Abstract
Cell adsorption and selective desorption for separation of microbial cells were conducted by using chitosan-immobilized silica (CIS). When chitosan was immobilized onto silica surfaces with glutaraldehyde, bacterial cells adsorbed well and retained viability. Testing of the adsorption and desorption ability of CIS using various microbes such as Escherichia coli, Aeromonas hydrophila, Pseudomonas aeruginosa, Bacillus subtilis, Micrococcus luteus, Staphylococcus aureus, Staphylococcus epidermidis, Lactobacillus casei, Streptococcus mutans, Streptococcus sobrinus, Streptococcus salivarius, Saccharomyces cerevisiae, Saccharomyces ludwigii, and Schizosaccharomyces pombe revealed that most microbes could be adsorbed and selectively desorbed under different conditions. In particular, recovery was improved when l-cysteine was added. A mixture of two bacterial strains adsorbed onto CIS could also be successfully separated by use of specific solutions for each strain. Most of the desorbed cells were alive. Thus, quantitative and selective fractionation of cells is readily achievable by employing chitosan, a known antibacterial material.
Infection and contagion are becoming increasing problems in our communities, for example, with nosocomial infections caused by antibiotic bacterial strains like methicillin-resistant Staphylococcus aureus, mass food poisoning caused by contaminating pathogenic bacteria like enteropathogenic Escherichia coli O157 in water or foods, and infections caused by the microscopic parasites Giardia and Cryptosporidium, which are found in water. Traditionally, these microorganisms can be found by culture-based methods, such as single-colony isolation and antibody utilization using an enzyme-linked immunosorbent assay (4, 5, 21). Recently, PCR has been commonly applied (9, 11, 12). Furthermore, various methods, such as differential staining, serological methods, flow cytometry, phage typing, protein analysis, and comparison of DNA nucleotide sequences, have been utilized. These methods enable us to detect microbes in small samples with high sensitivity (3).
At the industrial level, these approaches are not suitable due to limited time and the significant effort, cost, and scale involved. The culture-based method is restricted to viable target bacteria and is time-consuming. Therefore, it is of great interest to develop novel fast and inexpensive detection, separation, and isolation methods (1, 7, 10).
The purpose of the present study was to assess a new separation and concentration system for bacterial cells that is easy to apply and allows rapid processing by using chitosan as a functional ligand. Our final purpose in this study was to fractionate microbial cells with chitosan.
Chitosan is a polymeric β-1,4-N-acetylglucosamine derived from chitin. This major component of crustacean shells features organic adaptability and biodegradability and can adsorb metal ions, proteins, and bacterial cells (2, 8, 14). In particular, antibacterial activity of chitosan, due to adsorption and flocculation, has been well studied (16, 17). The polysaccharide has various molecular weights, and its polymers have higher antibacterial activity than chitosan oligomers, showing stronger effects on gram-positive than gram-negative bacteria (15). A key feature is its positive charges on the amino groups at C-2. Chitosan binds to the bacterial outer membrane because of its negatively charged carboxyl and phosphate groups. The outer membrane is thereby disrupted, leading to cellular damage or even death (6, 13). In solution, the material demonstrated antibacterial activity as expected, but we found that this could be suppressed by decreasing the amino groups by immobilizing chitosan onto silica with glutaraldehyde (2). As mentioned previously (6, 8, 13, 15-17), the high absorbing ability of chitosan also makes it useful for bacterial concentration while retaining viability.
Therefore, in this study, we attempted to develop a detection method from two points of view: separation and concentration. Experiments were conducted to study the adsorption, desorption, and fractionation of different microbial cell species, including both prokaryotes and unicellular eukaryotes. As a result, we were able to fractionate two types of bacteria with chitosan-immobilized silica (CIS).
MATERIALS AND METHODS
Immobilization of chitosan onto amino silica.
