Abstract
Maltose and maltotriose are the major sugars in brewer's wort. Brewer's yeasts contain multiple genes for maltose transporters. It is not known which of these express functional transporters. We correlated maltose transport kinetics with the genotypes of some ale and lager yeasts. Maltose transport by two ale strains was strongly inhibited by other α-glucosides, suggesting the use of broad substrate specificity transporters, such as Agt1p. Maltose transport by three lager strains was weakly inhibited by other α-glucosides, suggesting the use of narrow substrate specificity transporters. Hybridization studies showed that all five strains contained complete MAL1, MAL2, MAL3, and MAL4 loci, except for one ale strain, which lacked a MAL2 locus. All five strains also contained both AGT1 (coding a broad specificity α-glucoside transporter) and MAL11 alleles. MPH genes (maltose permease homologues) were present in the lager but not in the ale strains. During growth on maltose, the lager strains expressed AGT1 at low levels and MALx1 genes at high levels, whereas the ale strains expressed AGT1 at high levels and MALx1 genes at low levels. MPHx expression was negligible in all strains. The AGT1 sequences from the ale strains encoded full-length (616 amino acid) polypeptides, but those from both sequenced lager strains encoded truncated (394 amino acid) polypeptides that are unlikely to be functional transporters. Thus, despite the apparently similar genotypes of these ale and lager strains revealed by hybridization, maltose is predominantly carried by AGT1-encoded transporters in the ale strains and by MALx1-encoded transporters in the lager strains.
In all-malt brewer's worts, maltose accounts for ca. 60% of the total fermentable sugars and glucose and maltotriose each account for ca. 20%. Adjunct carbohydrates may be added to worts, particularly in so-called high gravity brewing, and often contain relatively more glucose. The sugars are fermented in the order glucose, maltose and maltotriose. This order results because glucose represses the synthesis of maltose and maltotriose transporters and of the α-glucosidases (maltases) that hydrolyze these sugars inside the cell (see references 9 and 14 and references therein). Glucose also inactivates preexisting maltose transporters (16, 21), a process that also occurs when brewer's yeast strains are added to wort (20). Maltose transport limits the rate of maltose fermentation (10, 15, 20). Significant amounts of maltotriose and maltose can remain in beer, lowering the yield of ethanol and causing flavor problems. Ale and lager yeasts appear to differ in their ability to utilize maltotriose, with lager strains utilizing maltotriose faster so that residual maltotriose is more common in ale fermentations (26). Maltose transport is more strongly inhibited by glucose in some ale strains than in some lager strains (5, 20). Maltotriose, sucrose, and trehalose were much stronger inhibitors of maltose transport by ale strains than by lager strains (20).
The five very similar, unlinked MAL loci (MAL1 to -4 and MAL6) each contain up to three different genes: each MALx1 (where x = 1 to 4 or 6) encodes a maltose transporter, MALx2, a maltase, and MALx3, a transcriptional activator of the other two genes (1, 3, 4, 10, 25). The MAL loci each map to the telomeric region of a different chromosome, MAL1 to chromosome VII, MAL2 to chromosome III, MAL3 to chromosome II, MAL4 to chromosome XI and MAL6 to chromosome VIII. Strains of Saccharomyces cerevisiae can contain maltose transporters encoded by several different MALx1 genes, as well as AGT1, MPH2, and MPH3 genes (see below). The substrate specificities of the different transporters are not well defined. Problems have included obtaining strains with defined, single transporters and obtaining sufficiently pure radioactively labeled substrates. The ability to ferment and grow on different sugars also has been used to indicate the substrate specificity of the maltose transporters, but the interpretation of these data can be ambiguous.
Chang et al. (2) were the first to obtain a yeast strain in which the maltose transporter was encoded by a single defined gene, MAL61 (2). This strain can transport and grow on maltose and turanose but not on maltotriose, α-methylglucoside or melezitose, suggesting that the Mal61 transporter is specific for maltose and turanose. The other Malx1 transporters are 97% identical to Mal61 and may have similar substrate specificity.
An allele of MAL11 (“MAL1g”) (3) with only 57% identity to MAL11 was characterized by Han et al. (11) and renamed AGT1 for α-glucoside transporter. Yeast strains carrying the AGT1 gene on a plasmid can ferment maltotriose, isomaltose, palatinose, and α-methylglucoside in addition to maltose and turanose, suggesting that Agt1 transporters have a relatively broad specificity, including, importantly, maltotriose. Subsequently, the Agt1 transporter was shown to carry at least maltose, maltotriose, sucrose, trehalose, melezitose, and α-methylglucoside, with trehalose the preferred substrate (23).
The hypothesis that Malx1 transporters are highly specific and unable to carry maltotriose has been challenged (7). Strains with MAL31, MAL61, or AGT1 as the sole maltose transporter gene grew well on maltotriose and transported radiolabeled maltose and maltotriose with maximum velocities close to 40 nmol min−1 mg of dry yeast−1 and Km values between 2.7 and 7.2 mM. The Agt1 transporter had a slightly lower Km for maltotriose (4.0 mM) than for maltose (5.1 mM), while the Mal31 and Mal61 transporters had lower Km values for maltose than for maltotriose. These results conflict with earlier findings that strains containing only Mal61 maltose transporter cannot grow on maltotriose (2, 11) and that Mal21p does not support H+-symport activity with maltotriose (24). Day et al. (7) discuss the possibility that small sequence differences between MALx1 genes from different strains could alter the substrate specificity of the encoded transporters. A further complication is that use of commercially available [14C]maltotriose without further purification can overestimate the rate of maltotriose transport by more than fourfold (8).
The S. cerevisiae genome contains two genes, YDL247w on chromosome IV and YJR160c on chromosome X, that are identical to each other, 75% identical to MAL61 and 53% identical to AGT1 and have been named, respectively, MPH2 and MPH3 (for maltose permease homologues). When introduced on single-copy plasmids to a strain lacking a functional maltose transporter, these genes confer the ability to grow on maltose, maltotriose, α-methylglucoside, and turanose, and to transport radiolabeled maltose and maltotriose (6). Km values were 4.4 and 7.2 mM for maltose and maltotriose, respectively, and the maximum rate was higher (49 U mg dry yeast−1) for maltotriose than for maltose (39 U mg dry yeast−1).
Jespersen et al. (12) mapped maltose transporter genes in 30 brewer's yeast strains by hybridization of specific probes to separated chromosomes. There were few differences between the ale strains compared to lager strains that might explain the different kinetics of maltose transport by ale strains and lager strains. In particular, all five ale strains and 22 of 25 lager strains contained MAL11, MAL31, MAL41, and AGT1 genes. MAL21 was present in 13 of 25 lager strains but none of the ale strains and MPH2 was present in 24 of 25 lager strains but only one ale strain. MAL61 and MPH3 were not detected in any strain.
The primary objectives of the present study were (i) to determine why maltose transport is strongly inhibited by other α-glucosides in some brewer's strains, but not in others (20), and (ii) to determine why brewer's strains differ in their ability completely to ferment maltose and maltotriose (26). Competition between maltose and maltotriose for transporters could be partially responsible for their incomplete consumption during fermentations of high-gravity worts. Our results show that in some brewer's yeast strains some of the apparently multiple maltose transporter genes do not code functional transporters. This can account for some marked differences between strains in α-glucoside transport and wort fermentation.
MATERIALS AND METHODS
Materials.
d-[U-14C]maltose was from Amersham Biosciences (Espoo, Finland). Maltose for uptake experiments (minimum purity, 99%), maltotriose (minimum purity, 95%) and trehalose were from Sigma-Aldrich (Helsinki, Finland). Maltose for growth media and α-methylglucoside (methyl-α-glucopyranoside) were from Fluka (Helsinki, Finland).
Strains.
Four industrial lager strains (A15, A24, A64, and A72) and two industrial ale strains (A60 and A179) from VTT's collection were used as typical representatives of strains in current industrial use. Strains A180 and A181 were isolated as single cell clones from A179 and appeared identical to A179. The laboratory strains were S288C, S150-2B, CEN.PK2-1D (VW-1B), and RH144-3A and the chromosome marker strain, YNN295, from Bio-Rad (Espoo, Finland). To determine the Mal-phenotypes, strains were grown in media with 1% yeast extract—2% Difco (Sparks, MD) Bacto peptone (YP) containing 2% maltose for 2 days at 25°C. All strains reached an optical density at 600 nm (OD600) of 10 to 12, except for the three laboratory strains S288C, S150-2B, and RH144-3A, whose OD600 remained below 1.4. These three laboratory strains were defined as Mal-negative.
