Abstract
The surface protein internalin A (InlA) contributes to the invasion of human intestinal epithelial cells by Listeria monocytogenes. Screening of L. monocytogenes strains isolated from human clinical cases (n = 46), foods (n = 118), and healthy animals (n = 58) in the United States revealed mutations in inlA leading to premature stop codons (PMSCs) in L. monocytogenes ribotypes DUP-1052A and DUP-16635A (PMSC mutation type 1), DUP-1025A and DUP-1031A (PMSC mutation type 2), and DUP-1046B and DUP-1062A (PMSC mutation type 3). While all DUP-1046B, DUP-1062A, DUP-16635A, and DUP-1031A isolates (n = 76) contained inlA PMSCs, ribotypes DUP-1052A and DUP-1025A (n = 72) contained isolates with and without inlA PMSCs. Western immunoblotting showed that all three inlA PMSCs result in the production of truncated and secreted InlA. Searches of the Pathogen Tracker database, which contains subtype and source information for more than 5,000 L. monocytogenes isolates, revealed that the six ribotypes shown to contain isolates with inlA PMSCs were overall more commonly isolated from foods than from human listeriosis cases. L. monocytogenes strains carrying inlA PMSCs also showed significantly (P = 0.0004) reduced invasion of Caco-2 cells compared to isolates with homologous 3′ inlA sequences without PMSCs. Invasion assays with an isogenic PMSC mutant further supported the observation that inlA PMSCs lead to reduced invasion of Caco-2 cells. Our data show that specific L. monocytogenes subtypes which are common among U.S. food isolates but rare among human listeriosis isolates carry inlA mutations that are associated with, and possibly at least partially responsible for, an attenuated invasion phenotype.
Listeria monocytogenes is a facultative intracellular food-borne pathogen that may cause encephalitis, meningitis, septicemia, or spontaneous late-term abortions in susceptible individuals (34). Although the incidence of listeriosis is low, listeriosis has a high case fatality rate (20 to 30%), which makes this disease a considerable public health concern (19). While L. monocytogenes can be classified into 13 different serotypes, only a few specific serotypes (4b, 1/2a, and 1/2b) are responsible for the majority (approximately 90%) of human listeriosis cases (18). Molecular subtyping studies have shown that L. monocytogenes isolates can be grouped into three genetic lineages (reviewed in reference 41). While different studies have suggested that L. monocytogenes genetic lineages and subtypes differ in their epidemiological associations with specific hosts and nonhost environments (6, 12, 18, 22, 26, 39), very limited information is available on underlying genetic mechanisms that may be responsible for virulence differences among L. monocytogenes subtypes. A better understanding of genetic differences and their relationship to virulence differences among L. monocytogenes strains is critical to allow the development of accurate science-based L. monocytogenes risk assessments.
Internalin (InlA), which is encoded by inlA, is an 800-amino-acid (aa) surface protein that facilitates the entry of L. monocytogenes into epithelial cells that express specific forms of E-cadherin. InlA appears to contribute to the invasion of intestinal epithelial cells by L. monocytogenes, an important first step in the pathogenesis of systemic listeriosis (17). While InlA may also contribute to crossing of the placental barrier by L. monocytogenes (16), a role for InlA in the pathogenesis of listerial abortions could not be confirmed in animal experiments using a guinea pig model (1). Interestingly, a series of studies on L. monocytogenes strains isolated in France identified at least six distinct nonsense mutations in inlA upstream of the region encoding the InlA membrane anchor (11, 13, 29, 31), resulting in the expression of a truncated form of InlA which is secreted rather than anchored to the bacterial cell wall (13). No studies have characterized the presence of L. monocytogenes isolates with nonsense mutations in inlA from countries other than France.
A previous study (24) by our group on the molecular phylogeny and evolution of 120 L. monocytogenes isolates obtained from human and animal clinical cases and foods in the United States revealed three unique mutations in inlA leading to premature stop codons (PMSCs), which appear to differ from those previously described for L. monocytogenes isolates from France. The current study was thus conducted to (i) identify additional mutations leading to PMSCs in inlA by using a larger collection of L. monocytogenes isolates collected in the United States, (ii) define the prevalence of L. monocytogenes isolates harboring inlA PMSC mutations among human, animal, and food-source populations in the United States, and (iii) determine the invasion phenotypes of L. monocytogenes strains with and without inlA PMSCs.
MATERIALS AND METHODS
Bacterial strains.
Six L. monocytogenes isolate sets (Table 1) were assembled from our culture collection of >5,000 L. monocytogenes isolates to screen for the presence of inlA PMSCs among L. monocytogenes strains isolated from different source populations in the United States. All isolates included in these six sets are described in detail in Supplemental Table 1 (http://www.foodscience.cornell.edu/wiedmann/Nightingale%20Supplementary2.txt). inlA sequencing data for these isolates have not been reported previously.
TABLE 1.