Fifty milligrams of amino silica (NH-DM1020; Fuji Silysia Chemical, Ltd., Japan) was washed with 190 μl of 0.1 M acetic acid (pH 5) and degassed in a microtube (2-ml microtube; Greiner Bio-One Co., Ltd.). Chitosan was dissolved in 0.1 M acetic acid (pH 5) supplemented with 0.25 mM sodium azide (Wako Pure Chemical Industries, Ltd., Japan), and 1.5 ml of a 0.05% (wt/vol) chitosan solution and 0.2 ml of 25% glutaraldehyde (Wako Pure Chemical Industries, Ltd., Japan) were mixed and incubated for 3 h at room temperature in tubes. The CIS was then rinsed twice with 0.1 M acetic acid (pH 5) and then three times with distilled water.
SEM.
The surface morphologies of CIS and the state of B. subtilis adsorbed onto CIS were studied using scanning electron microscopy (SEM) (SEM S-800; Hitachi High-Technologies, Inc., Japan). The sample was dehydrated, critical point dried, and coated with gold before undergoing SEM. Before dehydrating cells bound to CIS, CIS was washed five times with 10 mM MOPS (3-morpholinopropanesulfonic acid) (pH 7).
Bacterial strains and media.
E. coli (XL1-Blue) was purchased from Stratagene Japan. Pseudomonas aeruginosa (IAM 1514), Bacillus subtilis (IAM 12118), Micrococcus luteus (IAM 1056), S. aureus (IAM 12544), Saccharomyces cerevisiae (IAM 4512), Saccharomyces ludwigii (IAM 12243), and Schizosaccharomyces pombe (IAM 4863) were obtained from the Institute of Applied Microbiology Culture Collection. Aeromonas hydrophila (NRBC 12978) was obtained from the Biological Resource Center, National Institute of Technology and Evaluation. Staphylococcus epidermidis (JCM 2414) was obtained from the Japan Collection of Microorganisms, Institute of Physical and Chemical Research (RIKEN). Lactobacillus casei (NRIC1046) was obtained from the NODAI Research Institute Culture Collection, Tokyo University of Agriculture. Streptococcus salivarius (ATCC 9222) was obtained from the American Type Culture Collection. Streptococcus mutans and Streptococcus sobrinus were original isolates from the oral cavity obtained from Nihon University School of Dentistry of Matsudo (Japan).
The cultivation medium for E. coli, B. subtilis, M. luteus, and S. aureus was Luria-Bertani broth (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl, 1,000 ml of distilled water, pH 7), and cultures were incubated at 30°C (M. luteus and S. aureus) or 37°C (E. coli and B. subtilis) overnight. A. hydrophila was grown in 3% NaCl nutrient broth (Difco Laboratories) at 25°C for 16 h. P. aeruginosa was grown in nutrient broth at 30°C for 16 h. S. mutans, S. sobrinus, and S. salivarius were grown anaerobically in brain heart infusion broth (Nissui, Japan) overnight at 37°C. L. casei was grown on the following medium: 10 g of peptone, 10 g of yeast extract, 10 g of glucose, 3 g of CH3COONa, and 1,000 ml of deionized water, pH 6.5.
S. cerevisiae, S. ludwigii, and S. pombe were grown on MY medium (3 g of yeast extract, 3 g of malt extract, 5 g of peptone, 10 g of glucose, 1,000 ml of deionized water, pH 7.0) at 30°C for 1 day.
The starting cultures were transferred to solid agar plates prepared with the above-described media. In the stationary growth phase, they were harvested by centrifugation (3,000 rpm, 15 min, 4°C) and washed twice with 10 mM MOPS (pH 7.0), and pellets of cells were obtained.
Adsorption and desorption of E. coli.
Various adsorption conditions were tested. Cell suspensions were adjusted to mili Q water as the control or 50 mM cysteine and 0.01% (wt/vol) bovine serum albumin (BSA; Wako Pure Chemical Industries, Ltd., Japan). With the optical density (OD) at 600 nm for each suspension adjusted to approximately 0.7 (1.5 × 108 cells/ml), aliquots of 1.8 ml were incubated with 50 mg of CIS, amino silica (NH-DM1020), or pure silica (MB-5D; Fuji Silysia Chemical, Ltd., Japan) in microtubes for 1 h at room temperature on a rotary mixer. Amino silica has amino groups and pure silica has silanol groups on their surfaces. (Pure silica was modified with the silane-coupling reagent 3-aminopropyltriethoxysilane and changed to amino silica.) The adsorption amount was determined by the absorbance of the supernatants at 600 nm. A 1 M NaCl solution was used for desorption. After incubation for 15 min, the absorbance of the supernatant was measured. Recovery of cells was calculated as follows: desorption ratio (%) = amount of desorbed cells/amount of adsorbed cells × 100.