Maltose transport.
Yeasts were grown in 100 ml of YP-4% maltose in 250-ml flasks shaken at 150 rpm and 25°C to an OD600 of 6 to 12, corresponding to about 2 to 4 mg of dry yeast/ml. Under these conditions residual maltose was between 2 and 0.5%. The yeast were harvested by centrifugation (10 min, 9,000 × g, 0°C), washed with ice-cold water and then with ice-cold 0.1 M tartrate-Tris (pH 4.2), and finally suspended in the same buffer to 200 mg of fresh yeast/ml. Zero-trans [14C]maltose uptake rates were immediately determined essentially as described by Lucero et al. (17), and 1 U catalyzes the uptake of 1 μmol of maltose min−1 (5 mM maltose, pH 4.2, 20°C).
PFGE.
Yeast strains were propagated in YP-2% glucose for 2 days at 30°C and then harvested by centrifugation (3,000 × g, 5 min, 4°C). Supernatants were decanted, and cells were resuspended in 10 ml of 4°C 50 mM EDTA (pH 8). Cell concentrations were determined by OD600 measurements, and 8 × 106 to 60 × 106 cells were placed in each sample plug. Sample plugs were prepared with the CHEF Genomic DNA Plug Kit for Yeast (Bio-Rad). Cells were centrifuged (3 min, 5,000 × g, 4°C) and resuspended in the kit's Cell Suspension Buffer. Lyticase was added to a 150-U/ml final concentration, followed immediately by melted 2% Clean Cut agarose to a final concentration of 0.75%. These mixtures were dispensed into molds and allowed to solidify to produce plugs that could be loaded into the sample wells of the pulsed-field gel electrophoresis (PFGE) apparatus. After the agarose solidified, the sample plugs were pushed out of the molds into the kit's lyticase buffer containing 170 U of lyticase/ml. Plugs were incubated in the lyticase solution for 2 h at 37°C. The lyticase buffer was removed, and the plugs were rinsed with sterile water. Plugs were incubated in the kit's proteinase K reaction buffer containing 240 U of proteinase K/ml for 18 h at 50°C. After that the plugs were washed four times in the kit's washing buffer (1 h per wash) with gentle agitation and stored at 4°C in the same buffer.
Sample plugs were loaded into the wells of a 1.0% UltraPure agarose (Bio-Rad) gel. PFGE was performed at 14°C in 0.5× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA [pH 8]). A CHEF Mapper XA Pulsed Field Electrophoresis system (Bio-Rad) was used with the following settings: 6 V/cm in a 120° angle, pulse length increasing linearly from 26 to 228 s, and total running time of 22 h 52 min. A commercial chromosome marker preparation from S. cerevisiae strain YNN295 (Bio-Rad) was used for molecular mass calibration. After electrophoresis, the gels were stained with ethidium bromide and scanned and quantified with a Typhoon imager (Amersham Biosciences, Espoo, Finland) to estimate the total amount of DNA in each lane.
Chromosome blotting and hybridization.
Chromosomes separated by PFGE were partially depurinized by soaking the gels in 0.25 M HCl for 20 min. After that the gels were treated with 0.5 M NaOH-1.5 M NaCl for 30 min at room temperature and neutralized in 1.5 M NaCl-0.5 M Tris buffer (pH 7.5) for 30 min. The DNA was then transferred to a nylon filter (Hybond-N; Amersham Biosciences) by capillary blotting in 20× SSC (3 M NaCl-0.3 M sodium citrate [pH 7.0]). After blotting, DNA was UV cross-linked to the membrane. Prehybridization was performed at 48°C for 2 h in a hybridization mixture containing 5× SSPE (3 M NaCl-0.2 M sodium phosphate-0.1 M EDTA [pH 7.4]), 5× Denhardt solution, 50% formamide, 100 μg of single-strand salmon sperm DNA, and 1 μg of poly(A) DNA/ml. The hybridization mixture was then replaced with a fresh lot containing a labeled DNA probe at 0.5 × 106 cpm/ml. The probes were labeled with 32P by the random primer protocol using Hexalabel Plus DNA Labeling Kit (Fermentas Life Sciences, Hanover, MD). Unless stated otherwise, hybridization was carried out at 42°C overnight. After hybridization, filters were washed twice with 2× SSC at room temperature for 5 min and then in 0.1× SSC-0.1% sodium dodecyl sulfate (SDS) at 65°C for 40 min. Filters were exposed overnight to a phosphorimager screen, which was then scanned and signals quantified with ImageQuant software (Amersham Biosciences). Hybridization signals were normalized to the total amount of DNA in each gel lane before blotting.
Expression studies.
Yeasts were pregrown on YP-2% glucose, inoculated into 50 ml of YP containing 2% maltose, 2% glucose, or 1% maltose-1% glucose to give an initial OD600 of 0.5 and incubated with shaking (225 rpm) at 24°C in 250-ml flasks. At intervals, samples were withdrawn and centrifuged (1,800 × g, 2 min, 4°C). RNA was isolated from the yeast pellets with TRIzol Reagent (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Glucose, maltose, and ethanol in the supernatants were analyzed by high-pressure liquid chromatography, using a Waters 2690 Separation Module (Waters Corp., Milford, MA) and a Waters System Module liquid chromatography coupled to a Waters 2414 differential refractometer and Waters 2487 dual wavelength absorbance detector. The columns were a 100-by-7.8-mm Fast Acid column and a 300-by-7.8-mm HPX87H column from Bio-Rad. The columns were equilibrated and run with 2.5 mM H2SO4. Elution was at 0.5 ml/min and 60°C.
RNA was isolated by adding 1 ml of TRIzol reagent and 500 μg glass beads to 30 to 50 μg of yeast cells and homogenizing them with a Bead Beater (Howard Industries, Milford, IL) five times for 35 s. Homogenized samples were incubated for 5 min at room temperature (20 to 25°C). Chloroform (0.2 volumes) was added, and the tubes were shaken vigorously by hand for 15 s, incubated for 2 min at room temperature, and centrifuged (12,000 × g, 15 min, 4°C). The aqueous phase was collected, and RNA was precipitated by adding a 0.5 volume of isopropanol, followed by incubation for 10 min at room temperature and then centrifugation (12,000 × g, 10 min, 4°C). The RNA pellet was washed once with 75% ethanol and then dissolved in water.
RNA samples were fractionated on formaldehyde-agarose gels and blotted onto nitrocellulose filters by capillary blotting in 20× SSC. Filters were prehybridized for 1 h at 42°C in 15 ml of a hybridization solution containing 50% deionized formamide, 10% dextran sulfate, 1 M NaCl, 1% SDS, and 125 μg of herring sperm DNA/ml. After prehybridization, the probe (see Table 2) was added to 0.5 × 106 cpm/ml, and the incubation continued overnight. Probes were labeled with 32P by using the Hex Label Plus DNA Labeling kit (Fermentas Life Sciences). To correct for variations in loading, SYBR Green II (BMA)-stained gels were scanned with a Typhoon fluorescence imager before blotting and rRNA bands were quantified. Blots were washed once in 5× SSPE for 15 min at 42°C, twice in 1× SSPE-0.1% SDS for 15 min at 42°C and twice in 0.1× SSC-0.1% SDS for 15 min at 42°C. Blots were visualized by mounting them on a phosphorimager screen overnight (unless stated otherwise) and scanned and quantified with a Typhoon phosphorimager and ImageQuant software.
TABLE 2.
PCR primers used to make probes
| Gene | Directiona | Primer sequence | Sequence detectedb |
|---|---|---|---|
| AGT1c | F | 5′-TTGCTTTACAATGGATTTGGC-3′ | 842-1828 |
| R | 5′-CTCGCTGTTTTATGCTTGAGG-3′ | ||
| MAL13(AGT1)d | F | 5′-GACTTTAACTAAGCAAACATGC-3′ | 3-580 |
| R | 5′-CGTTCGATATTTGTGCAAAGCT-3′ | ||
| MAL61c | F | 5′-GGAGCCTTTCTATGCCCTGC-3′ | 361-1140 |
| R | 5′-TAATGATGCACCACAGGAGC-3′ | ||
| MAL62 | F | 5′-GCGTTGATGCTATTTGGGTT-3′ | 155-933 |
| R | 5′-GAAAAATGGCGAGGTACCAA-3′ | ||
| MAL63 | F | 5′-GTATTGCGAAACAGTCTTGC-3′ | 5-886 |
| R | 5′-CATCGACACAGTTAGTAGCC-3′ | ||
| MAL33 | F | 5′-ATAGCTCCACCTCAGCCAGA-3′ | 772-1313 |
| R | 5′-TGATTGCAATGTTTCAGGGA-3′ | ||
| MPHxe | F | 5′-TCCGTGGATCTTGTTGGAAA-3′ | 505-1294 |
| R | 5′-GTCCAAAAGCGTAAAGGTCA-3′ |
F, forward; R, reverse.
