Abstract
Multilocus sequence typing (MLST) identifies and groups bacterial strains based on DNA sequence data from (typically) seven housekeeping genes. MLST has also been employed to estimate the relative contributions of recombination and point mutation to clonal divergence. We applied MLST to the plant pathogen Xylella fastidiosa using an initial set of sequences for 10 loci (9.3 kb) of 25 strains from five different host plants, grapevine (PD strains), oleander (OLS strains), oak (OAK strains), almond (ALS strains), and peach (PP strains). An eBURST analysis identified six clonal complexes using the grouping criterion that each member must be identical to at least one other member at 7 or more of the 10 loci. These clonal complexes corresponded to previously identified phylogenetic clades; clonal complex 1 (CC1) (all PD strains plus two ALS strains) and CC2 (OLS strains) defined the X. fastidiosa subsp. fastidiosa and X. fastidiosa subsp. sandyi clades, while CC3 (ALS strains), CC4 (OAK strains), and CC5 (PP strains) were subclades of X. fastidiosa subsp. multiplex. CC6 (ALS strains) identified an X. fastidiosa subsp. multiplex-like group characterized by a high frequency of intersubspecific recombination. Compared to the recombination rate in other bacterial species, the recombination rate in X. fastidiosa is relatively low. Recombination between different alleles was estimated to give rise to 76% of the nucleotide changes and 31% of the allelic changes observed. The housekeeping loci holC, nuoL, leuA, gltT, cysG, petC, and lacF were chosen to form the basis of a public database for typing X. fastidiosa (www.mlst.net). These loci identified the same six clonal complexes using the strain grouping criterion of identity at five or more loci with at least one other member.
The ability to identify distinct bacterial strains is critically important in understanding the processes involved in the evolution of pathogenicity and in communicating the emergence and spread of disease-causing strains. Classical bacteriological techniques used for strain typing include Gram staining, sugar utilization analysis, and serological testing (6). The main problem with such tests is that the phenotypic expression of a trait may not necessarily reflect the underlying genetics involved. For this reason, DNA-based methods of strain typing, such as multilocus enzyme electrophoresis (MLEE) (38) and pulsed-field electrophoresis (PFGE) (28), have been employed increasingly. However, these methods have several drawbacks, including poor reproducibility between and within labs and an inability to quantitate genetic relationships. While PFGE can be highly discriminating, it is unsuitable for global or long-term epidemiology as it lacks the ability to uncover genetic relationships between strains at deeper phylogenetic levels (25).
Multilocus sequence typing (MLST) is a recently devised method for identifying strains of bacteria based solely on nucleotide sequence differences in a small number of genes (25). For MLST, each allele of a gene is given a number, and each strain characterized (for n loci) is represented by the set of n numbers defining the alleles at each locus. This defines the sequence type (ST). In contrast to the results of prior DNA-based methods, MLST sequence data are unambiguous, can be easily interpreted and replicated between labs, and is generally made available in a public database.
MLST typically has higher resolution than previous methods used to group bacterial strains. By convention, a seven-locus MLST data set is generally used, and such a set has been estimated to give a level of discrimination comparable to that of an MLEE study using 15 to 20 loci (16). A single nucleotide difference always produces a new allele in an MLST data set, while as many as eight amino acid substitutions may be required to produce a new allozyme in an MLEE data set (4). MLST also gives a comparable or even slightly better level of discrimination than PFGE (18, 24, 27, 29).
MLST is used to group closely related strains into clonal complexes. The definition of a clonal complex has been modified since the first use of MLST to maximize its effectiveness in defining meaningful groups. Initially, Enright and Spratt (10) defined a clonal complex as a group of strains having the same ST (i.e., the same allelic profile). This definition was later amended to any frequent group of strains (defined as the ancestral or “consensus” genotype) plus their single-locus variants (12, 16). Single-locus variants differ from the ancestral genotype at only one of the test loci. However, defining the ancestral type as the allelic profile or sequence type most commonly present in the clonal complex is subject to sampling bias. Feil et al. (13) used a parsimony-based approach and redefined the ancestral type as the sequence type with the most single-locus variants in the clonal complex. More recently, the definition of a clonal complex has been further relaxed, so that a clonal complex is a group in which every strain shares at least five identical alleles out of seven with at least one other genotype in the group (13) or has four alleles out of seven that are identical to alleles in a consensus or ancestral clone (8). Finally, Feil et al. (14) suggested that the definition of a clonal complex should be flexible depending upon the characteristics of the species in question.
One of the major factors influencing the nature and diversification of clonal complexes is the recombination rate. As the recombination rate increases, the phylogeny of individual descent within a bacterial species becomes increasingly randomized. Thus, a priori, we would expect the occurrence of well-defined clonal complexes to be more probable in species with low recombination rates. Indeed, the contribution of recombination relative to that of point mutation can profoundly affect variation and host adaptation within a bacterial species and is an important determinant of the evolutionary trajectory (26). This ratio inevitably varies among species, since the necessary precursor of recombination, lateral gene transfer, varies considerably (15). Recent studies that have estimated recombination in bacteria have revealed a wide range of values, from zero in Mycobacterium tuberculosis (40) to midrange values in Escherichia coli and Haemophilus influenzae (13) to high estimated values in Streptococcus pneumoniae, in which an allele is about 10 times more likely to change by recombination than by point mutation (13).
MLST data sets have been used to estimate the relative contributions of recombination and point mutations in the formation of new alleles within a clonal complex (12, 13, 15, 17, 34). Recombination can be identified by a mosaic structure in a particular gene, reflecting the different evolutionary histories of different regions of the gene. In contrast, a point mutation results in a novel allele distinguished by a single base change. To distinguish between these two sources of genetic variability, Feil et al. (15, 17) adopted the criterion that if a variant allele within a single-locus variant in a clonal complex differs at only one site from the “ancestral” allele of the complex, then it is considered a point mutation. If it differs at more than one site, it is considered to have originated from a recombination event. This classification assumes that it is unlikely that strains that are identical at several other loci have accumulated more than one mutational difference at a single remaining locus and allows measurement of the “effective” rate of recombination (i.e., recombination which results in novel alleles within a clonal complex).
