serotype A and C strains contained DNA sequence that is homologous to the 5′ region of the lgt cluster in 7169, as depicted in all three clusters in Fig. 1A to C. Additional sequence analysis of the cluster in serotype A-expressing strains revealed a region of sequence divergence located at the 3′ end of lgt2A, depicted in Fig. 1A. A ClustalW alignment with the translated sequence of Lgt2A revealed 61% identity and 75% similarity to Lgt2B/C (formally Lgt2), a β(1-4) galactosyltransferase in M. catarrhalis 7169 (6). A ClustalW alignment with the translated sequences of Lgt1 and Lgt3 from both serotypes A and C revealed ≥98% identity to the respective homologous Lgt enzymes identified in M. catarrhalis 7169 (6). These observations were confirmed by sequencing the entire lgt cluster in a second serotype C strain, CCUG 26391, and another serotype A strain, 27335 (Table 2) (data not shown).
FIG. 1.
Organization of lgt clusters in strains of M. catarrhalis expressing different LOS serotypes and binding sites of the primers used in the multiplex PCRs. (A) Representative (putative) lgt cluster in a LOS serotype A genome from M. catarrhalis strain ATCC 25238 (NCBI accession number DQ137417). (B) Representative lgt cluster in a LOS serotype B genome from M. catarrhalis strain 7169 (Edwards, 2005). (C) Representative (putative) lgt cluster in a LOS serotype C genome from M. catarrhalis strain RS10 (NCBI accession number DQ137418). The 5′ end of the cluster in serotype A and C strains contained DNA sequence that is homologous to the 5′ region of the lgt cluster in 7169, as depicted by gray shading in all three clusters (A to C). Black sequence portion in panel A represents a region of sequence divergence at the 3′ end of lgt2A.
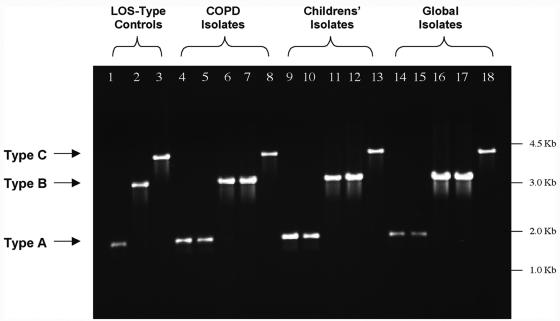
Due to the homologous DNA sequence 5′ of the lgt cluster in all three serotype strains, we implemented a strategy of multiplex PCR in which primers 406 and 649 were the forward primers for serotype B/C strains and serotype A strains, respectively, and primer 408 was the reverse primer for all three serotypes, as depicted in Fig. 1. This single PCR produces amplicons of 1.9 kb, 3.3 kb, and 4.3 kb for serotypes A, B, and C, respectively. Figure 2 (lanes 1 to 3) is a representative agarose gel depicting the amplicons resulting from this multiplex PCR using chromosomal DNA from M. catarrhalis LOS serotypes A, B, and C, the control strains ATCC 25238, 7169, and RS10, respectively (Table 2) (3, 4, 6). Other M. catarrhalis clinical isolates (Table 2) were also tested, some representing a geographically diverse panel (Fig. 2, lanes 14 to 18), as well as isolates from a local population of adults with COPD (Fig. 2, lanes 4 to 8) and children with otitis media with effusion (OME) (Fig. 2, lanes 9 to 13). Overall, 152 M. catarrhalis strains were typed using this system, and the results of these reactions are summarized in Table 3. The percentage of strains expressing serotype A, B, or C LOS was 64%, 30%, and 6%, respectively, which is consistent with the previous data (13, 28). However, the percentage of M. catarrhalis isolates expressing serotype A LOS was relatively high for the COPD isolates (81%) and relatively low for the diverse global isolates (46%). At this time it is difficult to determine the significance of this observation, and further analyses are needed to determine whether LOS serotype is a factor for tissue tropism (i.e., inner ear or lung), colonization, or subsequent infection.
FIG. 2.
Representative multiplex PCR test using primers 406, 408, and 649 (Table 1) with chromosomal DNA isolated from various M. catarrhalis isolates. Lanes 1 to 3 show amplicons from the three LOS serotype control strains: LOS serotype A, ATCC 25238 (lane 1); LOS serotype B, 7169 (lane 2); and LOS serotype C, RS10 (lane 3). Lanes 4 to 8 contain amplicons from five COPD patients' isolates: 3P3B1 (lane 4), 51P9B1 (lane 5), 5P26B1 (lane 6), 18P6B1 (lane 7), and 12P80B1 (lane 8). Lanes 9 to 13 contain amplicons from five children's OME isolates: HF-039 (lane 9), HF-2246 (lane 10), HF-165 (lane 11), HF-218 (lane 12), and HF-006 (lane 13). Lanes 14 to 18 contain amplicons from five geographically diverse (global) isolates: 2951 (lane 14), 035E (lane 15), CCUG 3292 (lane 16), ATCC 43617 (lane 17), and CCUG 26391 (lane 18).