The numbering is from the first nucleotide of the translational start.
AGT1 and MAL61 primers were the same as in reference 12.
Primers were designed with the Saccharomyces Genome Database primer design program (http://seq.yeastgenome.org/cgi-bin/web-primer).
MPHx primers were the same as in reference 6.
Sequencing of AGT1 genes.
AGT1 gene sequences of each strain were amplified by PCR with the specific primers AGT1 frw (5′-ATGAAAAATATCATTTCATTGGT-3′) and AGT1 rev (5′-TTAACATTTATCAGCTGCATTT-3′) from both ends of the gene. Genomic DNA of four brewer's strains (A15, A24, A60, and A179) was used as the template. The PCR-generated fragments were cloned by using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) and propagated in Escherichia coli. Plasmid DNA was isolated from independent E. coli clones and sequenced. Whole gene sequences were obtained with eight sequencing primers; universal M13 Forward and Reverse primers to sequence the start and end of the pCR-TOPO plasmid-ligated AGT1 gene and six internal primers from the coding strand. In addition to these clones when the whole gene was sequenced, an additional 20 independent clones from each strain were sequenced with only one of the internal sequencing primers (Sekv4, 5′-AAAGCAGATTGAATTGAC-3′). This primer, starting at nucleotide 1011, gave a readable sequence from approximately nucleotides 1150 to 1500, which includes the region where the genes from some strains appeared to have a frame shift. The model 3100 Genetic Analyzer sequencer was used (Applied Biosystems, Foster, CA). Multiple alignments were performed with the Multalin program at http://prodes.toulouse.inra.fr/multalin/multalin.html.
RESULTS
We characterized the maltose uptake kinetics of several brewer's yeast strains and then determined the (apparent) MAL genotypes of these strains by hybridization studies. Because brewer's strains are aneuploid and, in the case of lager strains, alloploid (13), we attempted to quantitate the MALx1 gene doses in these strains. We then examined the expression levels of the MALx1, AGT1, and MPHx genes and compared the AGT1 gene sequences in ale and lager strains.
Inhibition of maltose transport by trehalose and maltotriose.
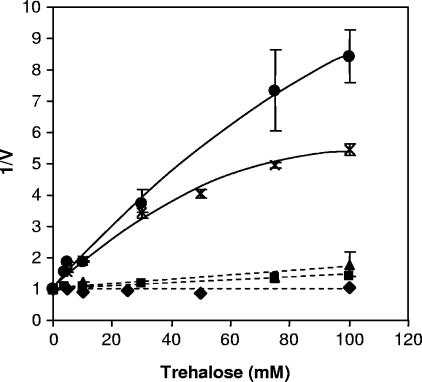
Trehalose is the preferred substrate of the Agt1 transporter but is not a substrate for the Malx1 and Mphx transporters (6, 23). Therefore, trehalose is expected to inhibit competitively maltose transport by the Agt1 transporters but not necessarily to affect transport by the Malx1 and Mph1 transporters. Maltose transport in both ale strains was strongly inhibited (50% at 10 mM trehalose and over 80% at 100 mM trehalose). For ale strain A60, inhibition leveled off at ∼82% (1/V = 5.5 in Fig. 1), suggesting that inhibition is partial, with ∼18% of the maltose transport capacity insensitive to trehalose. For ale strain A179, inhibition was 88% (1/V = 8.3) and still rising at 100 mM trehalose, suggesting that a smaller fraction (<10%) of the maltose transport capacity is insensitive to trehalose. All three lager strains were less sensitive to trehalose (Fig. 1). For A24 and A64, inhibition reached 30 and 55% (1/V values of 1.4 and 1.8, respectively) at 100 mM trehalose, whereas maltose transport by lager strain A15 was not affected by up to 100 mM trehalose. This result suggests that maltose-grown A15 does not express maltose transporters capable of carrying trehalose, whereas in A24 and A64 these transporters can account for 30 to 50% of the maltose transport capacity at 5 mM maltose.
FIG. 1.
Inhibition of maltose transport by trehalose. Rates of zero-trans uptake of maltose were measured at 5 mM [14C]maltose in the presence of the indicated concentrations of trehalose. For each strain, the rate in the absence of trehalose was set at 1.00. The figure shows the reciprocal rates (1/V) plotted against trehalose concentration for two ale yeasts, A60 (×) and A179 (•) and three lager yeasts: A15 (⧫), A24 (▪), and A64 (▴). Error bars show the range between two or three independent experiments.
Also maltotriose and α-methylglucoside strongly inhibited maltose transport by both ale strains but only weakly inhibited transport by the lager strains (Table 1). Maltotriose is a substrate of Agt1, Mphx, and Mal31 transporters, and α-methylglucoside is a substrate of Agt1 and Mphx but not of the Mal31 transporter (see reference 6).
TABLE 1.
Inhibition by maltotriose and α-methylglucoside of [14C]maltose transporta
| Inhibition type | Inhibition with strain:
|
||||
|---|---|---|---|---|---|
| A15 | A24 | A64 | A60 | A179 | |
| Avg inhibition by maltotriose (%) ± SD | 12 ± 7 | 16 ± 11 | 24 ± 4 | 66 ± 3 | 59 ± 25 |
| Avg inhibition by α-methyglucoside (%) ± range | 23 ± 3 | 10 ± 10 | 13 | 74 ± 3 | 41 |
Transport of 5 mM [14C]maltose was measured with the indicated strains (grown on maltose) in the presence or absence of 50 mM maltotriose or 170 mM α-methylglucoside. Where errors are shown, results for maltotriose are averages ± standard deviations (n = 3 to 6) and results for α-methylglucoside are averages of duplicates ± ranges.
These kinetic results suggest that when these brewer's strains are grown on maltose, high-specificity maltose transporters, such as Malx1p, account for 0 to 15% of the maltose transport capacity of ale strains but for 40 to 80% of the maltose transport capacity of lager strains. Correspondingly, low-specificity transporters, such as Agt1p and Mphp, are predicted to be important in the ale strains.
MAL genotypes of the strains.
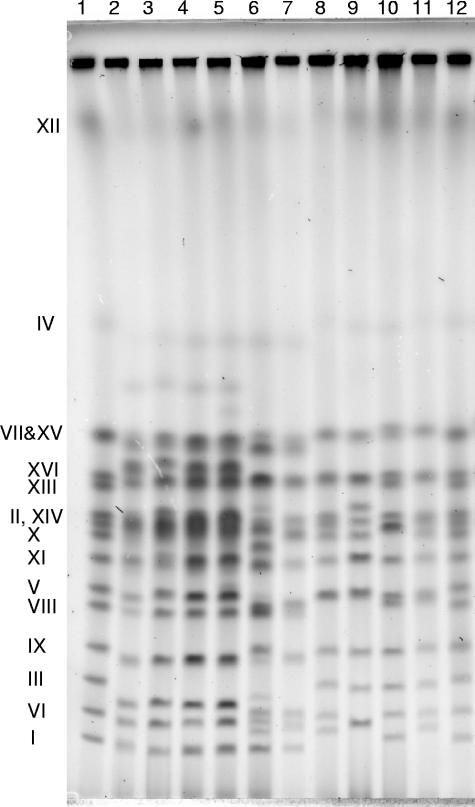
To test this hypothesis, we first identified which MAL loci were present in the strains. At low DNA loads, PFGE resolved 15 chromosome bands from the marker yeast strain, YNN295 (Fig. 2). Chromosomes VII and XV were presumed to comigrate as a duplex. Chromosome IV, faint in Fig. 2, is clearly visible in the brewer's yeasts at higher DNA loads (Fig. 3A). The four laboratory strains all differed from each other and from the marker strain in the size of one or more chromosomes. For the brewer's strains, greater differences appeared, including bands not present in the marker strain. Bands at or near the positions of chromosomes that often carry MAL loci—i.e., chromosomes VII, III, II, XI, and VIII—could be tentatively identified, although chromosome III was smaller in the lager strains than in the YNN295 marker strain. Chromosome III could not be identified in the ale strains based on mobility alone. Blots of these PFGE gels were hybridized sequentially with specific probes for the transporter, maltase, and activator genes at MAL loci (Table 2). Previously sequenced MALx1 genes are 97% identical, and the MAL61 probe is expected to hybridize to all of them. In some MAL1 loci the transporter gene is the AGT1 allele instead of MAL11 (11), and we used a specific AGT1 probe to detect this allele. MALx2 genes are 97% identical and the MAL62 probe is expected to hybridize to all of them. There is greater variation among MALx3 genes: MAL13, MAL23, and MAL43 are 91 to 94% identical to MAL63, but MAL33 and an allele of MAL13, found in MAL1 loci containing AGT1 and referred to in the present study as MAL13(AGT1), share only ca. 75% identity to each other and to other MALx3 sequences. We used specific probes [MAL33 and MAL13(AGT1)] to detect these two genes.