Description of L. monocytogenes isolate sets used for this study
| Isolate set no. | Description | Purpose and use of isolate seta |
|---|---|---|
| 1 | 36 food isolates representing all 36 EcoRI ribotypes found among food isolates collected in the United States, as described by Gray et al. (6) | Isolates were screened by sequencing an ∼1,600-bp 3′ fragment of inlA to identify premature stop codon mutations associated with food isolates |
| 2 | 58 isolates representing L. monocytogenes EcoRI ribotypes commonly detected in feces from healthy ruminants, selected from a previously described collection of 108 L. monocytogenes fecal isolates from healthy ruminants (25) | Isolates were screened by sequencing an ∼1,600-bp 3′ fragment of inlA to identify premature stop codon mutations associated with isolates from healthy cattle |
| 3 | 62 ribotype DUP-1062A isolates, representing all available DUP-1062A isolates from human listeriosis infections (n = 12), and 50 DUP-1062A food isolates selected from the 151 DUP-1062A food isolates described by Gray et al. (6) to represent a variety of different ready-to-eat foods | Isolates were screened by PCR-RFLP to identify the presence of premature stop codon mutation type 3; all DUP-1062A isolates characterized by serotyping (n = 11) were serotype 1/2a |
| 4 | 54 ribotype DUP-1052A isolates, representing all 30 DUP-1052A isolates from human listeriosis infections that were not previously characterized and 24 food isolates selected from the 58 DUP-1052A food isolates described by Gray et al. (6) to represent a variety of different ready-to-eat foods | Isolates were screened by sequencing an ∼800-bp 3′ fragment of inlA to identify the presence of premature stop codon mutation type 1; all DUP-1052A isolates characterized by serotyping (n = 10) were serotype 1/2b |
| 5 | Eight ribotype DUP-1025A isolates, representing all four DUP-1025A isolates from human listeriosis infections that were not previously characterized and selected food isolates (n = 4) | Isolates were screened by sequencing an ∼800-bp 3′ fragment of inlA to identify the presence of premature stop codon mutation type 2; all DUP-1025A isolates characterized by serotyping (n = 9) were serotype 1/2b |
| 6 | All DUP-16635A isolates from foods (n = 4) that were not previously characterized | Isolates were screened by sequencing an ∼800-bp 3′ fragment of inlA to identify the presence of premature stop codon mutation type 1; all DUP-16635A isolates (n = 5) were serotype 1/2b |
The number of isolates characterized by serotyping included 8 DUP-1052A, 2 DUP-1025A, 1 DUP-1031A, 3 DUP-1062A, and 2 DUP-1046B isolates serotyped by Nightingale et al. (24) as well as 24 isolates that were not previously reported.
inlA sequencing.
L. monocytogenes isolates representing molecular subtypes commonly isolated from foods and fecal samples from healthy ruminants (isolate sets 1 and 2; Table 1) were screened for the presence of PMSCs in inlA by first sequencing the 3′ end of inlA (approximately 800 bp) and subsequently sequencing an additional, approximately 800-bp fragment (5′ of the initial 3′ 800-bp region sequenced) if no PMSC mutations were detected in the initial 3′ fragment. To specifically screen for the presence of PMSC mutation types 1 and 2, which were previously mapped to the 3′ end of inlA (24), DNA sequencing of the 3′ fragment of inlA (approximately 800 bp) was conducted. Primer sequences used for PCR amplification and/or DNA sequencing as well as PCR conditions are detailed in Supplemental Tables 2 and 3 (http://www.foodscience.cornell.edu/wiedmann/Nightingale%20Supplementary2.txt). PCR was performed using Taq polymerase (Promega, Madison, WI) as described previously (24). PCR products were also purified, quantified, and sequenced using forward and reverse PCR primers as described previously (24). Nucleotide sequences were proofread and aligned with Seqman and MegAlign (Lasergene software suite; DNAStar, Madison, WI), respectively.
TABLE 2.
L. monocytogenes strains and isolates used for Western blot analysis and invasion assays
| Strain (previous name) | Genotype or ribotype | Source or reference |
|---|---|---|
| 10403S | Wild type | 2 |
| FSL K4-006 (EJL12, HEL-202) | 10403S ΔinlA | 1 |
| FSL K4-007 (HEL-137) | 10403S ΔinlB | 15 |
| FSL K4-009 | 10403S ΔinlAB | 14 |
| FSL W3-084 (HEL-399) | 10403S inlA with stop codon at aa 700 (PMSC mutation type 3) | This paper |
| FSL F2-245a | DUP-1052A, full-length inlA | This paper |
| FSL F2-563a | DUP-1052A, PMSC mutation type 1 | This paper |
| FSL F2-897b | DUP-1025A, full-length inlA | This paper |
| FSL R2-074b | DUP-1025A, PMSC mutation type 2 | This paper |
| FSL F2-539 (EGD-e)c | Full length inlA | 5 |
| FSL F2-515c | DUP-1062A, PMSC mutation type 3 | This paper |
FSL F2-245 and FSL F2-563 have identical 3′ inlA sequences, except for premature stop codon mutation type 1 in isolate FSL F2-563.
FSL F2-897 and FSL R2-074 have identical 3′ inlA sequences, except for premature stop codon mutation type 2 in isolate FSL R2-074.
FSL F2-539 (EGD-e) and FSL F2-515 have identical 3′ inlA sequences, except for premature stop codon mutation type 3 in isolate FSL F2-515.
TABLE 3.
Occurrence of premature inlA stop codons among six EcoRI ribotypes found to contain these mutations
| Ribotype | Mutation type | Incidence of premature inlA stop codon (no. of isolates with mutation [no. of isolates screened]) among isolates obtained from:
|
Prevalence (%) of premature inlA stop codon among all isolates | ||
|---|---|---|---|---|---|
| Humans | Food | Total | |||
| DUP-1052Aa | 1 | 7 (35) | 13 (27) | 20 (62) | 32.3 |
| DUP-16635Ab | 1 | 0 (0) | 5 (5) | 5 (5) | 100.0 |
| DUP-1025Ac | 2 | 0 (5) | 2 (6) | 2 (11) | 18.2 |
| DUP-1031Ad | 2 | 0 (0) | 1 (1) | 1 (1) | 100.0 |
| DUP-1062Ae | 3 | 12 (12) | 55 (55) | 67 (67) | 100.0 |
| DUP-1046Bf | 3 | 0 (0) | 3 (3) | 3 (3) | 100.0 |
Data represent 54 isolates from isolate set 4 (Table 1), one food isolate with ribotype DUP-1052A from isolate set 1, and five human and two food isolates from the work of Nightingale et al. (24).
Data represent four isolates from isolate set 6 (Table 1) and one food isolate with ribotype DUP-16635A from isolate set 1.
Data represent eight isolates from isolate set 5 (Table 1), one food isolate with ribotype DUP-1025A from isolate set 1, and one human and one food isolate from the work of Nightingale et al. (24).
Data represent one food isolate with ribotype DUP-1031A from isolate set 1 (Table 1).
Data represent 62 isolates from isolate set 3 (Table 1), one food isolate with ribotype DUP-1062A from isolate set 1, and four food isolates from the work of Nightingale et al. (24).
