Abstract
The expression of nitrogenase genes of Herbaspirillum sp. B501 associated in shoot (leaf and stem) of wild rice, Oryza officinalis, was studied by means of reverse transcription-PCR (RT-PCR) targeted at the nifH gene. RT-PCR analyses indicate that nifH transcript was detected exclusively from nitrogen-fixing cells of gfp-tagged strain B501gfp1 in both free-living and endophytic states by using a constitutive gfp gene transcript as a positive control. Transcription of nifH and nitrogen fixation in free-living cells were induced maximally at a 2% O2 concentration and repressed in free air (21% O2). nifH transcription was monitored in the endophytic cells by using total RNA extracted from B501gfp1-inoculated wild rice plants during daily light-dark cycles. The level of nifH transcription in planta varied dramatically, with a maximum during the light period. Moreover, the light radiation enhanced nifH expression even in free-living cells grown in culture. These results suggest that in planta nitrogen fixation by the endophyte shows a daily rhythm determined by the plant's light environment.
The availability of fixed nitrogen limits primary productivity in plant ecosystems (6). During their evolution, legumes have developed a symbiotic partnership with rhizobia that can fix atmospheric nitrogen and can thus attain the necessary fixed nitrogen. Among nonleguminous plants, several genera of diazotrophs have been isolated and characterized as nitrogen-fixing endophytes, including Gluconacetobacter (2, 4, 32), Azoarcus (7, 8, 13), Klebsiella (14), Herbaspirillum (2, 4, 9, 18), Azospirillum (9, 10), and Clostridium (23, 24).
To determine the contribution of fixed nitrogen to the host plants by these species (17), the long-term amount of endophyte-dependent nitrogen fixation has been studied by means of the incorporation of 15N2 gas (9, 13, 32) and the 15N isotope dilution method (14). Recently, expression of the nifH gene has been studied by means of reverse transcription-PCR (RT-PCR) to evaluate the nitrogen-fixing activity of endophytic Azoarcus sp. (13) and those of nitrogen-fixing microbial communities in the termite gut (25), soil (5), and marine water (37). The nifH gene encodes the Fe protein (dinitrogenase reductase) and is one of the nitrogenase structural genes (nifHDK) (27). Transcription of nif genes is strictly regulated by levels of molecular oxygen and fixed nitrogen to minimize unnecessary energy consumption (6). Thus, there are tight relationships between nitrogen-fixing activity and nif gene transcription (6, 7, 13).
In performing transcription analysis, the Northern and dot blot hybridization methods are insufficiently sensitive to detect low copy numbers of transcripts (11). Indeed, the densities of endophytic bacteria are usually <107 cells per gram (fresh weight) of the host plant (9, 18). In contrast, RT-PCR is highly sensitive and thus is probably suitable for transcription analysis of endophytes residing at low densities in plant tissues. In addition, real-time PCR systems using a fluorescent dye (SYBR Green I) that binds with double-stranded DNA are becoming popular methods to quantify mRNA by RT reaction (12).
Because endophytic microorganisms depend on an energy supply from their host plants, their metabolic functions should be affected by variations in the physiological properties of their hosts, such as rates of photosynthesis. However, plant photosynthesis might increase O2 concentrations around the endophyte and thereby repress expression of the endophyte's nif structural genes during the light period.
Shoot-associated Herbaspirillum sp. B501 is a diazotrophic endophyte that is capable of fixing nitrogen in leaves and stems of wild rice, Oryza officinalis W0012 (9). Thus, this bacterium represents a suitable system in which to examine whether nitrogen fixation by endophytes is subject to variations in the physiological status of the host plant. The aim of the present study was to understand the lifestyle of an endophyte, Herbaspirillum sp. B501gfp1, within its host plant in terms of the host's physiological functions and environments. To accomplish this, we developed a method for monitoring nifH transcription of the endophytic bacterium by means of RT-PCR.
MATERIALS AND METHODS
Bacterial strains, plasmids, plant material, and growth media.
The bacterial strains and plasmids used in our study are listed in Table 1. Herbaspirillum sp. B501 was previously isolated from wild rice species as a nitrogen-fixing endophyte and tagged with the gfp gene for observation of bacterial colonization in rice plants (9). Herbaspirillum sp. strains were grown in nutrient broth (NB) medium or modified Rennie (MR) medium, as described previously (9, 29). For growth under defined oxygen concentrations, strain B501gfp1 (a gfp-tagged strain of Herbaspirillum sp. B501) was grown in MR medium to an optical density at 660nm of 0.05, and the culture (1 ml) was added to 300 ml of fresh MR medium in a 500-ml flask. The medium was incubated at 30°C while being bubbled with gas filtered through a membrane filter with a pore size of 0.2 μm (Toyo Roshi Kaisha, Ltd., Tokyo, Japan): the gas was either free air (21.0% O2) or a mixture of O2 and N2 with O2 concentrations of 0.0, 0.2, 0.4, 0.6, 1.0, 2.0, 5.0, 8.0, or 12.0% (vol/vol; Tomoe Shokai Co., Ltd., Tokyo, Japan). For all gases, the flow rate was 100 ml/min. A species of wild rice, Oryza officinalis W0012, was used, as described previously (9). Semisolid agar containing nitrogen-free nutrients was used for rice growth as described previously (21). Cell numbers of strain B501gfp1 in culture were counted with a bacterial counting chamber by microscopy (9, 24). To estimate the cell-based nitrogen fixation and nif transcript concentration of endophytic strain B501gfp1, the cell number in plant macerate was determined by plate counts of green colonies by using selective NB medium containing kanamycin and a fluorescence stereomicroscope (9).
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| Herbaspirillum sp. strain B501 | Wild type | 9 |
| Herbaspirillum sp. strain B501gfp1 | Herbaspirillum sp. strain B501, gfp; Kmr | 9 |
| E. coli DH5 | recA; cloning strain | Toyobo Co., Ltd. |
| E. coli S17-1 | recA; Smr | 33 |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector; Apr | Takara Bio, Inc. |
| pB501nifH1 | pBluescript II SK(+) carrying a 2.9-kb EcoRI/PstI fragment of nifHD; Apr | This study |
Apr, ampicillin resistant; Kmr, kanamycin resistant; Smr, streptomycin resistant.
Cloning and sequencing of nifH.