FIG. 2.
PFGE separation of chromosomes from laboratory and brewer's strains. The gels were loaded with 8 × 106 cells/lane of the brewer's strains and 30 × 106 cells/lane of the laboratory strains. Strains were as follows: lane 1, chromosome marker strain YNN295; lane 2, A15; lane 3, A24; lane 4, A64; lane 5, A72; lane 6, A60; lane 7, A179; lane 8, S288C; lane 9, S150-2B; lane 10, CEN.PK2-1D; lane 11, RH144-3A; lane 12, chromosome marker strain YNN295. Chromosomes are identified on the left: chromosomes VII and XV are not resolved; chromosome II travels immediately above chromosome XIV.
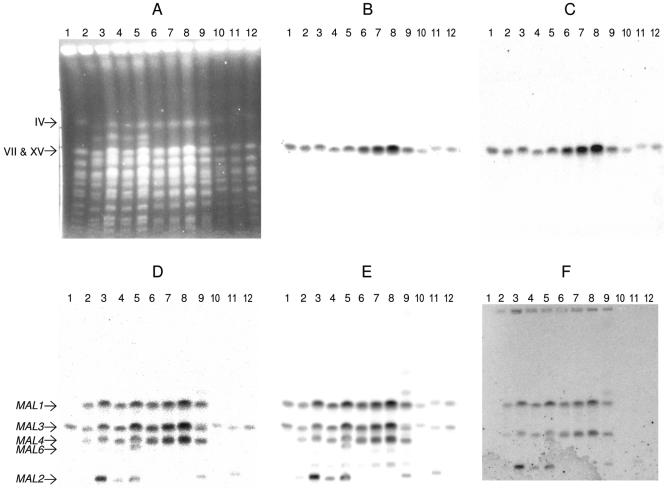
FIG. 3.
Detection of MAL loci. A PFGE gel was loaded with about 30 × 106 cells/lane. Strains were as follows: lane 1, Marker YNN 295; lane 2, A15; lane 3, A24; lane 4, A64; lane 5 A72; lane 6, A179; lane 7, A180; lane 8, A181; lane 9, A60; lane 10, S150-2B; lane 11, CEN.PK2-1D; and lane 12, RH144-3A. (A) Separated chromosomes stained with ethidium bromide. Chromosome IV and the duplex VII/XV are indicated. The gel was then blotted, and the blot was hybridized with the following probes: AGT1 (B), MAL13(AGT1) (C), MAL61 (D), MAL62 (E), and MAL63 (F). The MAL loci are identified on the left of panel D. The image for panel F was darkened to make the bands more visible.
The MAL61 and MAL62 probes hybridized in many lanes at the expected positions of the VII/XV duplex and chromosomes II, XI, and III, which carry, respectively, MAL1, MAL3, MAL4, and MAL2 loci (Fig. 3D). The AGT1 and MAL13(AGT1) probes hybridized to the VII/XV duplex of all of the strains in the present study, suggesting that they all carried (on chromosome VII) MAL1 loci containing the AGT1 allele of the transporter gene and the MAL13(AGT1) allele of the activator gene. These probes did not hybridize to any other chromosomes. The MAL61, MAL62, and MAL63 probes also bound to the VII/XV duplex of all six brewer's strains (Fig. 3D, E, and F). This pattern suggests that the brewer's strains each contained at least two kinds of chromosome VII, one with a MAL1 locus containing the AGT1 and MAL13(AGT1) alleles and another with a MAL1 locus containing MAL11 and MAL13 alleles. In contrast, MAL61 and MAL63 did not bind at this duplex in the laboratory strains (MAL62 did; Fig. 3E), suggesting that the single copy of chromosome VII in the (haploid) laboratory strains contained only the AGT1 and AGT1(MAL13) alleles.
Three lager strains (A24, A64, and A72), one ale strain (A60) and one laboratory strain (CEN.PK2-1D) contained complete MAL2 loci on chromosome III. Chromosome III is possibly missing from the other ale strain (see A179 in Fig. 2). The CEN.PK2-1D strain is known to have a MAL2-8c locus (22).
The MAL61 and MAL62 probes both bound to chromosome II of all strains, suggesting that all strains carry MAL3 loci with transporter and maltase genes (Fig. 3D and E). As expected, the MAL63 probe did not bind to chromosome II (Fig. 3F), which is consistent with the low identity (75%) between MAL63 and MAL33. However, the MAL33 probe bound to chromosome II of all strains (data not shown), indicating that these MAL3 loci are complete, i.e., they contain transporter, maltase, and activator genes.
In all of the brewer's strains, but none of the laboratory strains, the MAL61, MAL62, and MAL63 probes hybridized to chromosome XI, suggesting that the brewer's strains all carried complete MAL4 loci.
A72 was the only strain where the MAL61, MAL62, and MAL63 probes all hybridized to chromosome VIII, which carries the MAL6 locus. For all of the other strains the hybridization of the MAL61 probe to chromosome VIII was very faint, and hybridization of MAL62 and MAL63 probes could not be detected. None of the 30 brewer's yeasts studied by Jespersen et al. (12) contained a MAL6 locus.
Weak hybridization was sometimes observed to other chromosomes. For example, with ale strain A60, the MAL61, MAL62 and MAL63 probes hybridized to a chromosome of ∼1.3 Mbp (Fig. 3D, E and F). Binding of a MAL61 probe to an ∼1.3 Mbp chromosome also was observed by Jespersen et al. (12) in a lager yeast. For all brewer's strains, the MAL62 probe hybridized weakly to chromosome IX (immediately above the MAL22 bands in Fig. 3E).
Deduced differences in the MAL genotypes (Table 3) between the five brewer's strains were few. In particular, ale strain A60 was identical to lager strains A15, A24, and A64 except for the weakly observed MAL hybridization to the ∼1.3-Mbp chromosome of A60. Ale strain A179, and its derivatives, A180 and A181, were the only brewer's strains lacking a MAL2 locus. A72 was the only strain with a complete MAL6 locus. The maltose-negative laboratory strains S150-2B, RH144-3A, and YNN295 had complete MAL1 [with AGT1 and MAL13(AGT1) alleles] and MAL3 loci. Evidently, their MAL1 and MAL3 loci, even collectively, are insufficient for a maltose-positive phenotype. The maltose-positive laboratory strain, CEN.PK2-1D, had a genotype similar to the maltose negative strains plus a complete MAL2 locus.
TABLE 3.
MAL and MPH genotypes of the studied strainsa
| Locus | Genotype for strain(s):
|
||||||
|---|---|---|---|---|---|---|---|
| S150-2B, RH144-3A, YNN295 | CEN.PK2-1D | A60 | A179, A180, A181 | A15 | A24, A64 | A72 | |
| MAL1 | AGT1 MAL12 MAL13(A)b | AGT1 MAL12 MAL13(A)b | AGT1 MAL11 MAL12 MAL13 MAL13(A)b | AGT1 MAL11 MAL12 MAL13 MAL13(A)b | AGT1 MAL11 MAL12 MAL13 MAL13(A)b | AGT1 MAL11 MAL12 MAL13 MAL13(A)b | AGT1 MAL11 MAL12 MAL13 MAL13(A)b |
| MAL2c | MAL21-23 | MAL21-23 | MAL21-23 | MAL21-23 | ML21-23 | ||
| MAL3c | MAL31-33 | MAL31-33 | MAL31-33 | MAL31-33 | MAL31-33 | MAL31-33 | MAL31-33 |
| MAL4c | MAL41-43 | MAL41-43 | MAL41-43 | MAL41-43 | MAL41-43 | ||
| MAL6 | (MAL61)d | (MAL61)d | (MAL61)d | (MAL61)d | MAL61 | ||
| MAL62 | |||||||
| MAL63 | |||||||
| Other MAL loci | Chr IXe | Chr IXe | Chr IXe 1.3 Mbpf | Chr IXe | Chr IXe | Chr IXe | Chr IXe |
| MPHx | (MPH2)dMPH3 | (MPH2)d MPH3 | MPH2 | MPH2 | |||
| Other MPH loci | Chr VII/XV | ||||||
No entry means that the gene(s) was not detected in this yeast.