In this study, we developed an MLST system for the plant pathogen Xylella fastidiosa. X. fastidiosa is a gram-negative, xylem-limited eubacterium that is closely related to the xanthamonads (30). It is transported between plant hosts by xylem-feeding insect vectors (typically leaf hoppers belonging to the order Hemiptera). Different strains of the bacterium infect different species of plants throughout the Americas. These strains cause scorch diseases such as Pierce's disease (PD) in grapevine, almond leaf scorch (ALS) in almond, and oleander leaf scorch (OLS) in oleander in North America (21, 31) and citrus variegated chlorosis (CVC) in South America (5). Three distinct clades of X. fastidiosa have been identified in North America (37); these clades correspond to X. fastidiosa subsp. fastidiosa (renamed from the original subspecies, piercei) and X. fastidiosa subsp. multiplex (36) plus a third subspecies, X. fastidiosa subsp. sandyi, that so far has been found only in oleander (37). X. fastidiosa subsp. fastidiosa is found in grapevines, almond, and alfalfa, and X. fastidiosa subsp. multiplex consists of several plant host pathovars (e.g., almond, peach, plum, and oak pathovars).
In establishing an MLST system for identifying and classifying X. fastidiosa strains, we examined the effectiveness and robustness of the MLST method for detection of subspecies and plant host strains within the subspecies and for estimation of recombination rates. We were able to do this using the preexisting phylogeny of the strains (37), with which we could compare the clonal complexes. We investigated the extent to which the definition of a clonal complex influences the conclusions of an MLST study and the effect of choosing an MLST standard of 7 genes from a larger number (in our case 10 genes) on the clonal complexes identified and the estimated recombination rate.
Estimation of the recombination rate is very important for understanding bacterial evolution and the origin of new pathogenic strains. This is true for X. fastidiosa. The distinct phylogenetic clades of X. fastidiosa (37) suggest that recombination is limited; however, there is an opportunity for recombination between strains both within the insect vector, in which the X. fastidiosa strains are transported in the head and foregut (30), and in the plant host. For example, both strains of X. fastidiosa subsp. fastidiosa and strains X. fastidiosa subsp. multiplex have been isolated from symptomatic almond trees (1). We compared estimates of recombination obtained from the MLST data to independent estimates derived using other available methods (2, 35) and examined the extent to which reducing the size of the data set to the MLST standard of seven genes altered recombination estimates.
MATERIALS AND METHODS
X. fastidiosa strains.
Twenty-five strains of X. fastidiosa were used in this analysis; these strains originated from symptomatic individuals of five plant species, including grape with Pierce's disease (PD strains), oleander with oleander leaf scorch (OLS strains), almond with almond leaf scorch (ALS strains), oak with oak leaf scorch (OAK strains), and peach with phony peach scorch (PP strains) (Table 1). They included a PD strain (Temecula), an OLS strain (Ann-1), and an ALS strain (Dixon) used for genome sequencing (3, 41). Other strains reflected the geographic distribution of Pierce's disease (PD strains) and almond leaf scorch (ALS strains) within California and represented some of the larger-scale geographical distribution of PD strains (Florida) and OLS strains (Texas). To provide additional North American variation, we also included strains from other plant hosts in the eastern United States, including three strains from oak and two strains from peach. The sequenced CVC strain from South America (39) was used as the outgroup.
TABLE 1.
X. fastidiosa strains used in the MLST analyses plus the outgroup CVC18
| Designation | Strain | Host | Geographic origin | Reference or source |
|---|---|---|---|---|
| PD1 | Temecula | Grape | Temecula, CA | 41 |
| PD10 | I03 | Grape | Temecula, CA | 7 |
| PD7 | Traver | Grape | Tulare, CA | 20 |
| PD14 | Douglas | Grape | San Luis Obispo, CA | 20 |
| PD4 | STL | Grape | Napa, CA | 20 |
| PD6 | Conn Creek | Grape | Napa, CA | 20 |
| PD16 | 95-2 | Grape | Florida | 20 |
| OLS2 | Ann-1 | Oleander | Palm Springs, CA | A. Purcell |
| OLS9 | Riverside | Oleander | Riverside, CA | 7 |
| OLS8 | Texas | Oleander | Texas | 7 |
| OLS19 | Cathedral City | Oleander | Palm Springs, CA | A. Purcell |
| OLS20 | TR2 | Oleander | Orange, CA | A. Purcell |
| OLS21 | TS5 | Oleander | Orange, CA | A. Purcell |
| ALS3 | Dixon | Almond | Solano, CA | 20 |
| ALS15 | ALS-2 | Almond | San Joaquin, CA | 20 |
| ALS5 | Tulare | Almond | Tulare, CA | 20 |
| ALS11 | 237 | Almond | Temecula, CA | 7 |
| ALS12 | 276 | Almond | Temecula, CA | 7 |
| ALS13 | 187 | Almond | Temecula, CA | 7 |
| ALS22 | ALS-6 | Almond | San Joaquin, CA | 20 |
| OAK17 | Stucky | Oak | Georgia | 20 |
| OAK23 | 92-3 | Oak | Florida | 20 |
| OAK24 | Oak#2 | Oak | Georgia | 20 |
| PP27 | 5S2 | Peach | Georgia | 20 |
| PP28 | 5R1 | Peach | Georgia | 20 |
| PLS26 | 2#4 | Plum | Georgia | 20 |
| CVC18 | 9a5c | Citrus | Sao Paulo, Brazil | 39 |
Isolates were grown on PD3 agar medium. Bacterial DNA was extracted using the Chelex preparation (42) or by scraping bacterial cells and lysing the cells with distilled water.
Amplification and sequence determination for 10 genes.