TABLE 3.
LOS serotypes expressed by M. catarrhalis strains isolated from different populations of patients as determined by multiplex PCR
| Origin | No. isolates expressing serotype/ total isolates (%)
|
||
|---|---|---|---|
| A | B | C | |
| COPD patient isolates | 42/52 (81) | 8/52 (15) | 2/52 (4) |
| Children (OME isolates) | 32/50 (64) | 17/50 (34) | 1/50 (2) |
| Global (geographically diverse) strains | 23/50 (46) | 20/50 (40) | 7/50 (14) |
| Total | 97/152 (64) | 45/152 (30) | 10/152 (6) |
In order to confirm that the multiplex PCR method specifically correlated with the LOS serotype expressed by clinical isolates, compositional analysis and mass spectroscopy were performed as described previously (16). LOS was purified from a selected group of M. catarrhalis clinical isolates representing the three major serotypes, and oligosaccharides were isolated as described previously (6, 31). A portion (100 μg) of each OS was methanolyzed, trimethylsilylated, and analyzed by gas chromatography-mass spectrometry (MS). The glycosyl composition analyses for the OSs from M. catarrhalis strains 3P3B1 and 12P80B1 detected galactose, glucose, 3-deoxy-d-manno-octulosonic acid (KDO), and 2-acetamido-2-deoxy glucose as the major monosaccharide constituents, whereas the M. catarrhalis strain 43617 OS contained galactose, glucose, and KDO as the major monosaccharide constituents, with the absence of 2-acetamido-2-deoxy glucose residues (Table 4).
TABLE 4.
Ions observed from MALDI-TOF MS analyses of OSs from LOS isolated from different strains of M. catarrhalis and their proposed composition
| Oligosaccharide | Ion [M-H]− | Proposed composition |
|---|---|---|
| 3P3B1 (serotype A) | 1736 | Gal3Glc5GlcNAc1KDO |
| 1574 | Gal2Glc5GlcNAc1KDO | |
| 1412 | Gal1Glc5GlcNAc1KDO | |
| 43617 (serotype B) | 1858 | Gal4Glc6KDO |
| 1695 | Gal3Glc6KDO | |
| 1533 | Gal2Glc6KDO | |
| 1371 | Gal1Glc6KDO | |
| 12P80B1 (serotype C) | 1898 | Gal4Glc5GlcNAc1KDO |
| 1736 | Gal3Glc5GlcNAc1KDO | |
| 1574 | Gal2Glc5GlcNAc1KDO |
Methylation analysis of the strain 3P3B1 OS revealed the presence of terminal galactopyranose, terminal glucopyranose, 2-linked glucopyranose, 4-linked glucopyranose, 4-linked galactopyranose, 2,3,6-linked glucopyranose, and terminal 2-acetamido-2-deoxy glucopyranoside, indicating that 3P3B1 expresses serotype A LOS. Analysis of the strain 12P80B1 OS revealed the same profile as 3P3B1, except this sample contained 4-linked 2-acetamido-2-deoxy glucopyranoside rather than terminal 2-acetamido-2-deoxy glucopyranoside, indicating that 12P80B1 expresses serotype C LOS. Analysis of the strain 43617 OS revealed the presence of terminal galactopyranose, terminal glucopyranose, 2-linked glucopyranose, 4-linked glucopyranose, 4-linked galactopyranose, and 2,3,6-linked glucopyranose, with the absence of the 2-acetamido-2-deoxy glucopyranoside residues, indicating that 43617 expresses serotype B LOS. Analyses of the molecular masses of the OSs by negative-ion matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS, using dihydroxybenzoic acid as the matrix, as previously described (16), were heterogeneous and consistent with the composition and linkage analyses (data not shown). The results of the mass spectrometric analysis of the OSs are summarized in Table 4. These structural analyses were also performed with M. catarrhalis clinical isolates 035E (serotype A), 5P26B1 (serotype B), and CCUG 26391 (serotype C) (strains referenced in Table 1) and were consistent with the above analyses (data not shown).
The species specificity of the multiplex PCR assay was also evaluated with chromosomal DNA isolated from 20 strains representing 16 different species, within the genus Moraxella, as well as other mucosal pathogens and commensal organisms (Table 2). The LOS serotype-specific primers did not amplify products from any of the 20 heterologous strains tested in this study (data not shown).