Data represent one food isolate with ribotype DUP-1046B from isolate set 1 and two food isolates from the work of Nightingale et al. (24).
PCR-RFLP.
Partial inlA DNA sequence data for four DUP-1062A and all available DUP-1046B L. monocytogenes isolates from food samples previously uncovered the presence of a specific PMSC mutation (PMSC mutation type 3) in these isolates (24). A PCR-restriction fragment length polymorphism (PCR-RFLP) assay was developed to screen for the presence of this inlA PMSC mutation in DUP-1062A isolates from human clinical cases and from foods (Table 1). An analysis of inlA sequences with and without inlA PMSC mutation type 3 identified RsaI as a restriction enzyme that differentiates sequences with and without this mutation. PCR primers (Supplemental Table 2) were designed to amplify a 180-bp fragment of inlA encompassing PMSC mutation type 3. RsaI recognizes a sequence found in inlA sequences without PMSC mutation type 3, so L. monocytogenes isolates with PMSC mutation type 3 were expected to show a single 180-bp fragment after PCR-RFLP, while isolates without this mutation were expected to yield two fragments (117 and 63 bp). PCR was performed as described in Supplemental Table 3. PCR products were digested with RsaI (New England Biolabs, Beverly, MA) for 1 h at 37°C and subsequently analyzed by 3.0% agarose gel electrophoresis. Isolates confirmed by DNA sequencing to carry or not carry PMSC mutation type 3 were used as positive controls.
Caco-2 invasion assays.
Caco-2 invasion assays were performed essentially as previously described (28). Briefly, L. monocytogenes isolates were grown overnight at 30°C without shaking. Confluent Caco-2 monolayers were inoculated with 2 × 107 L. monocytogenes cells/well. Serial dilutions of the inoculum were plated on brain heart infusion (BHI) agar (Difco Laboratories, Detroit, MI) plates. Inoculated Caco-2 monolayers were incubated for 30 min at 37°C, followed by three washes with phosphate-buffered saline and the addition of fresh medium without antibiotics. Medium containing 150 μg/ml gentamicin was added at 45 min postinoculation to kill extracellular bacteria. At 90 min postinoculation, Caco-2 monolayers were washed three times with phosphate-buffered saline. Caco-2 cells were lysed by the addition of cold sterile distilled water and vigorous pipetting. Intracellular L. monocytogenes cells were enumerated by spread plating lysed Caco-2 cell suspensions on BHI agar plates. The invasion efficiency was reported as the percentage of the inoculum recovered by the enumeration of intracellular bacteria. A standard laboratory control strain (10403S) and uninoculated BHI broth were included as controls in each invasion assay. Three independent invasion assays were performed for each L. monocytogenes strain tested (Table 2).
Generation of an isogenic inlA PMSC mutant.
An isogenic mutant of laboratory strain 10403S (2) carrying inlA PMSC mutation type 3 (located at codon 700 of the inlA open reading frame) was generated. Site-directed mutagenesis was performed using SOE-PCR (splicing by overlap extension PCR) (10), using primers (5′ to 3′) Marq 147 (AACTGCAGCTGGGAATTTATTGACTGG), Marq 149 (GTAAATTGAGCCTACAGCGTAATG), Marq 148 (CATTACGCTGTAGGCTCAATTTAC), and Marq 150 (GCGGTACCTTGCTTGATTGGCGTTGGC). First, two fragments were amplified by PCR from strain 10403S, using primer pairs Marq 147/Marq 149 and Marq 148/Marq 150. Primers Marq 148 and 149 are complementary and introduce inlA PMSC mutation type 3 (TAG) at codon 700. The PCR products were purified and combined in a second PCR with primers Marq 147 and 150. The 1,098-bp product was digested with PstI and KpnI and ligated into pKSV7. The mutated inlA allele was verified by sequencing and recombined into the chromosome of L. monocytogenes 10403S by allelic exchange as previously described (3).
Western blotting.
Western immunoblot analysis of bacterial cell wall and supernatant fractions for the presence of InlA was performed on L. monocytogenes grown in LB supplemented with 50 mM morpholinepropanesulfonic acid (MOPS) adjusted to pH 7.3, 25 mM glucose-1-phosphate, and 0.2% activated charcoal. Chloramphenicol was added (10 μg/ml) 10 min prior to the harvest. Cell wall and supernatant fractions were prepared as previously described (36). Equivalent amounts of culture (in optical density [OD] units) were loaded per lane. Proteins were detected by Western immunoblotting using a mouse anti-InlA antiserum provided by T. Potter (National Jewish Medical and Research Center, Denver, CO) (15, 36).
Phylogenetic inference.
A multiple sequence alignment of the 3′ end of inlA (706 nucleotides) was created in MegAlign using the inlA sequences for all L. monocytogenes isolates (n = 160) sequenced for this study and sequences containing PMSCs from our previous study (24). This alignment was used to generate a data file for phylogenetic analyses containing a single representative isolate for each inlA allele (n = 38), using DnaSP, version 3.99 (32). A maximum likelihood phylogenetic tree was inferred using DNA substitution parameters selected by MODELTEST (30) and PAUP* (37), as previously described (24).
Serotyping.
Serotyping of L. monocytogenes was performed using the glass agglutination method (7, 38) and commercial Listeria antiserum (Seiken Listeria antiserum; Denka Seiken, Tokyo, Japan). Results were interpreted according to the serotyping scheme of Seeliger and Hohne (35).
Statistical analysis.
Chi-square tests were performed to describe the prevalence of PMSCs in food and human clinical isolates and to describe the distribution of ribotypes that carry inlA PMSCs among isolates from foods and human clinical listeriosis cases. Fisher's exact test was used if expected values were below 5 for one or more cells. A nonparametric equivalent to the t test (Wilcoxon rank sum test) was used to assess the difference in invasion efficiencies between the two groups of L. monocytogenes strains studied (i.e., strains with and without premature inlA stop codons). P values of <0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
A single sequence representing each unique inlA allele (n = 38) identified in this study was deposited in GenBank (accession no. DQ125375 to DQ125412). Accession no. DQ125408, DQ125406, and DQ125387 represent inlA sequences containing PMSC mutation types 1, 2, and 3, respectively. Sequences for all isolates characterized by partial inlA sequencing in this study (n = 160) are available through Pathogen Tracker (www.pathogentracker.net).