Ten genes were sequenced for the MLST analysis, and these genes represented a total of 9.3 kb. The genes were amplified using primers designed from four genomes using Oligo v.6 (33) and the methodology of Schuenzel et al. (37). The genes used occur in all four genomes sequenced (ALS strain Dixon [accession no. NC_002723], OLS strain Ann-1 [accession no. NC_002722], PD strain Temecula [accession no. AE009442], and CVC strain 9a5c [accession no. AE003849]). The genes were chosen for the MLST analysis based on a survey of the ALS and OLS genomes (Schuenzel et al., unpublished data). Because the X. fastiodiosa genomes have diverged only between 0.5% and 3.0% (3, 41; Schuenzel et al., unpublished data), we focused on genes that have diverged at least 1.0% between the ALS and OLS genomes. We also selected genes that represented a variety of biochemical functions and were distributed around the CVC genome (Table 2). The evolutionary pattern of each gene was characterized based on its rate of change and on its ratio of nonsynonymous substitutions to synonymous substitutions (dN/dS) (37).
TABLE 2.
Positions, functions, PCR primers, and lengths sequenced for the 10 X. fastidiosa genes
| Gene positiona | Used in seven-locus MLST | Genea | Gene functiona | Biochemical functiona | Primer sequences (forward/reverse) | Gene length (bp) |
|---|---|---|---|---|---|---|
| XF0136 | Yes | holC | DNA polymerase III holoenzyme, chi subunit | Replication | 5′-GATTTCCAAACCGCGCTTTC-3′/5′-TCATGTGCAGGCCGCGTCTCT-3′ | 342 |
| XF0257 | rfbD | dTDP-4-dehydrorhamnose-3, 5- epimerase | Surface polysaccharides | 5′-TTTGGTGATTGAGCCGAGGGT-3′/5′-CCATAAACGGCCGCTTTC-3′ | 429 | |
| XF0316 | Yes | nuoL | NADH-ubiquinone oxidoreductase, NQO12 subunit | Aerobic respiration | 5′-CATTATTGCCGGATTGTTAGG-3′/5′-GCGGGAAACATTACCAAGC-3′ | 1,821 |
| XF0318 | nuoN | NADH-ubiquinone oxidoreductase, NQO14 subunit | Aerobic respiration | 5′-GGGTTAAACATTGCCGATCT-3′/5′-CGGGTTCCAAAGGATTCCTAA-3′ | 1,311 | |
| XF0656 | Yes | gltT | Glutamate symport protein | Transport of amino acids | 5′-TTGGGTGTGGGTACGTTGCTG-3′/5′-CGCTGCCTCGTAAACCGTTGT-3′ | 951 |
| XF0832 | Yes | cysG | Siroheme synthase | Biosynthesis of heme, porphyrin | 5′-GGCGGCGGTAAGGTTG-3′/5′-GCGTATGTCTGTGCGGTGTGC-3′ | 1,170 |
| XF0910 | Yes | petC | Ubiquinol cytochrome c oxidoreductase, cytochrome c1 subunit | Electron transport | 5′-CTGCCATTCGTTGAAGTACCT-3′/5′-CGTCCTCCCAATAAGCCT-3′ | 531 |
| XF1632 | pilU | Twitching motility protein | Surface structures | 5′-CAATGAAGATTCACGGCAATA-3′/5′-ATAGTTAATGGCTCCGCTATG-3′ | 873 | |
| XF1818 | Yes | leuA | 2-Isopropylmalate synthase | Amino acid biosynthesis | 5′-GGGCGTAGACATTATCGAGAC-3′/5′-GTATCGTTGTGGCGTACACTG-3′ | 1,218 |
| XF2447 | Yes | lacF | ABC transporter sugar permease | Transport of carbohydrates | 5′-TTGCTGGTCCTGCGGTGTTG-3′/5′-CCTCGGGTCATCACATAAGGC-3′ | 642 |
Information based on information reported in reference 39.
Identification of clonal complexes.
Each unique allelic profile was assigned an ST number, and the STs were grouped into clonal complexes using eBURST (14). The program also identified clonal complexes, each clonal complex's putative ancestral genotype (identified as the ST with the most single-locus variants), and its associated single-locus variants. A criterion of seven shared alleles out of 10 alleles examined was used to delineate the clonal complexes; this criterion was similar to the criterion of five shared alleles out of seven alleles examined (13). Thus, within each complex, the STs must share seven or more alleles with at least one other ST of the clonal complex. We also investigated how modifying this criterion changed the assignment of STs to clonal complexes.
The similarities among allelic profiles were visualized with an unweighted pair-group method with arithmetic averages (UPGMA) dendrogram. The UPGMA dendrogram was based on the percentage of pairwise differences between the allelic profiles of the 25 strains and was constructed using START (22). The UPGMA dendrogram was compared to a maximum-likelihood tree for the same sequence data (37).
Recombination in clonal complexes.
The alleles within a clonal complex were compared to the alleles of the ancestral ST, and the number that differed by only 1 bp was used to approximate the number of point mutations (13). The number that differed at multiple sites was used as a measure of recombination events. The role of recombination relative to the role of mutation in creating clonal diversity was measured by determining the ratio of recombination to mutation (r/m ratio) per allele (where each event results in one change) and per nucleotide (where each mutation results in one change, but each recombination results in more than one change).
Rather than restrict our recombination analysis to single-locus variants of the ancestral type (which differ from the ancestral type at only one locus) (13), we expanded our sample to included STs that differ from the ancestral form at multiple loci. This modification could have led to an overestimate of the recombination rate, since it allowed for a longer evolutionary separation of sequences that increased the probability of a pair of point mutations occurring in the same gene. For this reason the occurrence of each 2-bp “recombination” was examined in detail.
In closely related clonal complexes, a single ancestral sequence type carrying all of the ancestral alleles is often difficult to identify. In such cases, we did not use a single ancestral sequence type but instead used a parsimony-based approach, in which the least derived allele at each locus was used as a basis for comparing variant alleles.