In conclusion, the results in this study indicate that this multiplex PCR test is a specific and highly effective assay for the identification of LOS serotypes expressed by M. catarrhalis strains. In addition, this system may have the added advantage of identifying a clinical isolate as an M. catarrhalis strain, although more data are needed. A single-step PCR method may prove to be a useful tool for basic research studies designed to determine the role of LOS in relation to colonization and pathogenesis, as well as provide a more specific means for clinical investigation of the type of M. catarrhalis strains infecting specific patients.
Acknowledgments
This research was supported by Public Health Service research grants AI46422 and DC005837 (A.A.C.). K.J.E. is also supported as a graduate student fellow by NIH training grant AI07614.
We thank Timothy Murphy for supplying the M. catarrhalis clinical isolates from the COPD clinic at the VA Medical Center in Buffalo, N.Y. We also thank Howard Faden for supplying the children's M. catarrhalis isolates from the Women and Children's Hospital in Buffalo, N.Y.
REFERENCES
- 1.Bakri, F., A. L. Brauer, S. Sethi, and T. F. Murphy. 2002. Systemic and mucosal antibody response to Moraxella catarrhalis after exacerbations of chronic obstructive pulmonary disease. J. Infect. Dis. 185:632-640. [DOI] [PubMed] [Google Scholar]
- 2.Campagnari, A. A., R. Karalus, M. Apicella, W. Melaugh, A. J. Lesse, and B. W. Gibson. 1994. Use of pyocin to select a Haemophilus ducreyi variant defective in lipooligosaccharide biosynthesis. Infect. Immun. 62:2379-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edebrink, P., P. E. Jansson, M. M. Rahman, G. Widmalm, T. Holme, and M. Rahman. 1995. Structural studies of the O-antigen oligosaccharides from two strains of Moraxella catarrhalis serotype C. Carbohydr. Res. 266:237-261. [DOI] [PubMed] [Google Scholar]
- 4.Edebrink, P., P. E. Jansson, M. M. Rahman, G. Widmalm, T. Holme, and M. Rahman. 1994. Structural studies of the O-polysaccharide from the lipopolysaccharide of Moraxella (Branhamella) catarrhalis serotype A (strain ATCC 25238). Carbohydr. Res. 257:269-284. [DOI] [PubMed] [Google Scholar]
- 5.Edebrink, P., P. E. Jansson, G. Widmalm, T. Holme, and M. Rahman. 1996. The structures of oligosaccharides isolated from the lipopolysaccharide of Moraxella catarrhalis serotype B, strain CCUG 3292. Carbohydr. Res. 295:127-146. [DOI] [PubMed] [Google Scholar]
- 6.Edwards, K. J., S. Allen, B. W. Gibson, and A. A. Campagnari. 2005. Characterization of a cluster of three glycosyltransferase enzymes essential for Moraxella catarrhalis lipooligosaccharide assembly. J. Bacteriol. 187:2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faden, H., J. Hong, and T. Murphy. 1992. Immune response to outer membrane antigens of Moraxella catarrhalis in children with otitis media. Infect. Immun. 60:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsgren, A., M. Brant, M. Karamehmedovic, and K. Riesbeck. 2003. The immunoglobulin D-binding protein MID from Moraxella catarrhalis is also an adhesin. Infect. Immun. 71:3302-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furano, K., and A. A. Campagnari. 2003. Inactivation of the Moraxella catarrhalis 7169 ferric uptake regulator increases susceptibility to the bactericidal activity of normal human sera. Infect. Immun. 71:1843-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helminen, M. E., I. Maciver, J. L. Latimer, J. Klesney-Tait, L. D. Cope, M. Paris, G. H. McCracken, Jr., and E. J. Hansen. 1994. A large, antigenically conserved protein on the surface of Moraxella catarrhalis is a target for protective antibodies. J. Infect. Dis. 170:867-872. [DOI] [PubMed] [Google Scholar]
- 11.Holm, M. M., S. L. Vanlerberg, I. M. Foley, D. D. Sledjeski, and E. R. Lafontaine. 2004. The Moraxella catarrhalis porin-like outer membrane protein CD is an adhesin for human lung cells. Infect. Immun. 72:1906-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holm, M. M., S. L. Vanlerberg, D. D. Sledjeski, and E. R. Lafontaine. 2003. The Hag protein of Moraxella catarrhalis strain O35E is associated with adherence to human lung and middle ear cells. Infect. Immun. 71:4977-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holme, T., M. Rahman, P. E. Jansson, and G. Widmalm. 1999. The lipopolysaccharide of Moraxella catarrhalis structural relationships and antigenic properties. Eur. J. Biochem. 265:524-529. [DOI] [PubMed] [Google Scholar]