DNA manipulations, including plasmid isolation, digestion, and transformation, were performed as described by Sambrook et al. (30). Total DNA was prepared from Herbaspirillum sp. B501 cells grown in NB (15). Southern hybridization analysis of B501 DNA was carried out as described previously (15). For the probe, a 0.4-kb DNA fragment was amplified from the B501 DNA by means of PCR using primers 19F and 407R, whose sequences are highly conserved in nifH genes from various organisms (34); these primers are described in Table 2. We then purified a 2.9-kb DNA fragment, which corresponded to the signal detected in the Southern hybridization, from the EcoRI/PstI-digested B501 DNA using the QIAEX II DNA extraction kit (QIAGEN GmbH, Hilden, Germany); it was then ligated to the EcoRI/PstI-digested pBluescript II SK(+) and transformed into Escherichia coli DH5α. The resulting colonies were lifted onto nylon membranes (Amersham Biosciences Corp., Piscataway, NJ), and screened by hybridization using the 0.4-kb DNA fragment of the nifH gene. As a result, we obtained a positive clone and designated the contained plasmid pB501nif1. The 2.9-kb insert was sequenced on both strands by using primers −21 M13, M13 Rev, nif370, nif370-1, nif370-2, nif370-3, nif370-4, nif270, nif270-1, nif270-2, nif270-3, and nif270-5 (Table 2). DNA sequencing was performed using the ABI PRISM Big Dye Terminator Cycle Sequencing kit and a 370 DNA sequencer (Applied Biosystems, Inc., Foster City, CA). The nucleotide sequence of the 2.9-kb DNA fragment appears in the DDBJ database under accession number AB196476.
TABLE 2.
Primers used in the present study
| Application | Primer and sequencea | Reference or source |
|---|---|---|
| Cloning | ||
| 19F | 5′-GCIWTYTAYGGIAARGGIGG-3′ | 34 |
| 407R | 5′-AAICCRCCRCAIACIACRTC-3′ | 34 |
| Sequencing | ||
| −21 M13 | 5′-TGTAAAACGACGGCCAGT-3′ | Applied Biosystems |
| M13 Rev | 5′-CAGGAAACAGCTATGACC-3′ | Applied Biosystems |
| nif370 | 5′-TGTAACACCAGAAAGAAGTG-3′ | Present study |
| nif370-1 | 5′-GGACACCATCCTGTCGCTGG-3′ | Present study |
| nif370-2 | 5′-ACAAGGAACTGGAACTGGCC-3′ | Present study |
| nif370-3 | 5′-TAGCGCAGGTTTGTTTACC-3′ | Present study |
| nif370-4 | 5′-ACATCAGCCATGGTCCGGT-3′ | Present study |
| nif270 | 5′-CTCGACCCAGGGAAATGCC-3′ | Present study |
| nif270-1 | 5′-CTCGTCGACGATCTTTTCCA-3′ | Present study |
| nif270-2 | 5′-TTTCTTCAACTGTGAGGCTC-3′ | Present study |
| nif270-3 | 5′-AGTTGGCGTACTTCAAAATG-3′ | Present study |
| nif270-5 | 5′-TTATTGGTCTTCGTCTCA-3′ | Present study |
| RT-PCR | ||
| nif270-3 | 5′-AGTTGGCGTACTTCAAAATG-3′ | Present study |
| nif370-1 | 5′-GGACACCATCCTGTCGCTGG-3′ | Present study |
| gfp553 | 5′-TCAACTAGCAGACCATTATCAACA-3′ | Present study |
| gfp681 | 5′-ACCATGTGGTCTCTCTTTTCG-3′ | Present study |
| Real-time RT-PCR | ||
| nifH238 | 5′-ATCGGCTACCAGAACATC-3′ | Present study |
| nifH366 | 5′-GTAATCGGTGTCATCATAGG-3′ | Present study |
I represents inosine, R represents A or G, W represents A or T, and Y represents C or T.
Rice cultivation and inoculation with Herbaspirillum.
Dehulled seeds of Oryza officinalis W0012 were surface sterilized with 70% ethanol for 1 min and then washed five times with sterile distilled water. Then seeds were shaken in 1% NaOCl solution for 1 min and then washed again five times with sterile distilled water.
For seed inoculation, Herbaspirillum sp. B501gfp1 was cultured in NB medium, the cells were harvested when the culture reached an optical density of 1.0 at 660nm by means of centrifugation at 17,742 × g for 5 min at 4°C. The cells were washed twice with sterile distilled water and resuspended in sterilized 0.85% (wt/vol) NaCl solution. Sterilized rice seeds were placed on semisolid medium in a 350-ml plant box (Cul-JAR300; Iwaki, Tokyo, Japan), and a suspension of Herbaspirillum sp. B501gfp1 was added at a concentration of 106 cells per seed. The seeds were incubated in a plant growth cabinet (LH300; NK System Co., Ltd., Osaka, Japan) that provided 65 μmol photons m−2 s−1 of photosynthetically active radiation (400 to 700 nm) under a daily cycle of 16 h of light and 8 h of dark at 25°C.
Acetylene reduction assay.
The nitrogen-fixing activity of the endophyte was examined by means of acetylene reduction activity. To determine the activity of free-living bacteria, Herbaspirillum sp. B501gfp1 was grown in NB or MR medium at 30°C for 24 h, and then 9 ml of the culture was transferred into a 27-ml test tube sealed with a rubber stopper. A total of 0.9 ml of acetylene gas (purity of 99.9999% [vol/vol]; Toho Acetylene Co., Tokyo, Japan) was injected to provide a concentration of 5% (vol/vol), and the test tube was incubated for 12 or 24 h at 30°C.
To determine the nitrogenase activity under defined O2 concentrations, 5 ml of B501gfp1 culture grown under the flow of different O2/N2 gas mixtures was transferred into a 29-ml L-type tube sealed with a double rubber stopper in the presence of the O2/N2 gas mixtures. After acetylene gas was injected to a concentration of 5% (vol/vol), the tube was shaken with 100 stroke min−1 at 30°C for 2 min by a L-tube shaker (Monod-mini; Taitec Co., Ltd., Koshigaya, Japan).
To determine the in planta nitrogenase activity, plants inoculated with B501gfp1 were washed with sterile distilled water and then transferred into a 33-ml bottle sealed with a rubber septum. Acetylene gas (1.65 ml) was injected to provide a concentration of 5% (vol/vol), and the bottle was incubated for 12 or 24 h at 30°C in the dark.