We also estimated homologous recombination using the method of Sawyer (35), as implemented in the START program (22). This method focuses on sites that exhibit silent (synonymous) polymorphism across the whole data set. For each gene, these sites are compared for all pairs of alleles. For each pair, the gene is partitioned into fragments, and a fragment is defined by the region between a site that is “discordant” (different) between the pair and either the next such discordant site or the end of the gene. The length of the fragment is determined by two methods. The length of a “condensed” fragment (in nucleotide base pairs [bp]) is the number of “concordant” sites that it contains, where a concordant site is a silent polymorphic site that is identical in the pair being compared. The length of an “uncondensed” fragment is the traditional length of a DNA sequence (i.e., the number of all nucleotide sites that it includes). If the length of the fragment is greater than expected by chance, recombination is indicated. Sawyer's test statistics are calculated by adding the squared lengths of the condensed fragments or the squared lengths of the uncondensed fragments (9, 35). Since two tests were performed, a sequential Bonferroni correction was applied, where a P value of <0.025 is necessary for the first significant result and a P value of <0.05 is necessary for the second significant result.
The method of Betran et al. (2), implemented in the DnaSP program (32), was also used to detect recombination events. This method is based on detecting regions of congruence between alleles in different designated subgroups (in this case, clonal complexes). The observed recombination length (L) (in nucleotides) is estimated as follows: L = TR − TL + 1, where TL and TR are the left and right site positions of the outermost informative nucleotide sites of a congruent recombination exchange, respectively.
Recombination events were also ascertained by visual inspection. Variant alleles in a clonal complex or closely related clonal complexes were compared to other alleles in the data set. If three or more changes in the variant allele were shared with an allele in a different clonal complex, then recombination was assumed to have occurred.
RESULTS
The allelic profiles of the 25 strains produced 19 different STs. The eBURST program assigned these STs to six clonal complexes (Table 3), where a clonal complex was defined by groups of STs which shared alleles at 7 out of the 10 loci with at least one other member of the complex. All strains from grapevine plus two strains from almond (ALS5 and ALS11) formed clonal complex 1 (CC1). Since ST1 had the most single-locus variants (Table 3), it was designated the founder pattern for the complex. The profiles of other strains in the complex differed from this profile at one to three loci. The six OLS strains formed a clonal complex, CC2, with three STs in which four strains had the ancestral allelic profile (ST7). In this case, either ST7 or ST8 is the potential ancestral ST since each is the other's only single-locus variant. Based on its high frequency, ST7 was designated the ancestral ST. This is a useful convention; however, since a higher frequency could be a product of sampling bias, the designation is not statistically significant. ALS3, ALS13, and ALL15 formed CC3, and the allelic profile of ALS15 (ST10) defined the founder type since it has two single-locus variants. The three OAK strains formed CC4, with OAK17 (ST13) representing the ancestral ST. The two PP strains formed CC5, sharing 8 of 10 alleles. Finally, the ALS12 and ALS22 strains formed CC6, sharing 7 of 10 alleles.
TABLE 3.
Allelic profiles of 25 X. fastidiosa isolates divided into six clonal complexes linking isolates sharing at least 70% of their alleles
| Isolate (ST) | Allelic profiles
|
No. of single-locus variants per ST | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| holCa | rfbD | nuoLa | nuoN | gltTa | cysGa | petCa | pilU | leuAa | lacFa | ||
| CC1 isolates | |||||||||||
| PD1 (ST1)b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 |
| PD4 (ST1)b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| PD6 (ST1)b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| PD10 (ST1)b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| PD7 (ST2) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 1 | 2 |
| ALS5 (ST3) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 2 |
| ALS11 (ST4) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 1 |
| PD16 (ST5) | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 0 |
| PD14 (ST6) | 1 | 1 | 1 | 4 | 1 | 4 | 1 | 6 | 1 | 1 | 0 |
| CC2 isolates | |||||||||||
| OLS8 (ST7)b | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 |
| OLS19 (ST7)b | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| OLS20 (ST7)b | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| OLS21 (ST7)b | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| OLS2 (ST8) | 2 | 2 | 5 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 |
| OLS9 (ST9) | 2 | 2 | 6 | 2 | 2 | 2 | 2 | 7 | 2 | 2 | 0 |
| CC3 isolates | |||||||||||
| ALS15 (ST10)b | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 2 |
| ALS3 (ST11) | 3 | 3 | 7 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 |
| ALS13 (ST12) | 3 | 4 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 |
| CC4 isolates | |||||||||||
| OAK17 (ST13)b | 4 | 4 | 7 | 3 | 4 | 5 | 3 | 3 | 3 | 5 | 2 |
| OAK23 (ST14) | 4 | 4 | 7 | 3 | 4 | 5 | 3 | 8 | 3 | 5 | 1 |
| OAK24 (ST15) | 5 | 4 | 7 | 3 | 4 | 5 | 3 | 3 | 3 | 5 | 1 |
| CC5 isolates | |||||||||||
| PP27 (ST16) | 6 | 4 | 7 | 3 | 5 | 3 | 4 | 3 | 3 | 5 | 0 |
| PP28 (ST17) | 6 | 4 | 7 | 3 | 6 | 3 | 4 | 3 | 5 | 5 | 0 |
| CC6 isolates | |||||||||||
| ALS12 (ST18) | 7 | 4 | 7 | 3 | 7 | 6 | 3 | 1 | 6 | 5 | 0 |
| ALS22 (ST19) | 7 | 4 | 8 | 5 | 7 | 7 | 3 | 1 | 6 | 5 | 0 |
Locus recommended for the seven-locus MLST data set.
Ancestral sequence type of clonal complex.