MAL13(A) indicates the MAL13(AGT1) allele.
The MAL2, MAL3, and MAL4 loci either were completely absent or contained all three genes MALx1, MALx2, and MALx3, shown as, e.g., MAL21-23.
Parentheses indicate that the hybridization detected was very weak.
The MAL62 probe (but not other probes) bound weakly near chromosome IX in all strains, stronger in brewer's strains.
In A60 MAL61, MAL62, and MAL63 probes bound weakly to a chromosome of ∼1.3 Mbp.
Quantitative analysis of MAL61 and MAL62 hybridization signals.
Brewer's yeasts are aneuploid. The copy number of each chromosome can vary between strains, and the same cell can carry different versions of each chromosome, including those that sometimes carry MAL loci. These differences will change the strengths of the hybridization signals. We normalized the intensity of the hybridization signals obtained with MAL61 and MAL62 probes at each MAL locus relative to the intensities obtained at the MAL3 locus (Table 4). For each locus of strain A15, the signal strengths normalized in this way were similar for both probes, suggesting that in each MAL locus of strain A15 the ratio of MALx1 and MALx2 genes is the same as that in its MAL3 locus. Since the MAL61 and MAL62 probes gave the same result, the different signal intensities probably reflect different copy numbers of the MAL loci in A15 (MAL1 > MAL3 > MAL4 > MAL2 > MAL6) rather than differences in sequence similarity to the probes.
TABLE 4.
Hybridization intensities of MAL61 and MAL62 probes at other MAL loci compared to MAL3a
| Strain | Hybridization intensity of MAL61 and MAL62 probes for MAL loci 1, 2, 4, and 6
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
MAL1
|
MAL2
|
MAL4
|
MAL6
|
|||||
| MAL61 | MAL62 | MAL61 | MAL62 | MAL61 | MAL62 | MAL61 | MAL62 | |
| A15 | 131 | 146 | 15 | 22 | 54 | 63 | 8 | 0 |
| A24 | 118 | 164 | 133 | 183 | 61 | 60 | 10 | 0 |
| A64 | 87 | 143 | 28 | 50 | 66 | 73 | 8 | 0 |
| A72 | 75 | 115 | 17 | 54 | 35 | 54 | 20 | 18 |
| A60 | 127 | 222 | 35 | 57 | 112 | 138 | 0 | 0 |
| A179 | 80 | 119 | 0 | 0 | 78 | 84 | 0 | 0 |
A PFGE gel blot was hybridized sequentially to MAL61 and MAL62 probes. Band intensities were normalized by setting the signals for each probe to 100 at the MAL3 locus.
For the other yeasts (Table 4), the normalized signals at MAL1 and MAL2 loci were markedly stronger with the MAL62 probe than with the MAL61 probe (see Discussion). Nevertheless, there was qualitative agreement between the two probes at all loci and some strain-specific differences in loci strengths were evident. A24 differed from other strains by having the strongest signal strengths at MAL2, suggesting that MAL2 is the most abundant MAL locus in this strain and that in A24 several copies of chromosome III carry a MAL locus. The other strains had weak signals at MAL2, suggesting that in these strains probably only one (or, for A179, no) copy of chromosome III carries a MAL locus. The signal strength at MAL4 was weakest in A72, suggesting that in this strain fewest copies of chromosome XI carry a MAL locus. The lager strains all had more copies of MAL3 loci than MAL4 loci, whereas for the ale strains (A60 and A179) hybridization to MAL3 and MAL4 were similar.
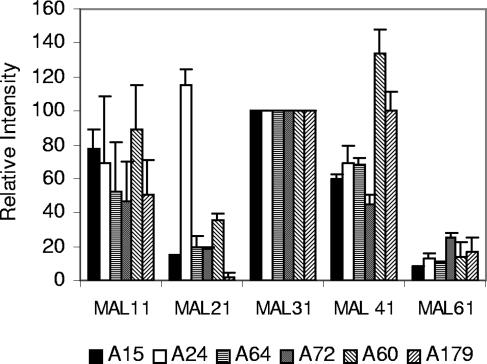
To confirm that these observations were not experimental artifacts of the particular filter probed, three independent filters were probed with MAL61 and the relative intensities of hybridization at different MAL loci compared (Fig. 4). The hybridization patterns were similar to those of Table 4. In particular, the stronger MAL21 signal with A24 than with the other yeasts and the stronger MAL41 signals with ale strains than lager strains were reproducible between gels.
FIG. 4.
MAL61 hybridization band intensities. Blots of three independent PFGE gels with different amounts of DNA (8 × 106 to 30 × 106 cells/lane) were hybridized with the MAL61 probe. Band intensities were normalized by setting MAL31 on each blot to 100. The bars represent the average relative intensity of each MALx1, and the error bars represent the standard deviations.
MPH genotypes.
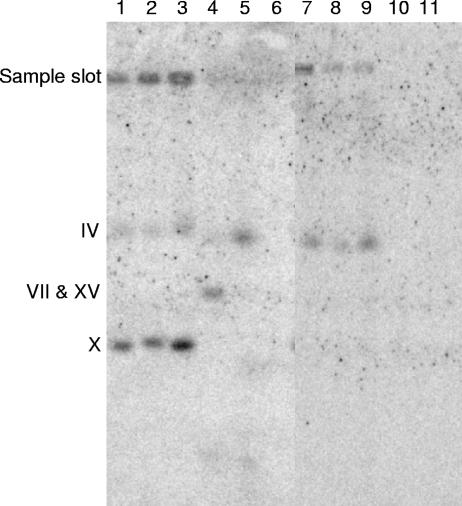
PFGE blots also were hybridized with the MPHx probe, which recognizes the MPH2 and MPH3 sequences found on chromosomes IV and X. The MPHx probe did not bind strongly, and long exposure times were needed to identify the hybridization sites (Fig. 5). Laboratory strains S150-2B, CEN.PK-1D, and RH144-3A (and also YNN295, not shown in Fig. 5) hybridized the MPHx probe to chromosome X and, rather weakly, to chromosome IV. The much weaker hybridization to chromosome IV observed with these haploid strains may indicate that the MPH2 genes, located on chromosome IV, have low sequence similarity to the probe. However, chromosome IV stained only weakly with ethidium bromide in our PFGE gels (Fig. 2 and 3A) and the MPHx probe also hybridized to the sample slot (Fig. 5). Thus, chromosome IV may have been partially trapped in the slot instead of migrating through the gel.
FIG. 5.
PFGE filters probed with MPHx. The two filters (lanes 1 to 6 and 7 to 11, respectively) were probed with MPHx and then exposed for 3 days. The samples were as follows: lane 1, S150-2B; lane 2, CEN.PK2-1D; lane 3, RH144-3A; lane 4, A15; lane 5, A64; lane 6,A60; lane 7, A24; lane 8, A64; lane 9, A72; lane 10, A179; and lane 11, A180. The positions of chromosomes IV and X, the duplex VII/XV, and the sample slot are shown.
The brewer's strains showed three different behaviors (Fig. 5). The ale strains, A60 and A179 (and A180), did not hybridize with the MPHx probe and presumably lack MPHx sequences. For lager strains A24, A64, and A72, the MPHx probe hybridized to chromosome IV and the sample slot, but not to chromosome X. Thus, these three strains carry MPH2 but not MPH3. With lager strain A15, the MPHx probe hybridized to the VII/XV duplex but not to chromosomes X or IV (the faint darkening near chromosome IV in Fig. 5, lane 4, is an artifact not seen on other filters). A15 is the strain from which our MPHx probe was cloned. This PCR was repeated and the sequences of the two independent probes were identical to the corresponding sequence of MPH in the Saccharomyces Genome Database (http://www.yeastgenome.org). Thus, A15 contains at least 790 bp of MPH sequence, and the hybridization signal locates this sequence to the chromosome VII/XV duplex.
Expression of maltose transporter genes during shake flask cultivations.
Six yeast strains were grown on YP-2% glucose, YP-2% maltose, or YP-1% glucose-1% maltose to test whether their maltose transporter genes were expressed. Glucose and maltose (and maltotriose, which we did not study) are the sugars of practical importance in brewery fermentations. In all cases, glucose was exhausted at 9 to 11 h, maltose was exhausted by 25 h, and ethanol peaked at about 10 h and was completely reconsumed by 33 h (data not shown). The incubations were continued up to 48 h.