Gas samples (each, 1 ml) from each tube and bottle were analyzed for the presence of ethylene with a Shimadzu GC-18A gas chromatograph equipped with a flame ionization detector and a Porapack R column as described previously (9).
Light and dark cultivation of free-living cells.
Herbaspirillum sp. B501gfp1 was grown in MR medium to an optical density of 0.05 at 660 nm, and the culture (3 ml) was added to 300 ml of fresh MR medium in a 500-ml flask. The flask was bubbled with a gas, a mixture of 2.0% (vol/vol) O2, under light and dark conditions at 25°C. A light-treated flask was illuminated by 65 μmol photons m−2 s−1 of photosynthetically active radiation (400 to 700 nm) using fluorescence lamps. A dark-treated flask was completely wrapped with black aluminum foil. The light and temperature were adjusted to the conditions where rice plants were cultivated. Culture was sampled 14 to 16 h after inoculation at an optical density at 660 nm of approximately 0.15 for RNA extraction.
RNA preparation.
To prepare total RNA of the cultured cells, 10 ml of culture was centrifuged at 17,742 × g for 10 min at 4°C. The bacterial pellet was homogenized by being vortexed in 1 ml of TRIzol reagent (Invitrogen Corp., Carlsbad, CA) for 1 min, and the solution was incubated at 60°C for 15 min. After 0.2 ml of chloroform was added, the mixture was centrifuged at 17,742 × g for 10 min. We collected the upper phase, added 0.2 ml of chloroform, and repeated the phase separation. The resulting upper phase was mixed with 0.5 ml of 2-propanol and the solution was centrifuged at 12,000 × g for 10 min at 4°C. The pellet was washed twice with 99.5% (vol/vol) ethanol, air dried, and then dissolved in 40 μl of sterile distilled water. We then added 5 μl of DNaseI (5 units/μl) and 5 μl of 10× buffer (Takara Bio, Inc., Otsu, Japan). The mixture was incubated at 37°C for 2 h, extracted with 50 μl of chloroform, and centrifuged at 14,700 × g for 10 min. RNA was recovered from the upper phase by 2-propanol precipitation, as described above. Each sample was examined for RNA concentration and purity using Gene Quant II (Amersham Bioscience, Piscataway, NJ) at 260, 280, and 320 nm and adjusted to the same RNA concentration for all samples.
To prepare the total RNA of an inoculated plant, we froze 0.03 to 0.10 g (fresh weight) of O. officinalis W0012 seedlings 7 days after inoculation with B501gfp1 in liquid nitrogen and ground the sample finely with a mortar and pestle. For time course experiments of nifH expression during light-dark cycles, we used the seedlings that were older than 7 days after inoculation. After 10 volumes of TRIzol reagent (Invitrogen Corp.) was added to the samples at room temperature, the solution was homogenized well using the mortar and pestle. The content was transferred into a fresh tube and incubated at 60°C for 15 min. The other procedures for RNA extraction and the DNaseI treatment were carried out as described above, except that we performed the chloroform extraction twice.
RT-PCR.
The RT reaction was carried out in a 20-μl solution containing 0.12 μg of total RNA with an Omniscript RT Kit (QIAGEN) according to the manufacturer's instructions. We then incorporated 2 μl of the resulting RT reaction mixture into the PCR. PCR was performed in a 50-μl solution with 30 to 40 cycles, eacho consisting of of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C using the HotStartTaq PCR kit (QIAGEN), following the manufacturer's instructions. Primer sets gfp533/gfp681 and nif370-1/nif270-3 (Table 2) were used to target the gfp and nifH genes, respectively. PCR products were analyzed by means of electrophoresis on Tris-borate-EDTA-agarose (2%) gels (Nippon Gene Co., Ltd., Toyama, Japan) stained with ethidium bromide (30).
Real-time PCR.
The primer set nifH238/nifH366 (Table 2) was designed to have the best performance in the real-time RT-PCR for amplifying the nifH gene with the Beacon Designer software (Premier Biosoft International, Palo Alto, CA.). We performed the real-time PCR with a QuantiTect SYBR Green RT-PCR kit (QIAGEN) and the i-Cycler optical system (Bio-Rad Laboratories, Inc., Tokyo, Japan), following the manufacturers' instructions. The 50-μl reaction mixture, containing 0.05 to 0.10 μg of total RNA, was incubated first at 50°C for 30 min to allow reverse transcription and then at 95°C for 15 min for initial activation of Taq polymerase, followed by 50 cycles, each consisting of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. At the final step, a melting-curve analysis of the PCR products was carried out to verify specific PCR amplification. The concentration of each nifH transcript was determined by plotting the threshold cycle value of its amplification against the standard curve, which we created by calibrating the values obtained from the EcoRI-digested pB501nif1 DNA used as templates at amounts ranging between 2.82 × 10−11 and 2.82 × 10−15 μmol.
RESULTS AND DISCUSSION
nifH sequence of Herbaspirillum sp. B501gfp1.
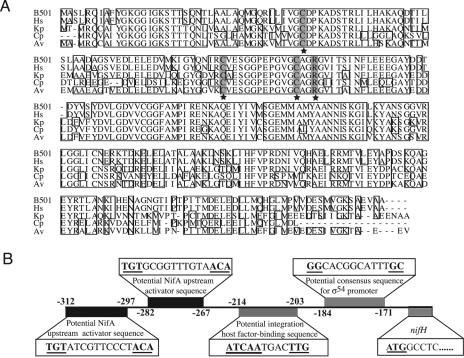
The 2.9-kb DNA fragment in pB501nifH1 (Table 1) contained nifH and truncated nifD genes with a nifH promoter region (accession number AB196476). The deduced amino acid sequence of nifH (292 amino acid residues) from Herbaspirillum sp. B501 was identical to that of Herbaspirillum seropedicae Z78 from sugarcane (Fig. 1A) (20). Functionally important residues were conserved (34): cysteine residues (Cys-40, Cys-87, and Cys-99) and one arginine residue (Arg-102) (Fig. 1A). The nifH promoter region of B501 had two potential NifA-binding sites, a potential integration host factor (IHF)-binding site, and a σ−54-like promoter (Fig. 1B), which are similar to those of H. seropedicae Z78 (20). Thus, regulation of the nif genes is probably mediated by nifA expression and NifA activation, which respond to the level of molecular oxygen and fixed nitrogen, as is the case with most other members of the Proteobacteria (6). Southern hybridization showed that the nifH gene is a single copy in strain B501 (data not shown). This indicates that nifH is a target gene suitable for monitoring nif transcription in the bacterium, because the expression of a single nifH gene is straightforwardly involved in functional nitrogenase activity.