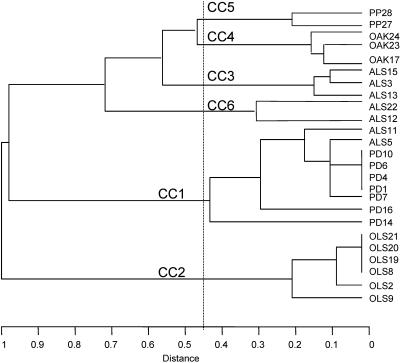
The UPGMA dendrogram, based on the alleles shared by strains, separated the six clonal complexes (Fig. 1). This topology is largely congruent with that of the maximum-likelihood phylogeny (37) shown in Fig. 2. The main difference is that on the UPGMA tree, CC2 (X. fastidiosa subsp. sandyi) is incorrectly placed at the base of the tree rather than grouping with CC1 (X. fastidiosa subsp. fastidiosa). This effect is not due to the fact that the UPGMA dendrogram is an unrooted tree. Adding the CVC strain (the outgroup in the maximum-likelihood analysis) resulted in the same topology since CC2 differs from all other strains at each of the 10 loci, whereas one CC1 allele appears in CC6 (Table 3). A minor difference is that CC4 groups with CC5 in the UPGMA analysis rather than with CC3.
FIG. 1.
Dendrogram showing the relationships between the clonal complexes based on UPGMA from the matrix of pairwise percentage differences between the allelic profiles of the 25 isolates. The clonal complexes (CC1 to CC6) were determined with the eBURST program, and 7 out of 10 alleles was the criterion for inclusion in a complex. The dotted line indicates the region demarcating the designation of six clonal complexes.
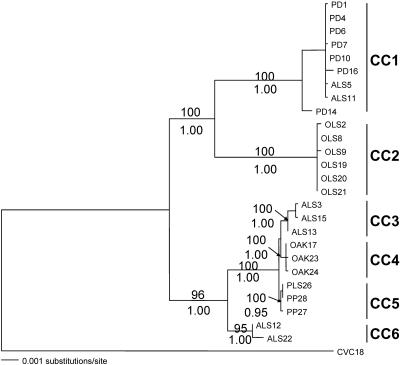
FIG. 2.
Maximum-likelihood phylogeny of 26 X. fastidiosa strains based on 9,307 bp using a general time reversible model with gamma distribution and invariant sites. The numbers above and below the lines at nodes indicate maximum-likelihood bootstrap support and Bayesian posterior probabilities, respectively. The clonal complexes (CC1 to CC6) were determined with the eBURST program, and 7 out of 10 alleles was the criterion for a complex.
The six clonal complexes identified are well supported in the maximum-likelihood tree (Fig. 2). Bayesian posterior probabilities and maximum-likelihood bootstrap analyses supported CC1 to CC4 with 100% support, while CC5 and CC6 received ≥95% support. Relaxing the stringency defining a clonal complex to 6 shared alleles out of 10 (instead of 7 out of 10) resulted in four clonal complexes (i.e., two fewer clonal complexes) since CC3 to CC5 form a single complex (corresponding to X. fatidiosa subsp. multiplex), while using 5 shared alleles out of 10 resulted in combination of CC6 with a multiplex for a total of three clonal complexes, corresponding to the three major groups shown in Fig. 2. The tendency of CC3 to CC5 and then CC3 to CC6 to collapse into single clonal complexes is apparent from both the UPGMA and maximum-likelihood trees (Fig. 1 and 2). CC3 to CC5 formed a clade with 100% support, while CC3 to CC6 received ≥96% support. Increasing the stringency to 8 out of 10 alleles created three singletons (ST6, ST18, and ST19) and five clonal complexes. Finally, using the criterion of Feil et al. (12) for defining a clonal complex as the ancestral type and its associated single-locus variants resulted in designation of four clonal complexes, CC1 (with ST5 and ST6 excluded), CC2 (with ST9 excluded), CC3, and CC4, plus seven singleton genotypes.
The effects of gene sampling were examined by limiting the number of loci chosen to seven (the suggested standard for MLST data sets). When a criterion of five shared loci out of seven was used to define a clonal complex, depending on the choice of genes, between three and six clonal complexes were identified. This variation was due to the erratic behavior of CC3 to CC6. With some choices these complexes remained distinct; with other choices CC4 and CC5, CC4 and CC6, or CC5 and CC6 combined or CC3 to CC6 combined. Some of these reduced data sets also resulted in ST6, ST18, and ST19 being assigned as singleton sequences.
A subset of seven genes that excluded the cell surface genes rfbD and pilU could be selected, which retained 16 of the 19 STs found in all 10 genes. These seven housekeeping genes are holC, nuoL, leuA, gltT, cysG, petC, and lacF (Table 3). When the clonal complex criterion of shared alleles for five out of seven genes was used, this set of seven genes continued to identify the same six clonal complexes. The remaining housekeeping gene, nuoN, was not used in the final seven-gene MLST set because including it would have split CC6 into two singletons.
The MLST set of seven genes exhibits a rate of evolution that varies symmetrically by a factor of about 2 above and below the mean rate of 1.99 (relative to the slowest) and has an average dN/dS of 0.169, with a relatively narrow range (0.08 to 0.32), all of which are well below the criterion for positive selection of >1.00 (Table 4). There is no indication that any of these genes are subject to unusual evolutionary behavior.
TABLE 4.
Rates of evolution of genes relative to each other, dN/dS ratios, and number of alleles identified at each locus
| Gene | Rate relative to slowest evolving gene | dN/dS | No. of alleles |
|---|---|---|---|
| holCa | 3.55 | 0.08 | 7 |
| rfbD | 2.20 | 0.29 | 4 |
| nuoLa | 1.79 | 0.14 | 8 |
| nuoN | 1.84 | 0.14 | 4 |
| gltTa | 1.41 | 0.09 | 7 |
| cysGa | 1.95 | 0.32 | 7 |
| petCa | 1.34 | 0.32 | 4 |
| pilU | 1.59 | 0.17 | 8 |
| leuAa | 1.00 | 0.08 | 6 |
| lacFa | 2.86 | 0.15 | 5 |
Locus recommended for the seven-locus MLST data set.
Given the strong support for combining CC3 to CC6 from the phylogenetic analyses and the number of alleles shared between these complexes, we combined them when we calculated the recombination estimates. CC1 and CC2 are more distantly related and do not have any alleles in common, and they were not combined for this purpose.