RESULTS AND DISCUSSION
While previous studies described inlA PMSCs among L. monocytogenes strains isolated in France from human fecal carriers and foods as well as, less commonly, from human clinical cases (11, 13, 29, 31), we were not aware of any data on the presence and distribution of inlA PMSCs in L. monocytogenes isolates from countries other than France. Our data reported here show that (i) multiple distinct inlA PMSC mutations are found in different L. monocytogenes EcoRI ribotypes, which overall are commonly isolated from foods and rarely cause human disease in the United States; (ii) isolates carrying inlA mutations resulting in PMSCs show attenuated invasiveness in human intestinal epithelial cells; and (iii) inlA PMSC mutations 3′ of the highly conserved leucine-rich repeat (LRR) region of InlA are rare among fecal isolates from healthy ruminants. Together with previous studies (11, 13, 29, 31), our results suggest that L. monocytogenes isolates with PMSCs in inlA are distributed worldwide and may represent a significant virulence-attenuated subpopulation of L. monocytogenes commonly isolated from foods. While the occurrence of multiple distinct ecologically successful inlA PMSC mutations suggests positive selection for the loss of cell wall-anchored InlA, the reservoir(s) of L. monocytogenes strains with secreted InlA and the selective advantages associated with these mutations remain to be established.
Multiple distinct inlA PMSCs are found in different L. monocytogenes EcoRI ribotypes that are commonly isolated from foods and rarely cause human disease in the United States.
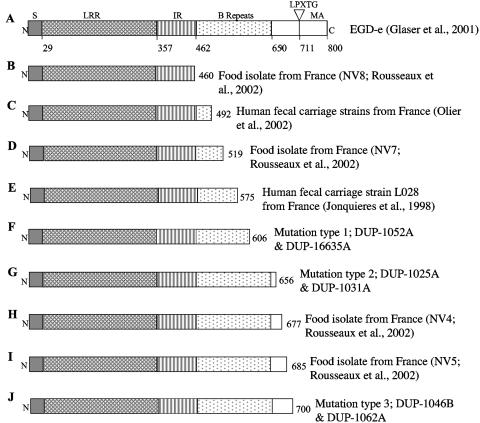
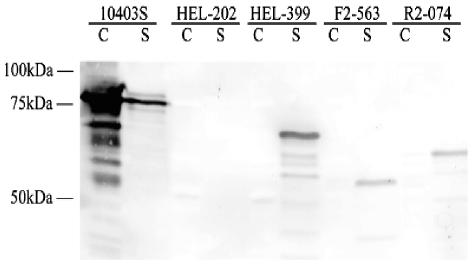
A previous study by our group (24) identified three unique inlA PMSC mutations in four L. monocytogenes ribotypes (ribotypes DUP-1052A, DUP-1062A, DUP-1046B, and DUP-1031A). Ribotypes DUP-1062A and DUP-1052A were not only previously shown to be the two most common L. monocytogenes ribotypes among more than 30,000 food samples collected in the United States (together representing 41.7% of 502 L. monocytogenes food isolates [6]) but were also found to be significantly more common among food isolates than among human clinical isolates (6). We thus hypothesized that inlA PMSCs might be associated with reduced human virulence among certain L. monocytogenes subtypes found in the United States. Since previous studies in France have also found multiple mutations leading to PMSCs in the 3′ region of inlA (11, 13, 29, 31), we initially sequenced the 3′ region of inlA (covering the region where PMSCs were previously identified [11, 13, 24, 29, 31]) in 36 L. monocytogenes isolates (isolate set 1; Table 1) representing all 36 ribotypes found among 502 previously collected U.S. food isolates (6). This isolate set included ribotypes commonly linked to human listeriosis cases (e.g., DUP-1038B, DUP-1042B, and DUP-1044A [6]). We identified two additional ribotypes, DUP-1025A and DUP-16635A, which carry inlA PMSCs. The inlA mutations identified here occurred in the same locations previously identified by our group (24) (Fig. 1). inlA PMSCs among the isolates screened thus represent three distinct mutation types (PMSC mutation types 1 through 3), with each occurring in two ribotypes (Table 3). All three inlA PMSC mutations observed here appear to be distinct from those reported in previous studies that provided DNA sequence data allowing us to map the specific mutations leading to PMSCs (13, 29, 31) (Fig. 1). We could not establish whether the mutations observed here were also distinct from those leading to truncated InlA in food and human clinical isolates collected in France (11), since these isolates were only characterized by Western blot analysis, which does not permit a definitive identification of specific mutations leading to PMSCs. All three inlA PMSCs observed here were located within 100 aa of each other and were positioned 3′ of the conserved LRR region and 5′ of the C-terminal LPXTG membrane-anchoring motif (Fig. 1). The occurrence of inlA PMSCs in this region was previously linked to the production of a truncated InlA protein that is secreted rather than anchored to the cell wall (11, 13). Western blot analysis confirmed that the inlA PMSCs reported here also result in the production of a truncated and secreted InlA protein (Fig. 2).
FIG. 1.
Full-length InlA (A) and locations of premature stop codons in InlA previously identified by research groups in France (B, C, D, E, H, and I) and reported here (F, G, and J). Full-length InlA represents the sequence for L. monocytogenes strain EGD-e. N, N-terminal end; S, signal sequence; LRR, leucine-rich repeat; IR, intergenic repeat; MA, membrane anchor; C, C-terminal end. Numbers below the EGD-e InlA sequence represent aa positions. Numbers at the right end for panels B to F represent the aa positions of the respective premature stop codons.
FIG. 2.