- 14.Hu, W. G., J. Chen, J. C. McMichael, and X. X. Gu. 2001. Functional characteristics of a protective monoclonal antibody against serotype A and C lipooligosaccharides from Moraxella catarrhalis. Infect. Immun. 69:1358-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jèonsson, I., T. Holme, A. Krook, M. Rahman, and M. Thorāen. 1992. Variability of surface-exposed antigens of different strains of Moraxella catarrhalis. Eur. J. Clin. Microbiol. Infect. Dis. 11:919-922. [DOI] [PubMed] [Google Scholar]
- 16.Kahler, C. M., A. Datta, Y. L. Tzeng, R. W. Carlson, and D. S. Stephens. 2005. Inner core assembly and structure of the lipooligosaccharide of Neisseria meningitidis: capacity of strain NMB to express all known immunotype epitopes. Glycobiology 15:409-419. [DOI] [PubMed] [Google Scholar]
- 17.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luke, N. R., S. Allen, B. W. Gibson, and A. A. Campagnari. 2003. Identification of a 3-deoxy-d-manno-octulosonic acid biosynthetic operon in Moraxella catarrhalis and analysis of a KdsA-deficient isogenic mutant. Infect. Immun. 71:6426-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luke, N. R., and A. A. Campagnari. 1999. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect. Immun. 67:5815-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luke, N. R., A. J. Howlett, J. Shao, and A. A. Campagnari. 2004. Expression of type IV pili by Moraxella catarrhalis is essential for natural competence and is affected by iron limitation. Infect. Immun. 72:6262-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy, T. F., A. L. Brauer, B. J. Grant, and S. Sethi. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172:195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy, T. F., C. Kirkham, E. DeNardin, and S. Sethi. 1999. Analysis of antigenic structure and human immune response to outer membrane protein CD of Moraxella catarrhalis. Infect. Immun. 67:4578-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng, D., B. P. Choudhury, R. S. Petralia, R. W. Carlson, and X. X. Gu. 2005. Roles of 3-deoxy-d-manno-2-octulosonic acid transferase from Moraxella catarrhalis in lipooligosaccharide biosynthesis and virulence. Infect. Immun. 73:4222-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman, M., and T. Holme. 1996. Antibody response in rabbits to serotype-specific determinants in lipopolysaccharides from Moraxella catarrhalis. J. Med. Microbiol. 44:348-354. [DOI] [PubMed] [Google Scholar]
- 25.Rahman, M., A. B. Jonsson, and T. Holme. 1998. Monoclonal antibodies to the epitope α-Gal-(1-4)-β-Gal-(1- of Moraxella catarrhalis) LPS react with a similar epitope in type IV pili of Neisseria meningitidis. Microb. Pathog. 24:299-308. [DOI] [PubMed] [Google Scholar]
- 26.Russo, T. A., J. E. Guenther, S. Wenderoth, and M. M. Frank. 1993. Generation of isogenic K54 capsule-deficient Escherichia coli strains through TnphoA-mediated gene disruption. Mol. Microbiol. 9:357-364. [DOI] [PubMed] [Google Scholar]
- 27.Russo, T. A., J. S. Thompson, V. G. Godoy, and M. H. Malamy. 1990. Cloning and expression of the Bacteroides fragilis TAL2480 neuraminidase gene, nanH, in Escherichia coli. J. Bacteriol. 172:2594-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaneechoutte, M., G. Verschraegen, G. Claeys, and A. M. Van Den Abeele. 1990. Serological typing of Branhamella catarrhalis strains on the basis of lipopolysaccharide antigens. J. Clin. Microbiol. 28:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace, R. J., Jr., V. A. Steingrube, D. R. Nash, D. G. Hollis, C. Flanagan, B. A. Brown, A. Labidi, and R. E. Weaver. 1989. BRO beta-lactamases of Branhamella catarrhalis and Moraxella subgenus Moraxella, including evidence for chromosomal beta-lactamase transfer by conjugation in B. catarrhalis, M. nonliquefaciens, and M. lacunata. Antimicrob. Agents Chemother. 33:1845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.York, W. S., A. G. Darville, M. McNeil, T. T. Stevenson, and P. Albersheim. 1985. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118:3-40. [Google Scholar]
- 32.Zaleski, A., N. K. Scheffler, P. Densen, F. K. Lee, A. A. Campagnari, B. W. Gibson, and M. A. Apicella. 2000. Lipooligosaccharide Pk (Galα1-4Galβ1-4Glc) epitope of Moraxella catarrhalis is a factor in resistance to bactericidal activity mediated by normal human serum. Infect. Immun. 68:5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]