A total of 10 allelic changes were putatively assigned as recombination events, compared to 22 changes that were assigned as point mutations (Table 5). At the nucleotide level, a total of 71 base pair changes were estimated to have occurred by recombination, compared to 22 changes that occurred by point mutation (Table 5). The ratio of the contribution to diversity by recombination to the contribution to diversity by point mutation (r/m ratio) is 0.45:1 at the allelic level and 3.23:1 at the individual nucleotide level.
TABLE 5.
Assignment of recombinants and point mutations for clonal groups on a per allele and per nucleotide basis
| Clonal complex(es) | No. of variant alleles | Allelic basis
|
Nucleotide basis
|
||
|---|---|---|---|---|---|
| No. of putative recombinations | No. of putative mutations | No. of putative recombinations | No. of putative mutations | ||
| CC1 | 8 | 2 | 6 | 19 | 6 |
| CC2 | 3 | 1 | 2 | 2 | 2 |
| CC3 to CC6 | 21 | 7 | 14 | 50 | 14 |
| Total | 32 | 10 | 22 | 71 | 22 |
If the data set is limited to seven loci, the estimated contribution of recombination to allelic and nucleotide diversity varies considerably. At the allelic level, the ratio of recombination to point mutation varies from 0.21:1 to 0.71:1, while at the individual nucleotide level, the ratio of recombination to point mutation varies from 1.07:1 to 5.07:1. If the recommended MLST subset of seven genes is used to estimate recombination, the r/m ratios are 0.53:1 at the allelic level and 3.20:1 at the nucleotide level. The three genes eliminated from this subset contain seven point mutations and two recombinations, which increases the estimated contribution of recombination to allelic diversity slightly. At the nucleotide level, the ratio is almost the same.
The Sawyer test (35) showed significant recombination in 1 of the 10 genes, cysG (Table 6). Two other genes, nuoL and rfbD, showed weak indications of recombination consistent across both tests (P < 0.10), and the pilU gene showed similar weak evidence (P < 0.10) based on the condensed fragment analysis but no indication of recombination based on the uncondensed analysis (Table 6).
TABLE 6.
Results of Sawyer's test for the genes with indications of homologous recombinationa
| Gene | Sum of squared lengths of condensed fragments
|
Sum of squared lengths of uncondensed fragments
|
||
|---|---|---|---|---|
| Observed sum (bp2) | P value | Observed sum (bp2) | P value | |
| holC | 603 | 0.1662 | 890,118 | 0.6321 |
| rfbD | 48 | 0.0837 | 734,090 | 0.0414 |
| nuoL | 3,025 | 0.0615 | 22,220,829 | 0.0474 |
| nuoN | 276 | 0.4417 | 5,115,662 | 0.2724 |
| gltT | 1,614 | 0.7731 | 9,410,055 | 0.4890 |
| cysG | 1,343 | 0.0170 | 10,804,998 | 0.0256 |
| petC | 38 | 0.1636 | 945,906 | 0.1647 |
| pilU | 1,960 | 0.0447 | 9,495,392 | 0.6445 |
| leuA | 777 | 0.5240 | 7,280,370 | 0.3365 |
| lacF | 750 | 0.1124 | 1,413,448 | 0.3391 |
See reference 35.
The DnaSP method suggested that there were five recombination events; four of these events were also indicated by visual inspection, and there was an additional short event that occurred in the leuA gene between CC6 and either CC1 or CC2 (Table 7).
TABLE 7.
Numbers of recombination events suggested by different methods
| Method | No. of recombination events | Genes featuring recombination events |
|---|---|---|
| Visual inspection | 4 | holC, cysG (2), pilU |
| MLST | 10 | holC, rfbD, nuoL (4), cysG (2), pilU, leuA |
| Sawyer's test | 1 | cysG |
| DnaSP | 5 | holC, cysG (2), pilU, leuA |
DISCUSSION
The MLST methodology divided the X. fastidiosa PD, OLS, ALS, OAK, and PP plant host strains into six clonal complexes. These clonal complexes corresponded to well-defined clades identified by maximum-likelihood phylogenetic analysis (37). They are also broadly consistent with previous studies based on randomly amplified polymorphic DNAs and biological traits of X. fastidiosa (1, 20). All PD and OLS strains were separated into CC1 and CC2 corresponding to X. fastidiosa subsp. fastidiosa and X. fastidiosa subsp. sandyi. The ALS strains were subdivided into three complexes (two distinct ALS groups, CC3 and CC6, plus the PD group, CC1), while the OAK and PP strains formed their own complexes, CC4 and CC5 (Fig. 2).
The six clonal complexes were based on a 70% criterion for grouping STs into clonal complexes; i.e., a clonal complex was defined as a network linking STs with allelic identity at 7 or more of 10 loci. Relaxing the identity criterion to 6 of 10 loci collapsed CC3 to CC5 (ALS, OAK, and PP strains) into a single group corresponding to X. fastidiosa subsp. multiplex. The remaining complex (CC6) was composed of a pair of ALS strains that Schuenzel et al. (37) set apart from the three subspecies since they include sequences characteristic of all three taxa.
The MLST approach allows each strain to be (i) defined by its allelic profile as a particular ST and (ii) grouped by a simple criterion of allelic identity among STs into clonal complexes. In the case of X. fastidiosa, it appears that 70% identity groups STs into plant host pathovars, while a 50 to 60% criterion groups STs at a broader subspecific level. The simplicity of MLST compared to a phylogenetic approach is a clear advantage in enabling communication of information when the spread of pathogenic strains is tracked and in facilitating rapid recognition of an unusual isolate. This simplicity has considerable practical value when, as is often the case, data sets involve hundreds of strains (10, 11, 23). The computational demands for analyzing such large data sets make the use of phylogenetic methods impractical.
The initial MLST analysis employed data for 10 loci. Reduction to a set of seven loci (the MLST standard) retained the same six clonal complexes when the criterion for grouping STs was kept at roughly 70% (five out of seven loci). All seven loci showed fairly homogeneous evolutionary characteristics, with dN/dS ratios typical of moderately constrained genes (Table 4).