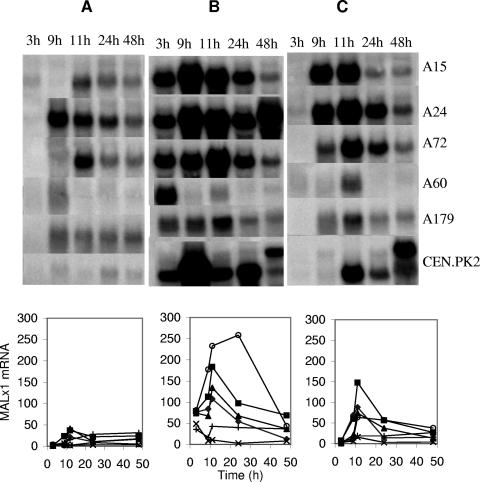
We probed the Northern filters with MAL61 (which recognizes all MALx1 genes), AGT1, MPHx and, as a reference, TDH1 probes (TDH1 encodes isoenzyme 1 of glyceraldehyde-3-phosphate dehydrogenase). Similar results were obtained with normalization to TDH1 or rRNA (data not shown). During and after growth on glucose, expression of MALx1 was low (less than 50 arbitrary units) for all strains (Fig. 6). During growth on 2% maltose, the maltose-positive laboratory strain CEN.PK2-1D and all three lager strains had rapidly increasing levels of MALx1 messengers, reaching 100 to 250 arbitrary units (AU). However, expression remained low in both ale strains. During growth on the 1% glucose-1% maltose mixture, expression of MALx1 by the lager strains and by CEN.PK2-1D was delayed until 9 or 11 h (glucose had decreased to between 0.4 and 0.1% at 9 h; data not shown) and then rose to 60 to 150 AU. Again, the ale strains showed low expression (under 30 AU). Thus, only the lager strains and CEN.PK expressed MALx1 genes strongly, and their expression was repressed by glucose and required maltose.
FIG. 6.
Expression of MALx1 during growths of three lager strains, two ale strains, and a maltose-positive laboratory strain on 2% glucose (A), 2% maltose (B), and 1% glucose-1% maltose (C). The upper panels show blots probed with MAL61. The lower panels show densitometric quantitation with normalization to TDH1. The normalized mRNA amounts are expressed in the same arbitrary units. The results from a single experiment are shown. Similar results were obtained in replicate growths on 2% maltose and 2% glucose. The strains are A15 (⧫), A24 (▪), A60 (×), A72 (▴), A179 (+), and CEN.PK2-1D (○).
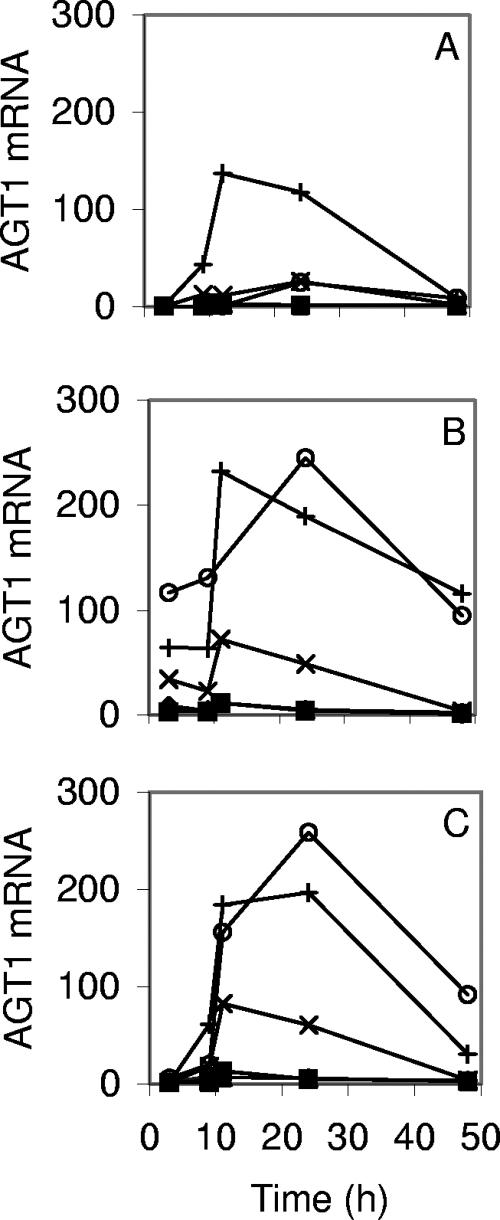
Figure 7 contains normalized plots of AGT1 expression during the same experiments. The lager strains did not express significant amounts of AGT1 under any of these growth conditions (normalized signals were below 13 AU). Ale strain A179 expressed AGT1 abundantly in all three media (140 to 240 AU), although on 2% glucose and 1% glucose-1% maltose expression was delayed until 9 h, by which time glucose had decreased to 0.4 and 0.1%, respectively. Ale strain A60 had a pattern qualitatively similar to that of A179, but the levels of AGT1 messenger were lower (reaching 26 to 80 AU). Although the AGT1 genes of the ale strains were repressed by glucose, those of A179, at least, did not require maltose for induction.
FIG. 7.
Expression of AGT1 during growths on 2% glucose (A), 2% maltose (B), and 1% glucose-1% maltose (C). The mRNA levels are expressed in the same arbitrary units in panels A, B, and C. Similar results were obtained in replicate growths on 2% maltose and 2% glucose. The strains are A15 (⧫), A24 (▪), A60 (×), A72 (▴), A179 (+), and CEN.PK2-1D (○).
Compared to MALx1 and AGT1, expression of the MPHx genes was very low for all strains in all growth conditions (data not shown). To obtain signals, the probed filters were exposed for 3 days instead of overnight. The highest level of apparent expression was with strain A179, which does not contain MPHx genes (Fig. 5 and Table 3). We assume the weak signals detected from these overexposed filters reflect cross-reactivity to a non-MPH messenger.
Sequence of AGT1 in two ale and two lager strains.
AGT1 genes were amplified from four brewer's yeasts in three (A15), two (A24), three (A60), and two (A179) independent PCRs, ligated into the pCR-TOPO plasmid and independent clones isolated and sequenced (Tables 5 and 6). Of the eight whole gene sequences from the two ale strains (A60 and A179), seven encoded full-length, 616-amino-acid proteins, similar to the sequence in the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org). AGT1 is entered as MAL11 because it is the allele of MAL11 present in the S288C strain of the SGD. However, one sequence (from A179) had a stop codon (TGA) at nucleotides 1183 to 1185, leading to a truncated, 394-amino-acid sequence. For the two lager strains, seven of eight whole gene sequences encoded this truncated polypeptide, but one (from A15) encoded the full-length protein. The nucleotide change leading to the stop codon was the insertion of an extra T at position 1183, which results in a stretch of eight consecutive T's instead of seven, the TGA stop codon at positions 1183 to 1185, and a frameshift in the rest of the nucleotide sequence.
TABLE 5.
Sequencing of AGT1 genes from ale and lager strains
| Strain | No. of PCRsa | No. of independent clonesb | No. of sequences
|
|
|---|---|---|---|---|
| 616-amino-acid proteinc | 394-amino-acid polypeptided | |||
| Whole gene sequences | ||||
| A15 | 3 | 5 | 1 | 4 |
| A24 | 2 | 3 | 0 | 3 |
| A60 | 3 | 5 | 5 | 0 |
| A179 | 2 | 3 | 2 | 1 |
| Nucleotides 1153 to 1500 | ||||
| A15 | 3 | 22 | 1 | 21 |
| A24 | 2 | 20 | 0 | 20 |
| A60 | 4 | 18 | 17 | 1 |
| A179 | 4 | 21 | 21 | 0 |
The number of independent PCRs.
The number of independent clones sequenced.
The number of sequences encoding full length, 616-amino-acid proteins.
The number of sequences encoding the truncated, 394-amino-acid polypeptide.
TABLE 6.