Western blot detection of InlA from broth-grown bacteria. Strains tested included (i) the laboratory control strain 10403S, which encodes full-length InlA (2,400 bp, 800 aa); (ii) an isogenic 10403S ΔinlA mutant (HEL-202, FSL K4-006); (iii) an isogenic mutant with PMSC mutation type 3 in a 10403S background (HEL-399, FSL W3-084); (iv) a wild-type isolate with PMSC mutation type 1 (FSL F2-563); and (v) a wild-type isolate with PMSC mutation type 2 (FSL R2-074). Proteins in the cell wall (C) and in the supernatant (S) fraction were detected by Western immunoblotting with a mouse anti-InlA antiserum.
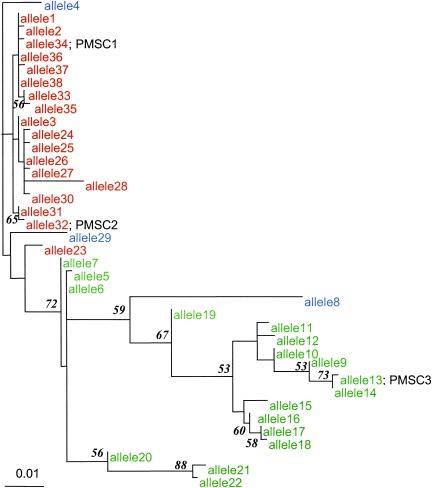
Phylogenetic analyses showed that the inlA PMSCs reported here represent three distinct evolutionary events (Fig. 3). The clustering of the inlA sequences containing mutations leading to PMSCs is also consistent with the lineage classification of the isolates carrying these mutations (Fig. 3). Specifically, inlA alleles carrying PMSC mutation types 1 and 2 group with other lineage I inlA sequences, and the PMSC mutation type 3 allele groups with lineage II isolates (Fig. 3). The lineage classification was also consistent with serotype classification (Table 3). Isolates with PMSC mutation types 1 and 2 were serotype 1/2b, which is typical for lineage I strains (23), while isolates with PMSC mutation type 3 were serotype 1/2a, which is typical for lineage II strains (23). In a previous study (24), sequencing the 3′ 800 bp of inlA failed to reveal any PMSC mutations among 36 lineage I L. monocytogenes isolates representing serotype 4b, a serotype associated with the majority of human listeriosis cases (18). The majority (n = 40) of the ruminant fecal isolates characterized in this study belonged to lineage II, which predominantly includes serotypes 1/2a and 1/2c (23). Thus, the lack of inlA premature stop codon mutations among animal isolates does not appear to represent an overrepresentation of isolates of serotype 4b, which is predominantly found among lineage I isolates and not found in lineage II isolates. We also recently identified a frameshift mutation in the 5′ portion of inlA, leading to a PMSC in the signal sequence of InlA (Fig. 1). This frameshift mutation was observed in L. monocytogenes isolates from foods that were characterized as ribotype DUP-1039C and serotype 1/2c (R. Orsi and M. Wiedmann, unpublished data). Interestingly, previous studies in France only identified inlA PMSC mutations 3′ of the LRR region of InlA in serotype 1/2c and 1/2a isolates (11), further confirming that the inlA PMSCs observed here represent genetic events independent from those leading to the emergence of the inlA PMSCs in France. The fact that at least nine independent evolutionary events (Fig. 1) in both of the major L. monocytogenes genetic lineages led to the generation of ecologically successful inlA alleles carrying PMSC mutations suggests the occurrence of positive selection and thus a distinct ecological advantage for a loss of cell wall-anchored InlA.
FIG. 3.
Phylogenetic tree depicting evolutionary relatedness of inlA alleles sequenced. The tree was constructed using a 706-bp 3′ fragment of inlA. Alleles representing premature stop codon mutation types 1, 2, and 3 are marked PMSC 1, 2, and 3, respectively. Genetic lineage I (as determined by EcoRI ribotyping) isolates are shown in red, lineage II isolates are shown in green, and lineage III isolates are shown in blue. inlA sequence data for lineage III isolates (n = 3), which have previously been shown to be distantly related to lineage I and II isolates (42), were used as an outgroup to root the inlA maximum likelihood tree. Bootstrap confidence measures above 50.0% for nodes in the inlA phylogram are shown at the nodes they describe. The inlA sequences used to construct this tree are available from GenBank (accession no. DQ125375 to DQ125412).
To further understand the prevalence and distribution of PMSC mutation types 1 through 3 among L. monocytogenes isolates in the United States, we assembled additional larger isolate sets (sets 3 through 6; Table 1) of human clinical isolates and food isolates representing four of the six ribotypes found here to carry inlA PMSC mutations (all isolates representing ribotypes DUP-1031A and DUP-1046B were characterized previously [24]). We screened these isolates using inlA sequencing or a newly developed RsaI PCR-RFLP assay targeting the single nucleotide polymorphism (SNP) responsible for PMSC mutation type 3, the most common mutation leading to truncated and secreted InlA. All isolates representing ribotypes DUP-16635A, DUP-1031A, DUP-1046B, and DUP-1062A carried the expected inlA PMSCs (Table 3). In contrast, ribotypes DUP-1025A and DUP-1052A contained isolates with and without inlA PMSCs (Table 3). While PMSC mutations were more common among food isolates than among human clinical isolates belonging to both ribotypes DUP-1025A and DUP-1052A, this association was only statistically significant (P < 0.05) for DUP-1052A isolates (Table 3). Ribotypes DUP-1025A and DUP-1052A contained isolates with partial inlA sequences that showed 100% sequence identity (within each ribotype), with the exception of their respective PMSC mutations, suggesting a recent emergence of these inlA allelic types.