The groups corresponding to the clonal complexes are found on both UPGMA and maximum-likelihood trees. However, the UPGMA approach for validating clonal complexes can be misleading. First, the complete set of six clonal complexes identified by the 7-out-of-10-shared-allele criterion can be recovered from the UPGMA dendrogram only in a very narrow window between allelic pairwise distances of 0.43 and 0.47 (Fig. 1). On one side of this window, CC4 and CC5 are combined, while on the other side, PD14 (ST6) would be removed from CC1 and designated a singleton. Second, the close relationship of X. fastidiosa subsp. fastidiosa (PD strains) and X. fastidiosa subsp. sandyi (OLS strains) could not be detected from the UPGMA tree because these taxa do not share any alleles. In contrast, at the nucleotide level, there are numerous synapomorphies linking the two clades, so the sequence-based maximum-likelihood method strongly recovers PD and OLS strains as sister clades. In general, support for the validity of clonal complexes should be based on a phylogenetic analysis of the sequence data. The phylogenetic approach always provides more information unless recombination rates are extremely high. Frequent recombination randomizes the phylogenetic signal; however, it also undermines the usefulness of the clonal complex since associations of alleles become fleeting.
Both the MLST analysis and the maximum-likelihood phylogeny identified one clonal complex (CC6, consisting of strains ALS12 and ALS22) that groups close to the multiplex strains but is distinct due to the presence of a number of recombination events with the other subspecies, X. fastidiosa subsp. fastidiosa and X. fastidiosa subsp. sandyi (37). Since recombination had not previously been reported in this species of bacteria, an initial estimate of the influence of recombination in X. fastidiosa was made. Despite the low number of strains used, our data suggest that X. fastidiosa is roughly one-half as likely to gain a new allele by recombination as by point mutation. However, an individual nucleotide is approximately three times more likely to change as a result of recombination than as a result of a point mutation, since in X. fastidiosa a single recombination results in, on average, about seven nucleotide changes (Table 5).
This estimate was based on classification of allelic changes due to single and multiple base differences as due to point mutation and recombination, respectively. A potential bias that diminishes the estimated role of recombination is the possibility that some single base changes may be due to recombination (13, 15, 17). On the other hand, two potential sources of error bias the estimate in favor of recombination. One of theses sources is the use of long genes. We used genes whose lengths varied from 345 bp to 1,824 bp. Longer gene segments are more likely to accumulate multiple point mutations that would be interpreted as marking recombination events. However, the bias due to length is quite small. To confirm this, we applied two kinds of correction: first, the criterion for recombination in longer genes was increased proportional to the length, and second, we reanalyzed the data with the longer genes divided into shorter independent segments of about 500 bp. Neither of these corrections significantly affected the results (data not shown). A second factor that could bias the results in favor of recombination was our use of multilocus variants rather than just single-locus variants in the comparison with the estimated ancestral type. This change had the effect of increasing the time scale of the comparisons (since multilocus variants are generally older than single-locus variants, particularly if the recombination rate is low). Increasing the time scale increases the risk of multiple point mutations. This problem decreases with sample size, since intermediate (single point mutation) alleles are likely to be observed. A possible example of this effect is a putative recombination in the leuA gene identified by the MLST and DnaSP methods. This case involves two shared changes separated by a single base pair in CC3 to CC5 compared with CC1, CC2, and CC6. These two changes could represent shared mutations that accumulated since the separation of CC3 to CC6 from CC1 and CC2, combined with recombination between CC6 and CC1 or CC2, or they could represent two shared mutations accumulating in the much shorter time since the common ancestor of CC3 to CC5 (Fig. 2).
Sawyer's test has the advantage of providing a statistical test for recombination events; however, it provided significant support for only one occurrence (corresponding to one of the four recombination events ascertained from visual inspection). Part of this lack of power arises because Sawyer's test cannot detect recombination when the recombined region is larger than the gene. For example, allele 1 of pilU is the ancestral allele for the PD clonal complex. This allele has completely recombined in ALS12 and ALS22 (Table 3). Also, Sawyer's test raises the problem of multiple testing. When the goal is to estimate the overall rate of recombination, each test is not strictly independent, so the significance values should be corrected in a table-wide manner (in this case for 20 tests [10 genes × 2 types of test]). This results in no value being returned as significant and illustrates the lack of power inherent in Sawyer's test.
Of the methods used for identifying recombination, the DnaSP method appears to be the most reliable, since it identifies all of the recombination events identified by visual inspection, plus one more (the leuA example discussed above) (Table 7). The multilocus variant method has the advantage that it provides a direct estimate of the relative contributions of mutation and recombination; however, the method may assign additional events as recombinations that could potentially be point mutations. Additional sampling should help uncover whether the alleles that differ from other alleles at only 2 to 4 bp represent cases of multiple point mutations alone or if recombination was involved. Finally, Sawyer's test appears to be much too conservative, failing to identify recombination events identified by visual inspection.
Compared to other bacteria, X. fastidiosa appears to have low ratios of recombination to point mutation on a per allele basis (0.46:1) and on a per nucleotide basis (3.23:1). For example, Streptococcus pneumoniae (ratios, 8.9:1 and 61:1) and Neisseria meningitidis (ratios, 4.75:1 and 100:1) (13) have ratios that are shifted more than 10-fold in favor of recombination. This strong bias toward recombination being the dominant force in the generation of new alleles persists even when Escherichia coli, a proteobacterium more closely related to X. fastidiosa, is considered. Guttman and Dykhuizen (19) found a recombination rate per nucleotide that was 50 times greater than the mutation rate for E. coli. The low rate of recombination in X. fastidiosa suggests that the phylogeny (Fig. 2) is the true evolutionary history. A similar, largely clonal pattern has been documented for Pseudomonas syringae (34).
The MLST method clearly offers an excellent opportunity for strain typing and cataloguing diversity within a bacterial species. Our database of 10 genes (9.3 kb) differentiated 19 STs from 25 strains, and a subset of five genes retained the same level of sequence type diversity. Since the suggested number of loci for an MLST data set is seven (16), we suggest that the holC, nuoL, leuA, gltT, cysG, petC, and lacF genes be used as the basis of MLST typing in this species. A database for this purpose has been established at http://www.mlst.net.