FIG. 1.
(A) Alignment of the amino acid sequence of the nifH product in our study with other homologues. B501, the nifH product of Herbaspirillum sp. strain B501 (AB196476); Hs, the nifH product of Herbaspirillum seropedicae (Z54207) (20); Kp, the nifH product of Klebsiella pneumoniae (V00631) (31); Cp, the nifH product of Clostridium pasteurianum (X07472) (36); and Av, the nifH product of Azotobacter vinelandii (M20568) (16). Three cysteine and one arginine residue (★) of the nifH product are conserved. (B) Structural organization of the nifH promoter region of the Herbaspirillum sp. B501 strain. Consensus sequences are underlined and in boldface type. Numbers are nucleotide positions from nifH gene of strain B501.
nifH transcription in free-living cells.
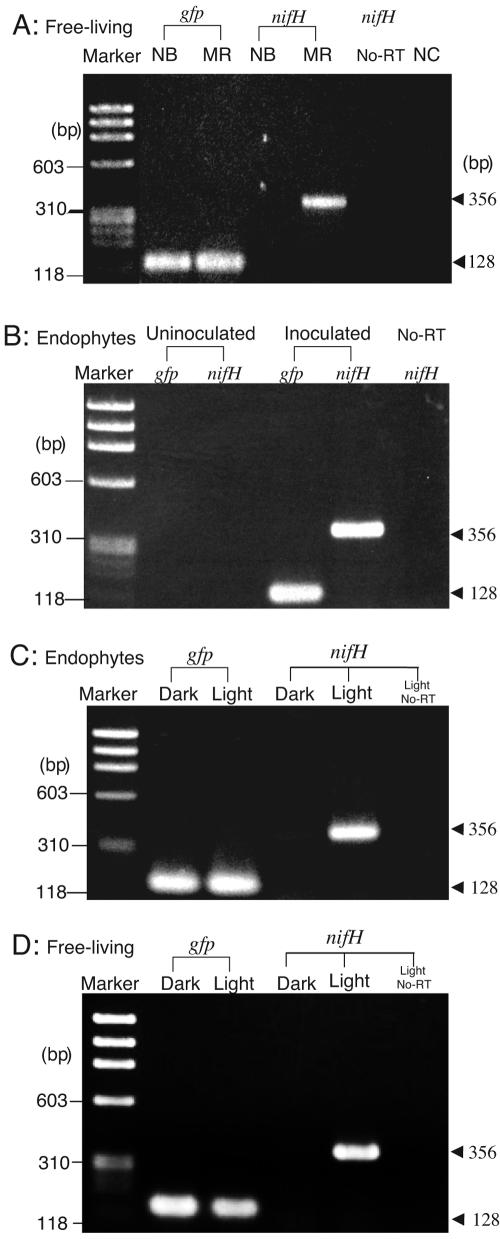
We first examined the correlation between nifH transcription and nitrogen fixation in free-living Herbaspirillum sp. B501gfp1. Nitrogen-fixing and non-nitrogen-fixing cells were prepared from cells cultured in MR or NB medium, respectively (Table 3). Because strain B501gfp1 harbored gfp genes with a constitutive promoter, PpsbA (9, 35), the gfp transcript was used as a positive control throughout this work. A band corresponding to the gfp transcript (128 bp) was observed in nitrogen-fixing and non-nitrogen-fixing cells grown in NB and MR media (Fig. 2A). In contrast, a band corresponding to the nifH transcript (356 bp)was detected exclusively in nitrogen-fixing cells cultured in MR medium (Fig. 2A). Exclusion of the RT reaction produced no PCR band for the nifH gene (Fig. 2A). These results indicate that nifH transcription was successfully detected using total RNA extracted from nitrogen-fixing cells by means of RT-PCR.
TABLE 3.
Nitrogen-fixing activity (estimated by means of acetylene reduction) in free-living and endophytic Herbaspirillum sp. strain B501gfp1
| Condition | Acetylene | Ethylene evolutiond | Acetylene reduction activity
|
|
|---|---|---|---|---|
| Total activitye | Sp act, nmol h−1 (108 cells)−1 | |||
| Culturea | ||||
| Modified Rennie (MR) | + | 65.5 ± 4.7 | 65.5 ± 4.7 | 6.4 ± 0.5 |
| Nutrient broth (NB) | + | NDb | NDb | |
| Plant (Oryza officinalis W0012)c | ||||
| + | 15.1 ± 3.0 | 14.8 | 7.6 | |
| − | 0.3 ± 0.1 | |||
Ethylene evolution from free-living cells grown in semisolid MR medium at 2% (vol/vol) O2 and in NB medium for 24 h. Values given are the means ± standard deviation for three determinations.
ND, not detected (<0.2 nmol h−1 tube−1).
Ethylene evolution from 7-day-old rice seedlings inoculated with Herbaspirillum sp. strain B501gfp1 in the presence and absence of 5% (vol/vol) acetylene gas for 24 h. Uninoculated plants emitted ethylene at a rate of 0.2 to 0.5 nmol h−1 g (fresh wt)−1. Values of ethylene evolution in the dark are the means ± standard deviation for three determinations. Acetylene reduction activity was calculated from the difference in ethylene evolution between rice seedlings in the presence and absence of 5% (vol/vol) acetylene gas.
Values for ethylene evolution for cultures are in nanomoles per hour per tube; for plants, values are in nanomoles per hour per gram (fresh weight).
Values for total acetylene activity are in nanomoles per hour per tube for cultures; for plants, values are nanomoles per hour per gram (fresh weight).
FIG. 2.