Adsorption of various strains.
Every cell pellet was resuspended in 50 mM l-cysteine (pH 5). With the OD at 600 nm for each suspension adjusted to approximately 0.7, aliquots of 1.8 ml were incubated with 50 mg of CIS, amino silica, or pure silica in tubes for 1 h at room temperature on a rotary mixer. The adsorption amount was determined by the absorbance of the supernatants at 600 nm.
Viability of cells.
After adsorption of each species onto CIS, the supernatant was exchanged five times with 10 mM MOPS (pH 7). Two microliters of CIS was spread onto agar plates prepared with nutrient media. The plates with CIS were incubated under the conditions described above. The viability of cells was judged from the observation of CFU.
Determination of zeta potential.
The zeta potentials of 14 bacterial strains were measured with NICOMP 380 ZLS (Particle Sizing System Co.) at room temperature. NICOMP 380 ZLS was equipped with a 20-mW laser diode and a photomultiplier tube detector with one optical fiber set at 19.8°. Bacteria from cell pellets were suspended in adsorption solution (50 mM cysteine, pH 5). The experimental reproducibility was tested five times.
Desorption of each strain.
Desorption of cells from CIS was attempted by incubation with various solutions listed in Table 1 for 15 min. All species were adsorbed onto CIS in the presence of 50 mM cysteine for 1 h at room temperature. Their supernatants were removed and examined by spectrometry at 600 nm. Next, 1.8 ml of various buffers (shown in Table 1) was introduced into the tubes followed by incubation for 15 min. The supernatants were removed, and the absorbance was determined at 600 nm to allow calculation of recovery.
TABLE 1.
Adsorption and desorption of E. coli with CIS, amino silica, and pure silica
| Adsorption condition | Adsorption medium | % Adsorption | % Recoverya |
|---|---|---|---|
| Control | CIS | 72.1 | 7.9 |
| Amino silica | 21.8 | 15.3 | |
| Pure silica | 0 | ND | |
| 50 mM cysteine | CIS | 88.6 | 63.0 |
| Amino silica | 56.8 | 13.5 | |
| Pure silica | 3.6 | ND | |
| 0.01% BSA | CIS | 0 | ND |
| Amino silica | 20.2 | 13.2 | |
| Pure silica | 3.8 | ND |
ND, not determined.
The following chemical agents were used for desorption experiments: 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) (DOJINDO, Ltd., Japan), polyoxyethylene 20 sorbitan monolaurate (Tween 20), sodium dodecyl sulfate (SDS), sodium polyphosphate, l-arginine, l-histidine, Tris, imidazole, citric acid, and acetic acid (Wako Pure Chemical Industries, Ltd., Japan).
Fractionation of cells in batches.
Two of the three species E. coli, B. subtilis, and S. cerevisiae were selected for fractionation. Suspensions of cells at 600 nm before adsorption were adjusted to 0.7 (B. subtilis, 1.3 × 108 cells/ml; S. cerevisiae, 0.9 × 107 cells/ml), and equal quantities of the two were mixed. A total of 1.8 ml of each suspension (each individual species and one mixed sample) was incubated with 50 mg of CIS for adsorption of cells for 1 h at room temperature. Fractionations of cells were conducted by exchanging various solutions in batches.
The following desorption solutions were used: solution A (desorption for E. coli) (0.3 M citric acid, 1% Tween 20, 50 mM cysteine [pH 3]), solution B (desorption for B. subtilis) (0.2 M arginine, 1% Tween 20, 50 mM cysteine [pH 9]), solution C (desorption for S. cerevisiae) (1 M NaCl, 50 mM cysteine [pH 5]), solution D (desorption for S. cerevisiae), (10 mM NaCl, 50 mM cysteine [pH 5]), and washing solution (50 mM cysteine, pH 5.0).