Two recent studies (6, 24) provided initial evidence that L. monocytogenes strains known to carry inlA PMSCs rarely cause human listeriosis in the United States. We conducted queries of the PathogenTracker database (www.pathogentracker.net), which contains information for >5,000 L. monocytogenes strains isolated predominantly in the United States and North America from various sources (e.g., human and animal clinical listeriosis cases, foods, healthy animals, and different environments), to probe the distribution of the ribotypes carrying inlA PMSCs on a larger scale. All L. monocytogenes ribotypes found here to carry inlA PMSCs were extremely rarely or never detected among animal isolates (Table 4), suggesting that a full-length InlA protein may play a critical role in animal listeriosis. L. monocytogenes ribotypes (DUP-1062A, DUP-16635A, DUP-1031A, and DUP-1046B) exclusively associated with PMSC mutations and one ribotype that was not exclusively associated with a PMSC (DUP-1052A) were more commonly isolated from foods than from human clinical cases (Table 4). A χ2 test of independence showed that ribotype DUP-1062A was significantly more common (P < 0.0001) among food isolates (n = 1,562) than among human clinical isolates (n = 1,009). While a total of 52 human clinical isolates grouped into one of the ribotypes found to be associated with PMSCs, only 19 of these isolates actually carried the inlA PMSC mutations associated with the respective ribotypes, as determined by sequencing of the 3′ end of inlA. Most of the human isolates carrying inlA PMSCs were obtained from blood (n = 16), while a single isolate each was obtained from pleural fluid and a skin graft site; no isolation site data were available for one isolate. This distribution of human isolates with inlA PMSC mutations is consistent with results from a recent study in France, which showed that none of 61 L. monocytogenes isolates from abortion cases carried a truncated InlA protein (11). While these data suggest that InlA may be critical for the ability of L. monocytogenes to cause abortions, consistent with a study that found a role for inlA in the invasion of trophoblasts (16), a recent study using a guinea pig model did not support an important role for InlA in crossing the placenta in vivo (1). We thus conclude that L. monocytogenes isolates with PMSCs in inlA represent a considerable subpopulation of L. monocytogenes isolates found in food which rarely cause human disease and may have a reduced ability to cause fetal infections. Further studies using L. monocytogenes isolates with inlA PMSCs will be necessary to evaluate the potential of these strains to cause abortions.
TABLE 4.
Frequency of L. monocytogenes ribotypes associated with premature inlA stop codons among isolates in the Pathogen Tracker database
| EcoRI ribotype | Mutation type | Lineagea | Serotype (no.)b | No. (prevalence [%]) of isolates in Pathogen Trackerc obtained from:
|
||||
|---|---|---|---|---|---|---|---|---|
| Humansd | Animalse | Foodsf | Otherg | Total | ||||
| DUP-1052A | 1 | I | 1/2b (10) | 35 (3.5) | 1 (0.08) | 78 (5.0) | 82 (6.8) | 196 (3.9) |
| DUP-16635A | 1 | I | 1/2b (5) | 0 | 0 | 5 (0.3) | 1 (0.1) | 6 (0.1) |
| DUP-1025A | 2 | I | 1/2b (9) | 5 (0.5) | 2 (0.15) | 6 (0.4) | 4 (0.3) | 17 (0.3) |
| DUP-1031A | 2 | I | 1/2b (1) | 0 | 0 | 1 (0.06) | 0 | 1 (0.02) |
| DUP-1062A | 3 | II | 1/2a (11) | 12 (1.2) | 0 | 170 (10.9)* | 37 (3.1) | 219 (4.3) |
| DUP-1046B | 3 | II | 1/2a (3) | 0 | 0 | 3 (0.2) | 0 | 3 (0.06) |
| Total | 1,009 | 1,297 | 1,562 | 1,213 | 5,081 | |||
Lineages were assigned based on the EcoRI ribotypes, as described by Wiedmann et al. (42).
Numbers reflect eight DUP-1052A, two DUP-1025A, one DUP-1031A, three DUP-1062A, and two DUP-1046B isolates serotyped by Nightingale et al. (24) as well as serotypes for 24 isolates not previously reported.
Numbers represent isolates present in Pathogen Tracker on 1 August 2004; detailed source information on isolates can be obtained at www.pathogentracker.net. Percentages represent the prevalence of isolates with a given ribotype among all isolates in a given category (e.g., humans).
The majority of human isolates (>90%) were from invasive listeriosis cases in North America.
Animal isolates included isolates from animals with invasive listeriosis as well as fecal isolates from animals without symptoms of listeriosis, the majority of which (>95%) were from North America.
The majority of food isolates (>95%) were from North America. *, statistically significantly (P < 0.0001) more common among food isolates than among human isolates.
Other isolates included isolates from various environments, including food processing plants, as well as isolates of unknown origin.
L. monocytogenes isolates with PMSCs show reduced invasion efficiencies.
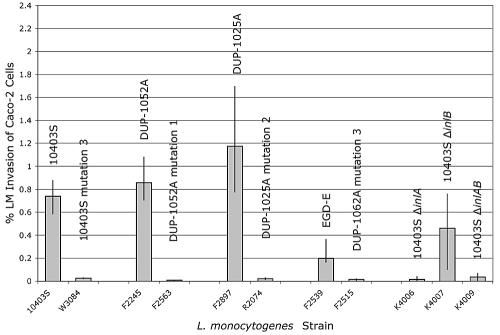
InlA has been shown to be sufficient for the invasion of human intestinal epithelial cells by L. monocytogenes (21), and previous studies of L. monocytogenes isolates from France carrying inlA PMSCs showed that these isolates displayed reduced invasion with the Caco-2 human intestinal epithelial cell line (27, 28). We paired L. monocytogenes isolates representing each unique inlA PMSC mutation identified here with isolates with homologous partial inlA sequences (except for each nonsense mutation) to assess the ability of these strains to invade Caco-2 cells (Table 2). While all DUP-1062A isolates were shown to harbor an inlA PMSC mutation, partial inlA sequences for DUP-1062A isolates were homologous to the partial L. monocytogenes EGD-e inlA sequence (5), with the exception of PMSC mutation type 3. We thus included EGD-e, which groups into lineage II, as do all DUP-1062A isolates, as an initial nonisogenic control for ribotype DUP-1062A. L. monocytogenes strains harboring inlA PMSCs showed significantly (P = 0.0004) reduced invasion efficiencies in Caco-2 cells compared to strains with full-length InlA (Fig. 4). Isogenic ΔinlA and ΔinlAB null mutants showed an invasion capacity similar to that of L. monocytogenes strains carrying inlA PMSCs, supporting the critical role of a cell wall-anchored InlA protein for the invasion of human intestinal epithelial cells. The invasion efficiency of the ΔinlB null mutant was lower than that of the isogenic wild-type strain (10403S), but higher than that of the corresponding isogenic ΔinlA mutant, consistent with previous reports (14, 15).