Products amplified by means of RT-PCR from total RNA extracted from various sources. (A) Free-living Herbaspirillum sp. B501gfp1. (B) Endophytic Herbaspirillum sp. B501gfp1; 7-day-old plants inoculated with B501gfp1 were sampled during the light period and then subjected to acetylene reduction (Table 3) and this RT-PCR assay. (C) nifH and gfp transcripts from endophytic Herbaspirillum sp. B501gfp1 in the wild rice plants between the dark and light periods. (D) nifH and gfp transcripts from free-living cells of Herbaspirillum sp. B501gfp1 grown in light and dark environments (see Materials and Methods). gfp, a gfp primer used for PCR; nif, a nifH primer used for PCR; NB, template from cells cultured in nutrient broth medium; MR, template from cells cultured in modified Rennie medium; No-RT, PCR amplification without reverse transcription; NC, negative control reaction without PCR template; Uninoculated, template from plants not inoculated with B501gfp1; Inoculated, template from plants inoculated with B501gfp1. Total RNA for RT-PCR was adjusted to the same amount within each panel.
Effects of oxygen concentrations on nifH transcription.
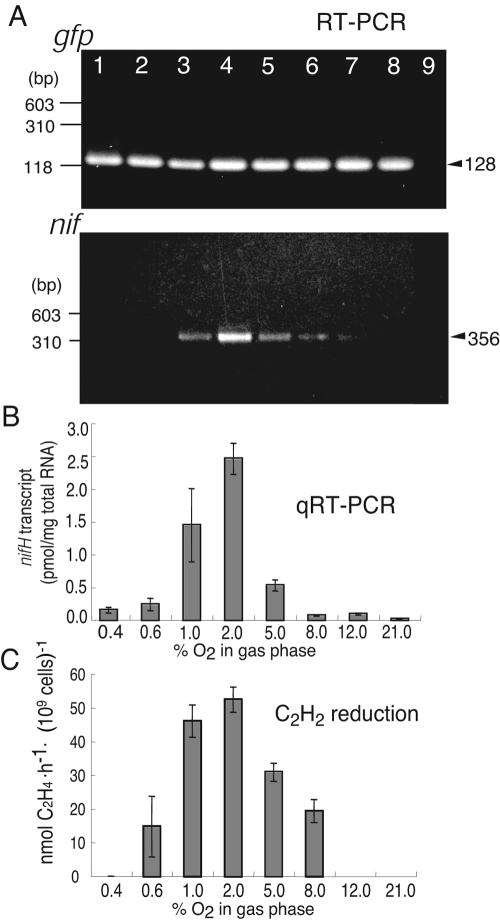
The nif promoter sequence of Herbaspirillum sp. B501 suggested that the nif genes are subjected to transcriptional activation by NifA (Fig. 1), which responds to the concentration of molecular oxygen (5). Thus, we examined the effect of O2 concentration on nifH transcription and nitrogenase activity in free-living cells of B501gfp1.
Although the gfp transcript showed a constant level regardless of the O2 regime and thus served as a positive control, the levels of nifH transcription changed drastically in response to changing O2 concentrations (Fig. 3A and B). At a 2% O2 concentration, nitrogenase activity (Fig. 3C) and nifH expression (Fig. 3A and B) reached their maximum levels in the bacterium. In contrast, the activity of the nifH gene and nitrogenase were strongly repressed under aerobic conditions (21% O2) (Fig. 3A to C). These results indicate that Herbaspirillum sp. B501 requires a microaerobic environment to be capable of expressing nitrogenase activity, as is the case for other diazotrophic endophytes (1, 8, 26) and nitrogen-fixing microbes (6, 28). RT-PCR analysis of nifH expression was an efficient method for monitoring the potential nitrogen-fixing activity of Herbaspirillum sp. B501gfp1.
FIG. 3.
(A) Products of nifH amplified by means of RT-PCR using total RNA extracted from free-living Herbaspirillum sp. B501gfp1 cultured under different oxygen concentrations. Lanes 1 to 8, RNA was extracted from cells cultured under 0.4, 0.6, 1.0, 2.0, 5.0, 8.0, 12.0, and 21.0% (vol/vol) oxygen concentrations, respectively; lane 9, negative control reaction without PCR template. gfp, a pair of gfp primers was used to amplify the gfp transcripts; nif, a pair of nifH primers was used to amplify the nifH transcripts. Total RNA was adjusted to the same amount for the respective RT-PCRs. (B) Quantification of the nifH transcripts of Herbaspirillum sp. B501gfp1, cultured under different oxygen concentrations, by means of real time RT-PCR. (C) Acetylene reduction activity of Herbaspirillum sp. B501gfp1 cultured under different oxygen concentrations.
nifH transcription in endophytic cells.
The 7-day-old seedlings of O. officinalis W0012 inoculated with B501gfp1 showed nitrogen-fixing activity (Table 3), which is similar to previously reported results (9). After the assay, total RNA was extracted from the surface-washed seedlings and subjected to nifH transcription analysis by means of RT-PCR. Bands corresponding to the gfp and nifH genes were observed in B501gfp1-inoculated plants at expected positions of 128 and 356 bp, respectively (Fig. 2B). Elimination of the RT reaction confirmed that the 356-bp band was derived from nifH mRNA but not from nifH DNA (Fig. 2B). Uninoculated plants showed no background of PCR amplification products for gfp and nifH transcripts (Fig. 2B). These results indicate that the gfp and nifH transcripts of endophytic Herbaspirillum sp. B501gfp1 in the plant tissues of wild rice could be detected by means of RT-PCR without interference from the plant RNA.
Photosynthesis by the host plant supplies photosynthate to the endophytes as their energy source (4), but during the light period, it also generates O2 that might inhibit nitrogen fixation by the endophytes (9, 22). Indeed, the activity of the nifH gene and nitrogenase in free-living B501 cells were strongly repressed under aerobic conditions (21% O2) (Fig. 3). Thus, we compared nifH expression by endophytic B501gfp1 in 7-day-old seedlings sampled at midnight (dark period) and at noon (light period). Surprisingly, we detected nifH transcription exclusively in the noon sample but observed similar levels of gfp transcription both at midnight and at noon (Fig. 2C). This result suggests that light and/or presumably photosynthesis in response to that light enhances rather than represses nifH transcription.
Daily rhythm of nifH transcription in endophytic cells.
Preliminary RT-PCR results (i.e., the detection of nifH transcription in the light period) (Fig. 2C) prompted us to follow the time course of nifH expression between the light and dark periods. We quantified nifH transcription in endophytic B501gfp1 by means of real-time RT-PCR using 7- to 10-day-old O. officinalis W0012 plants inoculated with the endophyte.