Amino acid changes between the proteins coded by the AGT1 genes in the Saccharomyces Genome Database and the ale and lager strains
| TMDb | Amino acida
|
|||||
|---|---|---|---|---|---|---|
| Position | SGD | A60 | A179 | A15 | A24 | |
| 40 | D | N | N | N | N | |
| 45 | K | R | R | |||
| 59 | T | I | ||||
| 74 | V | M | M | |||
| 78 | M | T | T | |||
| 80 | A | T | T | |||
| 83 | D | E | E | |||
| 102 | K | I | I | |||
| 128 | S | N | N | N | N | |
| I (164-184) | 164 | L | Q | Q | Q | Q |
| 168 | M | T | T | |||
| 175 | T | P | P | |||
| 198 | I | V | V | |||
| II (202-222) | 215 | I | V | V | V | V |
| 219 | I | M | M | M | M | |
| 226 | S | G | G | G | G | |
| 228 | A | T | T | T | T | |
| III (241-266) | 276 | G | D | D | ||
| IV (284-311) | 333 | V | I | I | ||
| 359 | N | D | D | D | D | |
| 375 | T | A | A | |||
| 381 | S | T | T | T | T | |
| 385 | V | C | C | C | C | |
| 392 | Y | C | ||||
| 395 | E | Stopc | Stopc | |||
| V (405-427) | 409 | L | V | V | V | V |
| VI (432-459) | 448 | I | V/Id | I/Vd | V | V |
| 459 | S | G | G | G | ||
| VII (461-489) | 488 | A | T | T | ||
| VIII (504-532) | 509 | L | I | |||
| 548 | V | A | ||||
| 556 | T | S | ||||
| 595 | A | D | ||||
Amino acids are shown at positions where the SGD and brewer's yeast protein sequences differ; no entry means the amino acid is the same as in the SGD.
The positions of transmembrane domains (TMDs) indicated in SGD are shown.
The sequences of the lager strains' hypothetical proteins after the Stop codons were obtained by correcting the frameshift.
Residue 448 was the same as in the SGD in 3 of 13 A60 sequences and 10 of 17 A179 sequences.
These results might mean that both A15 and A179 contain both correct and frameshifted versions of AGT1. Alternatively, A15 may contain only the frameshifted version and A179 only the correct version, but during PCR or sequencing reactions the sequences of seven or eight T’s were occasionally misread. To distinguish these hypotheses, we cloned and sequenced ∼20 independent pCR-TOPO clones containing the portion from nucleotides 1153 to 1500 of the AGT1 gene from each strain. For the two ale strains, 38 of 39 sequences encoded full-length proteins (Table 5), strongly suggesting that the single occurrence of the frameshifted sequence represents a PCR or sequencing error rather than the presence of both correct and defective versions of AGT1 in these ale strains. Conversely, 41 of 42 sequences from the two lager strains contain the extra T, strongly suggesting that all copies of AGT1 in these strains are defective. The AGT1 sequences from all four strains are deposited in the NCBI databases as DQ091763 (A60), DQ091764 (A179), DQ091765 (A15), and DQ091766 (A24).
DISCUSSION
The inhibition studies (Fig. 1 and Table 1) indicate that maltose transport by the two studied ale strains was mainly mediated by broad specificity α-glucoside transporters that also can carry trehalose, maltotriose, and other α-glucosides, whereas transport by the three lager strains used mainly transporters with higher specificity for maltose. Indeed, maltose transport by the A15 lager strain was not detectably (±7%) inhibited by 100 mM trehalose. Agt1p is the only transporter known to carry both maltose and trehalose (6, 23). We therefore expected that the Agt1 transporter would be absent from A15 and of minor importance in the other lager strains but would be the dominant maltose transporter of the ale strains.
Based on hybridization studies (Fig. 3), all of our brewer's strains contained both MALx1 and AGT1 genes, as has been previously reported for other brewer's yeasts (12). During growth on maltose, AGT1 genes were strongly expressed in both ale strains and weakly expressed in the three lager strains, whereas MALx1 genes were expressed strongly in the lager strains and weakly expressed in the ale strains (Fig. 6 and 7). The AGT1 sequences from the ale strains encoded full-length proteins expected to be active transporters (a property that has been confirmed by overexpression of these AGT1 genes in laboratory yeasts; V. Vidgren, M.-L. Vehkomäki, L. Ruohonen, and J. Londesborough, unpublished results). In contrast, AGT1 sequences from the two tested lager strains encoded truncated polypeptides that are unlikely to be functional transporters. Thus, the genotypes determined by hybridization were misleading, and the expression and sequence results support the hypothesis that Agt1 transporters dominate in ale strains and Malx1 transporters dominate in lager strains.
If Agt1 transporters are primarily responsible for maltose uptake in ale strains, then in the final stages of wort fermentations by ale strains maltose competes with an excess of maltotriose for the same transporters. This competition may partially explain why residual maltose (and maltotriose) are more common in ale fermentations than lager fermentations (26). The lack of functional Agt1 transporters in A15 and A24 raises the question of how they transport maltotriose. MPHx genes were present in all three lager yeasts, although they were absent from the two ale strains. However, these genes were not expressed in the lager strains during growth on maltose or glucose. Possibly, they are expressed under other conditions, e.g., in the presence of maltotriose. Recently, A15 and three other lager strains have been shown to carry MTT1 genes, which encode maltose transporters with relatively high activity toward maltotriose (8). The MTT1 gene is 98% identical to the Saccharomyces pastorianus mty1 gene (M. Salema-Oom, unpublished; NCBI accession number AJ491328). Another possible maltotriose transporter may be Mal31p. In contrast to early reports that Malx1 transporters cannot carry maltotriose, recent work suggests that Mal31p and Mal61p can carry maltotriose (7) and A15 and A24 both contain MAL31 genes.
The (hypothetical) proteins encoded by the AGT1 genes from the two lager strains were almost identical (Table 6). We have no other reason to think that A15 and A24 are closely related, which suggests that there may be other lager strains with similarly defective AGT1 genes. More sequences are needed to test whether this mutation is a general characteristic of lager strains.
The two ale protein sequences differ at one amino acid residue, 59, in the first 447 amino acids, and at five residues in the remaining 169 amino acids (Table 6). Apparently, two different copies of AGT1 were cloned from each ale yeast since in 3 of 13 sequences from A60 and 10 of 17 sequences from A179 nucleotide 1342 was A (leading to the same amino acid, I448, as in the SGD) and in the remaining sequences it was G (leading to V448). Thus, these strains probably contain at least three different versions of chromosome VII, one carrying MAL11 and two carrying different versions of AGT1. Approximately 30% of the amino acid changes were in transmembrane domains, which account for ∼30% of the total amino acid sequence (Table 6), suggesting that there was no bias for or against changes in the transmembrane domains. Altogether, there were 33 amino acid changes between the SGD sequence and the brewer's yeasts' sequences, 13 common to all of the brewer's strains, 5 specific for the lager strains, and 15 specific to one or both ale strains. The lager and ale strains are thus more similar to each other than either is to the laboratory strain, S288C, whose genome provided the SGD. S288C is maltose negative, and it is not known whether its AGT1 gene encodes a functional transporter.
For all strains except A15 the hybridization signal intensities at the MAL1 and MAL2 loci were markedly higher for the MAL62 probe than the MAL61 probe (Fig. 4A). Signal strengths were normalized by setting the MAL31 and MAL32 signals to 100 for each strain, so this result suggests that the ratio of MAL11 to MAL12 genes and of MAL21 to MAL22 genes is less than the ratio of MAL31 to MAL32 genes. Michels et al. (18) described MAL3 loci in which tandem partial repeats result in more copies of MAL31 genes than MAL32 genes. A tandem repeat could contribute to the unequal labeling by the MAL61 and MAL62 probes. However, for the MAL1 locus, unequal labeling also is expected, because the brewer's yeasts have some MAL loci with AGT1 sequences, which do not bind the MAL61 probe but are accompanied by MAL12 genes that do bind the MAL62 probe. Based on this argument the apparently equal binding of MAL61 and MAL62 probes to the MAL1 locus of strain A15 is unexpected and suggests that this strain may carry (defective) AGT1 sequences that are not accompanied by MAL12 sequences.
The ale strains differed from one another kinetically in that trehalose inhibition approached a limiting value of ∼82% for A60 but was possibly complete for A179 (Fig. 1). A60 possesses an intact MAL2 locus that is missing in A179. Mal21p may, therefore, be the trehalose-insensitive maltose transporter of A60. All three maltose-negative laboratory strains lacked MAL2, whereas the maltose-positive laboratory strain, CEN.PK2-1D, possessed this locus (Table 3). Although expression of MALx1 genes was low in both ale strains (Fig. 6) MALx1 was expressed in A60 during early growth on maltose. The lager strains also differed among themselves. Maltose transport by A15 was completely insensitive to inhibition by trehalose, whereas maltose transport by A24 (which also contains a defective AGT1 gene) and A64 (whose AGT1 gene was not sequenced) was reduced by 30 or 55%, respectively, at 100 mM trehalose. Thus, A24 and A64 must have a maltose transporter that either carries or is inhibited by trehalose, and, at least for A24, it cannot be Agt1p. Previously studied Malx1 and Mphx transporters do not carry trehalose (6, 7). Mtt1 (see above), which is present in some lager strains but is possibly defective in A15 (8) is another candidate protein for this function. Another possibility is that A24 and A64 contain maltose transporters that bind and are inhibited by trehalose but do not carry it.