FIG. 4.
Caco-2 cell invasion efficiency of L. monocytogenes isolates with inlA PMSC mutations. The isolates tested included L. monocytogenes 10403S and an isogenic mutant with inlA PMSC mutation type 3 (FSL W3-084) as well as three pairs of wild-type strains with homologous 3′ inlA sequences, with and without the respective PMSC mutation types 1, 2, and 3; in addition, four control strains (10403S isogenic ΔinlA, ΔinlB, and ΔinlA null mutants) were included (Table 2). The invasion efficiency of each L. monocytogenes strain was assayed in triplicate in each independent experiment, and three independent invasion experiments were performed for each strain. Bars represent median invasion efficiencies (% of intracellular bacteria relative to initial inoculum), and error bars indicate the minimum and maximum invasion efficiencies observed for each strain or isolate. A nonparametric approximation of a t test showed that L. monocytogenes isolates with inlA PMSC mutations have significantly (P = 0.0004) reduced invasion efficiencies in Caco-2 cells compared to isolates with an inlA gene encoding full-length InlA.
To further characterize the effect of inlA PMSCs on invasiveness without confounding other strain differences, we constructed an isogenic L. monocytogenes 10403S mutant (FSL W3-084; Table 2) with inlA PMSC mutation type 3. This isogenic mutant was shown to produce a truncated and secreted InlA protein (Fig. 2) and also showed reduced Caco-2 cell invasion efficiency, similar to natural L. monocytogenes isolates carrying PMSC types 1, 2, and 3. These data further support the observation that L. monocytogenes strains with inlA PMSCs are characterized by attenuated invasiveness and thus indicate that SNPs in inlA may be suitable targets for the development of rapid assays for the detection of virulence-attenuated L. monocytogenes. However, based on the data presented here, it is not possible to differentiate whether these inlA PMSC mutations are fully responsible for the attenuated virulence phenotype or whether these inlA PMSCs accumulated as a result of the evolution of environmentally adapted ecotypes with already reduced virulence, which did not require a full-length InlA protein for ecological success. Further studies using a carefully planned set of isogenic mutants (including a DUP-1062A isolate with a replacement of PMSC mutation type 3) and appropriate animal infection models (17) are necessary to more conclusively define the contribution of the inlA PMSC mutations observed here to virulence attenuation, particularly in light of recent data that showed that isogenic restoration of another inlA PMSC failed to completely restore virulence in a chicken embryo assay (27).
Truncated and secreted InlA is not associated with a healthy L. monocytogenes fecal carriage state in ruminants.
The occurrence of at least nine distinct evolutionary events that gave rise to ecologically successful inlA allelic variants carrying PMSC mutations supports the hypothesis that the production of a truncated and secreted InlA protein may provide a distinct selective advantage in at least some specific niche. Olier et al. (28) suggested that L. monocytogenes strains with truncated InlA may be associated with a human fecal carriage state. However, recent studies indicated that the prevalence of L. monocytogenes in human stool samples from the general population is often <1% (8, 9, 33). Since fecal L. monocytogenes carriage in healthy farm ruminants appears to be quite common (20, 25, 40), we hypothesized that a truncated and secreted InlA may be associated with L. monocytogenes fecal carriage in healthy ruminants. Screening by sequencing approximately 1,600 bp at the 3′ end of inlA of 58 L. monocytogenes fecal isolates from healthy ruminants (set 2; Table 1), representing ribotypes commonly isolated from ruminant fecal samples, showed that none of these 58 isolates carried PMSCs in the 3′ 1,600 bp of inlA. These results are consistent with Pathogen Tracker database query results, which showed a very low prevalence of ribotypes associated with inlA PMSCs among 1,297 animal isolates (Table 4). We thus conclude that ruminants, including animals with clinical listeriosis and asymptomatic fecal carriers, do not represent a reservoir for L. monocytogenes strains with inlA PMSCs.
Conclusions.
While several lines of evidence suggest that L. monocytogenes strains differ in their ability to cause human disease (6, 11), limited information on the specific genetic characteristics responsible for these virulence differences is available. In conjunction with a recent study of 150 food and 300 human isolates collected in France (11), our data strongly suggest that a considerable proportion of L. monocytogenes strains isolated from foods have a reduced ability to invade human intestinal epithelial cells. Our data also support that this attenuated invasiveness is linked to multiple distinct SNPs in inlA which lead to the production of truncated and secreted InlA molecules. Since L. monocytogenes strains with inlA PMSCs have been isolated from some human cases, further studies to determine whether human listeriosis cases caused by isolates with PMSCs in inlA are associated with specific host factors (e.g., extreme immunosuppression) are appropriate. Nevertheless, the data presented here have important implications for microbial risk assessments and regulations targeting L. monocytogenes by identifying genetically well-defined and biologically meaningful L. monocytogenes subpopulations that appear to have reduced human virulence. In addition to the significance of our findings for food safety and public health, our data contribute to our understanding of the ecology and evolution of L. monocytogenes. In particular, future efforts to elucidate the selective advantages associated with the production of a truncated and secreted InlA protein may provide a better understanding of the function of L. monocytogenes internalins in different hosts and nonhost environments.
Acknowledgments
This work was supported by NIH grant R01GM63259.
We thank Qi Sun (Computational Biology Service Unit, Cornell University) for setting up the computing system to perform evolutionary analyses. We also thank J. Miller for providing strain EJL12, H. Marquis for providing strain HEL-137 and for the construction of strain HEL-399, T. Potter for providing mouse anti-InlA antiserum, E. Fortes and A. Ho for expert technical assistance, and M. Samadpour for serotyping.