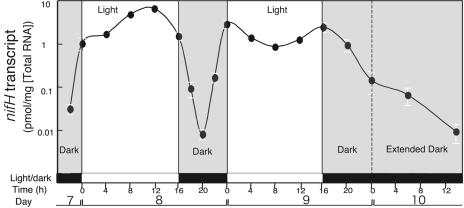
Significant fluctuations in the nifH transcription level were observed during light and dark cycles (Fig. 4). The level of nifH transcription during the light period was consistently higher than during the dark period. Based on the amount of total RNA extracted from the plants, transcription during the light period reached a maximum of 100 times the transcription level during the dark period (Fig. 4). From the curve for levels of nifH transcripts from 22:00 h on day 7 to 16:00 h on day 9, we observed a daily rhythm in nifH transcription: the amount of nifH transcription decreased to its minimum level by around midnight and reached its maximum level during the light period (Fig. 4).
FIG. 4.
Fluctuations in nifH transcription by endophytic Herbaspirillum sp. B501gfp1 during light and dark cycles. The nifH transcript of B501gfp1 was determined with three replications by means of real-time RT-PCR. Shaded zones represent the dark period, whereas white zones represent the light period; Time, time (in hours) during the course of one light-dark cycle; Day, days of growth of the O. officinalis W0012 seedlings after inoculation with Herbaspirillum sp. B501gfp1. Each point indicates the mean of three measurements using single RNA preparation extracted from three inoculated plants.
Since gfp transcription was detected during the night and day in the inoculated plants (Fig. 2C), the daily rhythm of nifH transcription was also not due to proliferation and death of the endophyte within the rice plants. Tight relationships exist between nitrogen-fixing activity and nif expression, as shown in Fig. 3 and as reported by other researchers (6, 7, 13). Thus, our results strongly suggest that nitrogen fixation by the endophytes fluctuated markedly in response to the daily rhythm of nifH expression during light and dark periods. The presence of a daily rhythm of nifH expression also indicated how to measure the nitrogen-fixing activity of endophytes in plants by the 15N2 and acetylene methods. The plant's light environment, the time of sampling, and the incubation duration probably all affect the endophyte's activity and should thus be accounted for by the sampling design.
To examine whether this daily rhythm represented a circadian rhythm (19), we preliminarily extended the dark period from days 9 and 10. This extension on day 10 showed no apparent increase in the level of nifH transcription; however, nifH mRNA began to accumulate prior to illumination in the experiment. Thus, we were not able to conclude that the daily rhythm was brought about by a circadian rhythm or light/dark stimulation so far.
Comparison of nifH expression between endophytic and free-living cells.
In the present study, we determined the amounts of nifH transcription in free-living and endophytic cells of Herbaspirillum sp. B501 by means of real-time RT-PCR. Thus, it is appropriate to compare transcription levels, although we assumed that the efficiencies of RNA extraction, the RT reaction, and the PCR were equal between both cell types. Expressed as cell numbers, the concentration of nifH transcripts in endophytic cells averaged 670 pmol/1010 cells during the light period and 5 pmol/1010 cells during the dark period (Table 4) versus only 10 pmol/1010 cells in free-living cells of B501gfp1 grown at an optimal O2 concentration (2%) in MR medium. If our assumption of comparability is correct, then the level of nifH transcription by endophytic cells during the light period was markedly higher (by a factor of 67) than that of free-living cells at the optimal O2 concentration.
TABLE 4.
Quantification of nifH transcription in the total RNA of free-living and endophytic Herbaspirillum sp. B501gfp1a
| Condition |
nifH transcription
|
|
|---|---|---|
| pmol (mg total RNA)−1 (A) | pmol (1010 cells)−1 (B)b,c | |
| Culturea | ||
| Modified Rennie (2% O2) | 2.5 ± 0.4 | 10 ± 2 |
| Nutrient broth | NDd | |
| Plant (Oriza officinalis W0012)a | ||
| Inoculated (light period) | 2.3 ± 0.6 | 670 ± 160 |
| Inoculated (dark period) | 0.03 ± 0.009 | 5 ± 2 |
| Uninoculated | NDd | |
Culture, the amount of nifH transcription in free-living B501gfp1 cells grown in semisolid modified Rennie (MR) medium at 2% (vol/vol) O2 and in nutrient broth (NB) medium for 24 h. Values given are the means ± standard deviation for three determinations. Plant, the amount of nifH transcription in endophytic B501gfp1 cells during the light and dark periods (Fig. 4). Values given are the means ± standard deviation for 10 data points at 0:00, 4:00, 8:00, 12:00, and 16:00 h on days 8 and 9 for the light period, and four data points at 18:00, 20:00, and 22:00 h on days 7 and 8 for the dark period (Fig. 4).
B = A × (μg of total RNA)/[(total cells or CFU)/1010].
Cell numbers were estimated by direct counting with a microscope for cultures and by plate counting for endophytic populations.
ND, not detected (<1.4 × 10−3 pmol (mg total RNA)−1).
nifH expression of free-living cells under light and dark conditions.
RT-PCR analyses demonstrated that the level of nifH transcription in Herbaspirillum sp. B501 during the light period was higher than that during the dark period (Fig. 2C and 4; Table 3). The light period-dependent nifH expression occurred in a complex biological system with the plant and endophyte. To address which organisms perceive light, we simply examined the nifH expression of free-living cells grown under light and dark conditions. Herbaspirillum sp. B501gfp1 was grown in MR broth medium in a flask at 2% (vol/vol) O2 under light and dark conditions: the light intensity and temperature were adjusted to the levels used for cultivation of the inoculated rice plants.
Similar levels of gfp transcription were observed in cells grown in both the light and the dark (Fig. 2D). Nevertheless, nifH transcription was detected exclusively in the cells grown inthe light but not detected in the cells grown in the dark (Fig. 2D), which is similar to results obtained with endophytic cells (Fig. 2C). This result implies that at least the endophyte is able to percept light and up-regulates nifH gene expression, although we cannot completely exclude the involvement of the plant into the light period-dependent nifH expression.