Acknowledgments
This study was supported by a USDA-CREES grant under the UC Pierce's Disease program to L.N. and R.S.
REFERENCES
- 1.Almeida, R. P. P., and A. H. Purcell. 2003. Biological traits of Xylella fastidiosa strains from grapes and almonds. Appl. Environ. Microbiol. 69:7447-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betran, E., J. Rozas, A. Navarro, and A. Barbadilla. 1997. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics 146:88-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharyya, A., S. Stilwagen, N. Ivanova, M. D'Souza, A. Bernal, A. Lykidis, V. Kapatral, I. Anderson, N. Larsen, T. Los, G. Reznik, E. Selkov, Jr., T. L. Walunas, H. Feil, W. S. Feil, A. Purcell, J. L. Lassez, T. L. Hawkins, R. Haselkorn, R. Overbeek, P. F. Predki, and N. C. Kyrpides. 2002. Whole-genome comparative analysis of three phytopathogenic Xylella fastidiosa strains. Proc. Natl. Acad. Sci. USA 99:12403-12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, E. F., K. Nelson, F. S. Wang, T. S. Whittam, and R. K. Selander. 1994. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 91:1280-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brlansky, R. H., C. L. Davis, L. W. Timmer, D. S. Howd, and J. Constera. 1991. Xylem-limited bacteria in citrus from Argentina with symptoms of citrus variegated chlorosis. Phytopathology 81:1210. [Google Scholar]
- 6.Clarke, S. C. 2002. Nucleotide sequence-based typing of bacteria and the impact of automation. Bioessays 24:858-862. [DOI] [PubMed] [Google Scholar]
- 7.Costa, H. S., E. Raetz, T. R. Pinckard, C. Gispert, R. Hernandez-Martinez, C. K. Dumenyo, and D. A. Cooksey. 2004. Plant hosts of Xylella fastidiosa in and near southern California vineyards. Plant Dis. 88:1255-1261. [DOI] [PubMed]
- 8.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drouin, G., F. Prat, M. Ell, and G. D. P. Clarke. 1999. Detecting and characterizing gene conversion between multigene family members. Mol. Biol. Evol. 16:1369-1390. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 11.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feil, E. J., M. C. Enright, and B. G. Spratt. 2000. Estimating the relative contribution of mutation and recombination to clonal diversification: a comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res. Microbiol. 151:465-469. [DOI] [PubMed] [Google Scholar]
- 13.Feil, E. J., E. C. Holmes, D. E. Bessen, M.-S. Chan, N. P. J. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feil, E. J., M. C. J. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 16.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feil, E. J., and B. G. Spratt. 2001. Recombination and population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 18.Grundmann, H., S. Hori, M. C. Enright, C. Webster, A. Tami, E. J. Feil, and T. Pitt. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 40:4544-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guttman, D. S., and D. E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266:1380-1383. [DOI] [PubMed] [Google Scholar]
- 20.Hendson, M., A. H. Purcell, D. Chen, C. Smart, M. Guilhabert, and B. Kirkpatrick. 2001. Genetic diversity of Pierce's disease strains and other pathotypes of Xylella fastidiosa. Appl. Environ. Microbiol. 67:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopkins, D. L. 1989. Xylella fastidiosa: xylem-limited bacterial pathogen of plants. Annu. Rev. Phytopathol. 27:271-290. [Google Scholar]
- 22.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 23.King, S. J., J. A. Leigh, P. J. Heath, I. Luque, C. Tarradas, C. G. Dowson, and A. M. Whatmore. 2002. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40:3671-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nallapareddy, S. R., R. W. Duh, K. V. Singh, and B. E. Murray. 2002. Molecular typing of selected Enterococcus faecalis isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noller, A. C., M. C. McEllistrem, O. C. Stine, J. M. Morris, Jr., D. J. Booxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulse-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peacock, S. J., G. D. de Silva, A. Justice, A. Cowland, C. E. Moore, C. G. Winearls, and N. P. Day. 2002. Comparison of multilocus sequence typing and pulsed-field gel electrophoresis as tools for typing Staphylococcus aureus isolates in a microepidemiological setting. J. Clin. Microbiol. 40:3764-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell, A. H., and D. L. Hopkins. 1996. Fastidious xylem-limited bacterial pathogens. Annu. Rev. Phytopathol. 34:131-151. [DOI] [PubMed] [Google Scholar]
- 31.Purcell, A. H., S. R. Saunders, M. Hendson, M. E. Grebus, and M. J. Henry. 1999. Causal role of Xylella fastidiosa in oleander leaf scorch. Phytopathology 89:53-58. [DOI] [PubMed] [Google Scholar]
- 32.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 33.Rychlik, W. 2002. OLIGO: primer analysis software, version 6.65. Molecular Biology Insights Inc., Cascade, CO.
- 34.Sarkar, S. F., and D. S. Guttman. 2004. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70:1999-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526-538. [DOI] [PubMed] [Google Scholar]
- 36.Schaad, N. W., E. Postnikova, G. Lacy, M. Fatmi, and C. J. Chang. 2004. Xylella fastidiosa subspecies: X. fastidiosa subsp. piercei, subsp. nov., X. fastidiosa subsp. multiplex subsp. nov., and X. fastidiosa subsp. pauca subsp. nov. Syst. Appl. Microbiol. 27:290-300. (Erratum 27: 763). [DOI] [PubMed] [Google Scholar]
- 37.Schuenzel, E. L., M. Scally, R. Stouthamer, and L. Nunney. 2005. A multi-gene phylogenetic study of clonal diversity and divergence in North American strains of the plant pathogen Xylella fastidiosa. Appl. Environ. Microbiol. 71:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittman. 1986. Methods of multilocus enzyme electrophoresis for bacterial genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson, A. J., et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 40.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Sluys, M. A., et al. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walsh, P. S., D. A. Metzger, and R. Higuchi. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10:506-513. [PubMed] [Google Scholar]