REFERENCES
- 1.Barkardjiev, A. I., B. A. Stacy, S. J. Fisher, and D. A. Portnoy. 2004. Listeriosis in the pregnant guinea pig: a model of vertical transmission. Infect. Immun. 72:489-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes: the influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 3.Camili, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of pclA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doumith, M., C. Cazalet, N. Simones, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusnoik, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvelin, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simones, A. Tierrez, J. A. Vaszquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 6.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Food and human isolates of Listeria monocytogenes form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray, M. L., and H. H. Killinger. 1966. Listeria monocytogenes and Listeria infections. Bacteriol. Rev. 30:309-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grif, K., G. Patscheider, M. P. Dierich, and F. Allerberger. 2003. Incidence of fecal carriage of Listeria monocytogenes in three healthy volunteers: a one-year prospective study stool survey. Eur. J. Clin. Microbiol. Infect. Dis. 22:16-20. [DOI] [PubMed] [Google Scholar]
- 9.Grif, K., I. Hein, M. Wagner, E. Brandi, O. Mpamugo, J. McLauchlin, M. P. Dierich, and F. Allerberger. 2001. Prevalence and characterization of Listeria monocytogenes in the feces of healthy Austrians. Wien. Klin. Wochenshr. 113:737-742. [PubMed] [Google Scholar]
- 10.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 11.Jacquet, C., M. Doumith, J. I. Gordon, P. M. V. Martin, P. Cossart, and M. Lecuit. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 12.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 13.Jonquieres, R., H. Bierne, J. Mengaud, and P. Cossart. 1998. The inlA gene of Listeria monocytogenes L028 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66:3420-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, H., H. Marquis, and K. J. Boor. 2005. σB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151:3215-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, H., K. J. Boor, and H. Marquis. 2004. Listeria monocytogenes sigma B contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72:7374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lecuit, M., D. M. Nelson, S. D. Smith, H. Khun, M. Huerre, M. C. Vacher-Lavenu, J. I. Gordon, and P. Cossart. 2004. Targeting and crossing of the human maternofetal barrier by Listeria monocytogenes: role of internalin interaction with trophoblast E-cadherin. Proc. Natl. Acad. Sci. USA 101:6152-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecuit, M., S. Vandormael-Pournin, J. Lefort, M. Huerre, P. Gounon, C. Dupuy, C. Babinet, and P. Cossart. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722-1724. [DOI] [PubMed] [Google Scholar]
- 18.McLauchlin, J. 1990. Distribution of serovars of Listeria monocytogenes isolated from different categories of patients with listeriosis. Eur. J. Clin. Microbiol. Infect. Dis. 9:210-213. [DOI] [PubMed] [Google Scholar]
- 19.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCraig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng, J., and M. P. Doyle. 1997. Emerging issues in microbiological food safety. Annu. Rev. Nutr. 17:255-275. [DOI] [PubMed] [Google Scholar]
- 21.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of Listeria monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 22.Mereghetti, L., P. Lanotte, V. Savoye-Marczuk, N. Marquet-Van Der Mee, A. Audurier, and R. Quentin. 2002. Combined ribotyping and random multiprimer DNA analysis to probe the population structure of Listeria monocytogenes. Appl. Environ. Microbiol. 68:2849-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nightingale, K. K., K. Windham, and M. Wiedmann. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187:5537-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nightingale, K. K., Y. H. Schukken, C. R. Nightingale, E. D. Fortes, A. J. Ho, Z. Her, Y. T. Grohn, P. L. McDonough, and M. Wiedmann. 2004. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70:4458-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norton, D. M., M. A. McCamey, K. L. Gall, J. M. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Molecular studies on the ecology of Listeria monocytogenes in the smoked fish processing industry. Appl. Environ. Microbiol. 67:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olier, M., D. Garmyn, S. Rousseaux, J. P. Lemaitre, P. Piveteau, and J. Guzzo. 2005. Truncated internalin A and asymptomatic Listeria monocytogenes carriage: in vivo investigation by allelic exchange. Infect. Immun. 73:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olier, M., F. Pierre, S. Rousseaux, J. P. Lamaitre, A. Rousset, P. Piveteau, and J. Guzzo. 2003. Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 71:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olier, M., F. Pierre, J. P. Lemaitre, C. Divies, A. Rousset, and J. Guzzo. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 30.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 31.Rousseaux, S., M. Olier, J. P. Lamaitre, P. Piveteau, and J. Guzzo. 2004. Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 70:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP: DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 33.Sauders, B. D., D. Pettit, B. Currie, P. Suits, A. Evans, K. Stellrecht, D. M. Dryja, D. Slate, and M. Wiedmann. 2005. Low prevalence of Listeria monocytogenes in human stool. J. Food Prot. 68:178-181. [DOI] [PubMed] [Google Scholar]
- 34.Schlech, W. F. 2000. Foodborne listeriosis. Clin. Infect. Dis. 31:770-775. [DOI] [PubMed] [Google Scholar]
- 35.Seeliger, H. P. R., and K. Hohne. 1979. Serotyping of Listeria monocytogenes and related species, p. 31-49. In T. Bergen and J. R. Norris (ed.), Methods in microbiology. Academic Press, London, United Kingdom.
- 36.Snyder, A., and H. Marquis. 2003. Restricted translocation across the cell wall regulates secretion of the broad-range phospholipase C of Listeria monocytogenes. J. Bacteriol. 185:5953-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swofford, D. 1997. Phylogenetic analysis using parsimony (and other methods). Sinauer Associates, Sunderland, Mass.
- 38.U.S. Food and Drug Administration. 2003. Serodiagnosis of Listeria monocytogenes. In Bacteriological analytical manual (BAM). U.S. Food and Drug Administration, Washington, D.C. [Online.] http://www.cfsan.fda.gov/∼ebam/bam-11.html.
- 39.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 15:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wesley, I. V. 1999. Listeriosis in animals, p. 39-73. In E. T. Ryser and E. H. Marth (ed.), Listeria listeriosis and food safety, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 41.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524-531. [PubMed] [Google Scholar]
- 42.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]