To our knowledge, our study represents the first observation of a daily rhythm of nif gene expression in endophytic microbes within plant tissues during the light-dark cycles. In addition, it is a preliminary but fascinating discovery that light radiation induces nifH gene expression in free-living Herbaspirillum sp. strain 501. So far, we do not yet know exactly how the daily rhythm of nifH expression in the endophyte was generated during the light-dark cycles and how light induces nifH gene expression even in the free-living bacterium.
Photosynthetic bacteria, such as cyanobacteria, have evolved the ability to sense light and show phototaxis to choose a more favorable position for optimal growth (3). Thus, we want to discuss the physiological and ecological implications of our findings: light period-dependent nifH expression in the endophytes. The plant probably will need more fixed nitrogen when it is photosynthetically active, while the bacterium would take advantage of the photosynthate that is produced during the light period. Therefore, one would expect the bacterium to have evolved a mechanism to up-regulate nif gene expression during the light reaction. The photosynthate-mediated interactions between plant and endophyte might be more important rather than the so-called oxygen paradox for nitrogen fixation (22).
ADDENDUM IN PROOF
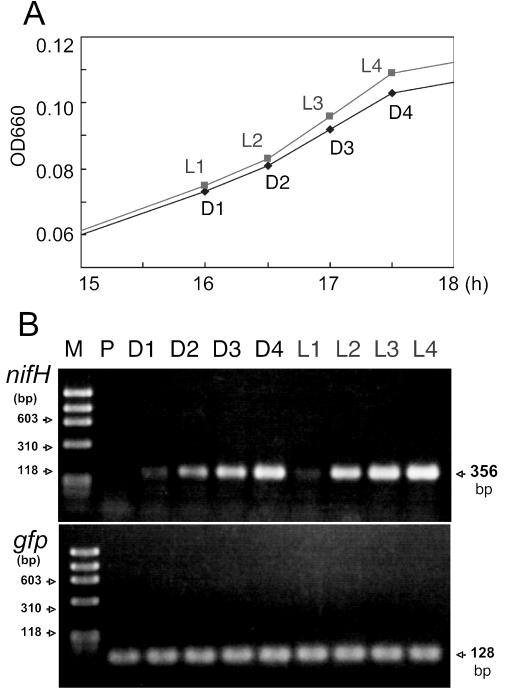
Additional experiments were performed after this paper was accepted for publication. In these experiments, time courses of nifH and gfp transcripts in free-living cells of Herbaspirillum sp. strain B51gfp1 grown in modified Rennie broth in light and dark environments were monitored. The cells exposed to the light radiation showed slightly higher levels of growth (Fig. A1A) and nifH gene expression (compared with gfp signal) than those in the dark (Fig. A1B). However, the nifH gene was expressed even in the free-living cells grown in the dark (Fig. A1B, lanes D3 and D4). In our experimental system, the expression of the nifH gene started around growth points L2 and D2 (Fig. A1). Because the previous result (Fig. 2D) was obtained at this growth stage, we erroneously concluded that the nifH gene was not expressed in the cells in the dark at all (Fig. 2D). The new finding (Fig. A1) does not affect the significance of this work, because the levels of in planta nifH transcription varied drastically, with a maximum during the light period (Fig. 2C and 4). However, this finding further clarifies the contribution of plants to light period-dependent nifH expression in endophytes.
FIG. A1.
Growth of and gene expression by free-living cells of Herbaspirillum sp. strain B501gfp1 grown in light and dark environments. (A) Tiem courses of optical density at 660 nm (OD660) for Herbaspirillum sp. strain B501gfp1 cells grown in light (L1, L2, L3, and L4) and dark (D1, D2, D3, and D4) environments (see text). (B) RT-PCR analysis targeted to nifH and gfp genes from total RNA from cultured cells of Herbaspirillum sp. B501gfp1. Lane M, size marker; lane P, preculture for inoculation. Cells of lanes D1, D2, D3, D4, L1, L2, L3, and L4 were sampled as indicated in the growth curves in panel A. Total RNA was adjusted to the same amount for each reaction in the RT-PCR analysis.
.
Acknowledgments
This work was supported in part by grants from the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and from the Ministry of Education, Science, Sports, and Culture of Japan (grant numbers 14360037 and 17658034) to K.M.
We thank T. Sato and T. Abe (Tohoku University) for help and advice on rice cultivation and for discussion.
REFERENCES
- 1.Baldani, J. I., V. L. D. Baldani, L. Seldin, and J. Döbereiner. 1986. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a root-associated nitrogen-fixing bacterium. Int. J. Syst. Bacteriol. 34:451-456. [Google Scholar]
- 2.Baldani, J. I., L. Caruso, V. L. D. Baldani, S. Goi, and J. Döbereiner. 1997. Recent advances in BNF with non-legume plants. Soil Biol. Biochem. 29:911-922. [Google Scholar]
- 3.Bhaya, D. 2004. Light matters: phototaxis and signal transduction in unicellular cyanobacteria. Mol. Microbiol. 53:745-754. [DOI] [PubMed] [Google Scholar]
- 4.Boddey, R. M., O. C. de Oliveira, S. Urquiaga, V. M. Reis, F. L. Olivares, V. L. D. Baldani, and J. Döbereiner. 1995. Biological nitrogen fixation associated with sugar cane and rice: contributions and prospects for improvement. Plant Soil 174:195-209. [Google Scholar]
- 5.Burgmann, H., F. Widmer, W. V. Sigler, and J. Zeyer. 2003. mRNA extraction and reverse transcription-PCR protocol for detection of nifH gene expression by Azotobacter vinelandii in soil. Appl. Environ. Microbiol. 69:1928-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon, R., and D. Kahn. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:612-631. [DOI] [PubMed] [Google Scholar]
- 7.Egener, T., T. Hurek, and B. Reinhold-Hurek. 1999. Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol. Plant-Microbe Interact. 12:813-819. [DOI] [PubMed] [Google Scholar]
- 8.Egener, T., T. Hurek, and B. Reinhold-Hurek. 1998. Use of green fluorescent protein to detect expression of nif genes of Azoarcus sp. strain BH72, a grass-associated diazotroph, on rice roots. Mol. Plant-Microbe Interact. 11:71-75. [DOI] [PubMed] [Google Scholar]
- 9.Elbeltagy, A., K. Nishioka, T. Sato, H. Suzuki, B. Ye, T. Hamada, T. Isawa, H. Mitsui, and K. Minamisawa. 2001. Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 67:5285-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elbeltagy, A., K. Nishioka, H. Suzuki, T. Sato, Y. Sato, H. Morisaki, H. Mitsui, and K. Minamisawa. 2000. Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil. Sci. Plant Nutr. 46:617-629. [Google Scholar]