Day et al. (7) found that strains with Mal31p, Mal61p, or Agt1p as the sole maltose transporters transported both [14C]maltotriose and [14C]maltose with similar maximum velocities (40 nmol min−1 mg dry mass−1) and Km values (2.7 to 7.2 mM) for both sugars. These results imply that 50 mM maltotriose should strongly inhibit (by ∼70%) the uptake of 5 mM [14C]maltose by each of these three transporters, but we found inhibitions of only 12 to 24% (Table 1) for our lager strains, in which Malx1 transporters were dominant. Each of these lager strains contains several MALx1 genes, including MAL31 and, for A72, MAL61 (Table 3), but we do not know which of these genes are functional. Even if MAL31 and MAL61 are functional in these strains, they may not be identical to the genes used by Day et al. (7). A change altering only one amino acid might abolish activity toward maltose or maltotriose or greatly change the relative Km values.
When more sequences from brewing strains are available, it will become clear whether most lager strains have defective AGT1 genes and most ale strains have AGT1 genes that differ from the sequence in the Saccharomyces Genome Database and resemble those of A60 and A179. Our results suggest that to understand fully the transport of maltose and other α-glucosides by brewer's strains it may be necessary to sequence each strain's transporter genes and kinetically characterize the transporters they encode. Some of these redundant genes are defective, but little is known about how small sequence changes may affect the activity and substrate specificity of the encoded transporters.
Acknowledgments
We thank Maija-Leena Vehkomäki, Marja Ilmén, and Peter Richard for useful discussions and Silja Home, Jukka Kronlöf, and Merja Penttilä for their interest and encouragement. We thank Eija Rintala for help with analyzing the sequence data and Aila Siltala, Pirjo Tähtinen, and Seija Rissanen for technical assistance. Strain RH144-3A was a gift from Johan Thevelein, Catholic University of Leuven, Leuven, Belgium.
This study was supported by the Finnish brewing and malting industry (Panimolaboratorio Oy).
REFERENCES
- 1.Barnett, J. A. 1981. The utilization of disaccharides and some other sugars by yeasts. Adv. Carbohydr. Chem. Biochem. 39:347-404. [DOI] [PubMed] [Google Scholar]
- 2.Chang, Y. S., R. A. Dubin, E. Perkins, C. A. Michels, and R. B. Needleman. 1989. Identification and characterization of the maltose permease in a genetically defined Saccharomyces strain. J. Bacteriol. 171:6148-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charron, M. J., C. A. Michels. 1988. The naturally occurring alleles of MAL1 in Saccharomyces species evolved by various mutagenic processes including chromosomal rearrangement. Genetics 120:83-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charron, M. J., E. Read, S. R. Haut, and C. A. Michels. 1989. Molecular evolution of the telomere-associated MAL loci of Saccharomyces. Genetics 122:307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crumplen, R. M., J. C. Slaughter, and G. G. Stewart. 1996. Characteristics of maltose transport activity in ale and lager strains of the yeast Saccharomyces cerevisiae. Lett. Appl. Microbiol. 23:448-452. [DOI] [PubMed] [Google Scholar]
- 6.Day, R. E., V. J. Higgins, P. J. Rogers, and I. W. Dawes. 2002. Characterization of the putative maltose transporters encoded by YDL247w and YJR160c. Yeast 19:1015-1027. [DOI] [PubMed] [Google Scholar]
- 7.Day, R. E., P. J. Rogers, I. W. Dawes, and V. J. Higgins. 2002. Molecular analysis of maltotriose transport and utilization by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 68:5326-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietvorst, J., J. Londesborough, and H. Y. Steensma. 2005. Maltotriose utilization in lager yeast strains: MTT1 encodes a maltotriose transporter. Yeast 22:775-788. [DOI] [PubMed] [Google Scholar]
- 9.Federoff, H. J., T. R. Eccleshall, and J. Marmur. 1983. Carbon catabolite repression of maltase synthesis in Saccharomyces carlsbergensis. J. Bacteriol. 156:301-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldenthal, M. J., M. Vanoni, B. Buchferer, and J. Marmur. 1987. Regulation of MAL gene expression in yeast: gene dosage effects. Mol. Gen. Genet. 209:508-517. [DOI] [PubMed] [Google Scholar]
- 11.Han, E. K., F. Cotty, C. Sottas, H. Jiang, and C. A. Michels. 1995. Characterization of AGT1 encoding a general α-glucoside transporter from Saccharomyces. Mol. Microbiol. 17:1093-1107. [DOI] [PubMed] [Google Scholar]
- 12.Jespersen, L., L. B. Cesar, P. G. Meaden, and M. Jakobsen. 1999. Multiple α-glucoside transporter genes in brewer's yeast. Appl. Environ. Microbiol. 65:450-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joubert, R., P. Brignon, C. Lehmann, C. Monribot, F. Gendre, and H. Boucherie. 2000. Two-dimensional gel analysis of the proteome of lager brewing yeasts. Yeast 16:511-522. [DOI] [PubMed] [Google Scholar]
- 14.Klein, C. J. L., L. Olsson, B. Rønnow, J. D. Mikkelsen, and J. Nielsen. 1996. Alleviation of glucose repression of maltose metabolism by MIG1 disruption in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 62:4441-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kodama, Y., N. Fukui, T. Ashikari, Y. Shibano, K. Morioka-Fujimoto, Y. Hiraki, and K. Nakatani. 1995. Improvement of maltose fermentation efficiency: constitutive expression of MAL genes in brewing yeasts. J. Am. Soc. Brew. Chem. 53:24-29. [Google Scholar]
- 16.Lucero, P., M. Herweijer, and R. Lagunas. 1993. Catabolite inactivation of the yeast maltose transporter is due to proteolysis. FEBS Lett. 333:165-168. [DOI] [PubMed] [Google Scholar]
- 17.Lucero, P., É. Peñalver, E. Moreno, and R. Lagunas. 1997. Moderate concentrations of ethanol inhibit endocytosis of the yeast maltose transporter. Appl. Environ. Microbiol. 63:3831-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michels, C. A., E. Read, K. Nat, and M. J. Charron. 1992. The telomere-associated MAL3 locus of Saccharomyces is a tandem array of repeated genes. Yeast 8:655-665. [DOI] [PubMed] [Google Scholar]
- 19.Naumov, G. I., E. S. Naumova, and C. A. Michels. 1993. Genetic variation of the repeated MAL loci in natural populations of Saccharomyces cerevisiae and Saccharomyces paradoxus. Genetics 136:803-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rautio J., and J. Londesborough. 2003. Maltose transport by brewer's yeasts in brewer's wort. J. Inst. Brew. 109:251-261. [Google Scholar]
- 21.Riballo, E., M. Herweijer, D. H. Wolf, and R. Lagunas. 1995. Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J. Bacteriol. 177:5622-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodicio, R., and F. K. Zimmermann. 1985. Cloning of maltase regulatory genes in Saccharomyces cerevisiae. Curr. Genet. 9:539-545. [Google Scholar]
- 23.Stambuk, B. U., M. A. da Silva, A. D. Panek, and P. S. de Araujo. 1999. Active α-glucoside transport in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 170:105-110. [DOI] [PubMed] [Google Scholar]
- 24.Stambuk, B. U., and P. S. de Araujo. 2001. Kinetics of active α-glucoside transport in Saccharomyces cerevisiae. FEMS Yeast Res. 1:73-78. [DOI] [PubMed] [Google Scholar]
- 25.Vanoni, M., P. Sollitti, M. Goldenthal, and J. Marmur. 1989. Structure and regulation of the multigene family controlling maltose fermentation in budding yeast. Proc. Nucleic Acids Res. Mol. Biol. 37:281-322. [DOI] [PubMed] [Google Scholar]
- 26.Zheng, X., T. D'Amore, I. Russel, and G. G. Stewart. 1994. Factors influencing maltotriose utilization during brewery wort fermentations. J. Am. Soc. Brew. Chem. 52:41-47. [Google Scholar]