- 11.Gary, G., S. Perrin, K. Blanchard, and H. F. Bunn. 1990. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc. Natl. Acad. Sci. USA 87:2725-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giulietti, A., L. Overbergh, D. Valckx, B. Decallonne, R. Bouillon, and C. Mathieu. 2001. An overview of real-time quantitative PCR: Applications to quantify cytokine gene expression. Methods 25:386-401. [DOI] [PubMed] [Google Scholar]
- 13.Hurek, T., L. L. Handley, B. Reinhold-Hurek, and Y. Piché. 2002. Azoarcus grass endophytes contribute fixed nitrogen to the plant in an unculturable state. Mol. Plant-Microbe Interact. 15:233-242. [DOI] [PubMed] [Google Scholar]
- 14.Iniguez, A. L., Y. Dong, and E. W. Triplett. 2004. Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Mol. Plant-Microbe Interact. 17:1078-1085. [DOI] [PubMed] [Google Scholar]
- 15.Isawa, T., R. Sameshima, H. Mitsui, and K. Minamisawa. 1999. IS1631 occurrence in Bradyrhizobium japonicum highly reiterated sequence-possessing strains with high copy numbers of repeated sequences RSα and RSβ. Appl. Environ. Microbiol. 65:3493-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson, M. R., K. E. Brigle, L. T. Bennett, R. A. Setterquist, M. S. Wilson, V. L. Cash, J. Beynon, W. E. Newton, and D. R. Dean. 1989. Physical andgenetic map of the major nif gene cluster from Azotobacter vinelandii. J.Bacteriol. 171:1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.James, E. K. 2000. Nitrogen fixation in endophytic and associative symbiosis. Field Crops Res. 65:197-209. [Google Scholar]
- 18.James, E. K., P. Gyaneshwar, N. Mathan, W. L. Barraquio, P. M. Reddy, P. P. M. Lannetta, F. L. Olivares, and J. K. Ladha. 2002. Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol. Plant-Microbe Interact. 15:894-906. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, C. H. 2004. As time glows by in bacteria. Nature 430:23-24. [DOI] [PubMed] [Google Scholar]
- 20.Machado, I. M. P., M. G. Yates, H. B. Machado, E. M. Souza, and F. O. Pedrosa. 1996. Cloning and sequencing of the nitrogenase structural genes nifHDK of Herbaspirillum seropedicae. Braz. J. Med. Biol. Res. 29:1599-1602. [PubMed] [Google Scholar]
- 21.Mae, T., and K. Ohira. 1981. The remobilization of nitrogen related to leaf growth and senescence in rice plants (Oryza sativa L.). Plant Cell Physiol. 22:1067-1074. [Google Scholar]
- 22.Marchal, K., and J. Vanderleyden. 2000. The “oxygen paradox” of dinitrogen-fixing bacteria. Biol. Fertil. Soils 30:363-373. [Google Scholar]
- 23.Minamisawa, K., K. Nishioka, T. Miyaki, B. Ye, T. Miyamoto, M. You, A. Saito, M. Saito, W. L. Barraquio, N. Teaumroong, T. Sein, and T. Sato. 2004. Anaerobic nitrogen-fixing consortia consisting of clostridia isolated from gramineous plants. Appl. Environ. Microbiol. 70:3096-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyamoto, T., M. Kawahara, and K. Minamisawa. 2004. Novel endophytic nitrogen-fixing clostridia from the grass Miscanthus sinensis as revealed by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 70:6580-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noda, S., M. Ohkuma, R. Usami, K. Horikoshi, and T. Kudo. 1999. Culture-independent characterization of a gene responsible for nitrogen fixation in the symbiotic microbial community in the gut of the termite Neotermes koshunensis. Appl. Environ. Microbiol. 65:4935-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan, B., and J. K. Vessey. 2001. Response of the endophytic diazotroph Gluconacetobacter diazotrophicus on solid media to change in atmospheric partial O2 pressure. Appl. Environ. Microbiol. 67:4694-4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters, J. W., K. Fisher, and D. R. Dean. 1995. Nitrogenase structure and function: a biochemical-genetic perspective. Annu. Rev. Microbiol. 49:335-366. [DOI] [PubMed] [Google Scholar]
- 28.Poole, R. K., and S. Hill. 1997. Respiratory protection of nitrogenase activity in Azotobacter vinelandii—roles of the terminal oxidase. Biosci. Rep. 17:303-317. [DOI] [PubMed] [Google Scholar]
- 29.Rennie, R. J. 1981. A single medium for the isolation of acetylene-reducing (dinitrogen-fixing) bacteria from soils. Can. J. Microbiol. 27:8-14. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Scott, K. F., B. G. Rolfe, and J. Shine. 1981. Biological nitrogen fixation: primary structure of the Klebsiella pneumoniae nifH and nifD genes. J. Mol. Appl. Genet. 1:71-81. [PubMed] [Google Scholar]
- 32.Sevilla, M., R. H. Burris, N. Gunapala, and C. Kennedy. 2001. Comparison of benefit to sugarcane plant growth and 15N2 incorporation following inoculation of sterile plants with Acetobacter diazotrophicus wild-type and nif− mutant strains. Mol. Plant-Microbe Interact. 14:359-366. [DOI] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:789-791. [Google Scholar]
- 34.Ueda, T., Y. Suga, N. Yahiro, and T. Matsuguchi. 1995. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J. Bacteriol. 177:1414-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unge, A., R. Tomboline, M. E. Davey, F. J. Debruijin, and J. K. Jansson. 1998. Molecular microbial ecology manual, p. 6.1.13:6.1-16. Kluwer Academic Publishers, Amsterdam, The Netherlands.
- 36.Wang, S. Z., J. S. Chen, and J. L. Johnson. 1988. The presence of five nifH-like sequences in Clostridium pasteurianum: sequence divergence and transcription properties. Nucleic Acids Res. 16:439-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zani, S., M. T. Mellon, J. L. Collier, and J. Zehr. 2000. Expression of nifH genes in natural microbial assemblages in Lake George, New York, detected by reverse transcriptase PCR. Appl. Environ. Microbiol. 66:3119-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]