The supernatant obtained after incubation for adsorption for 60 min was designated fraction 1 (see Fig. 5a). CIS with cells was then washed with washing solution three times for 5 min. (fractions 2 to 4). Fractions 5 to 7, which were eluted by desorption solution A, and fractions 10 to 12, which were eluted by solution B, were incubated for 15 min. Fractions 8 and 9 were washed with washing solution.
FIG. 5.
Fractionation of microbial cells in batches. The combinations of cells were (a) E. coli and B. subtilis, (b) S. cerevisiae and B. subtilis, and (c) S. cerevisiae and E. coli. The mixtures of two species comprised equal quantities of the two. The addition of the first desorption solution was for fractions 5, 6, 7, and the second one was for fractions 10, 11, and 12. Other fractions were washed with 50 mM cysteine.
Similarly, fractions 5 to 7 eluted by desorption solution C and fractions 10 to 12 eluted by solution B were incubated for 15 min (see Fig. 5b). Fractions 5 to 7 eluted by desorption solution D and fractions 10 to 12 eluted by solution A were incubated for 15 min (see Fig. 5c). Other fractions (fractions 2 to 4, 8, and 9) were washed with washing solution.
The obtained supernatants of the peaks (fractions 5 and 10) were centrifuged (5,000 rpm, 4°C, 5 min) and concentrated to 100 μl for microscopic observation (BX51; Olympus Co., Tokyo, Japan).
RESULTS AND DISCUSSION
In this study, we conducted fractionation of microbial cells with CIS by selective desorption. This research aimed to develop a concentration and selective separation system by using very familiar materials such as chitosan or silica.
(i) Conditions for the adsorption and desorption of E. coli cells. Adsorption and desorption of E. coli.
An investigation of the conditions necessary for good adsorption and desorption of E. coli was conducted. Control samples were suspended in mili Q water. This condition showed good adsorption of cells, but cell recovery was low after treatment with 1 M NaCl or even with a higher-ionic-strength solution (4 M NaCl). This result demonstrated very tight binding between bacteria and CIS, so several blocking agents were tested in order to weaken the binding force. Although BSA has been the most popular blocking agent, cells did not adsorb under these conditions (Table 1). Chitosan adsorbed BSA strongly (20). We tried several other agents and found that l-cysteine was the most effective agent (Table 1). We found that when cysteine was added as a blocking agent, not only was adsorption still efficient but desorption was also facilitated. Therefore, we used 50 mM cysteine as the adsorption buffer for experiments shown in Fig. 1 and for the later part of this study.
FIG. 1.
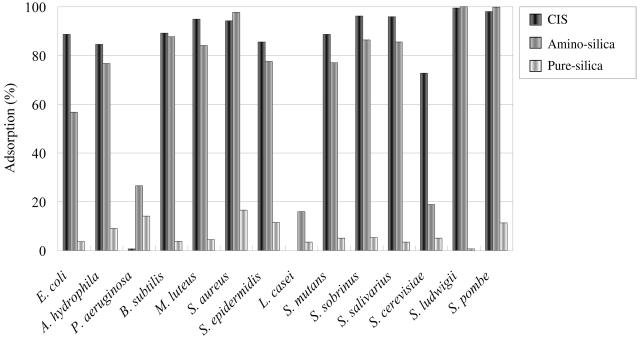
Adsorption of various strains onto CIS, amino silica, and pure silica. Microbial cells of E. coli, A. hydrophila, P. aeruginosa, B. subtilis, M. luteus, S. aureus, S. epidermidis, L. casei, S. mutans, S. sobrinus, S. salivarius, S. cerevisiae, S. ludwigii, and S. pombe were incubated with 50 mg of CIS, amino silica, or pure silica for 1 h in 50 mM cysteine. Bacterial cells were resuspended to an OD at 600 nm of 0.7. After incubation, the amount of adsorption was calculated by determining the OD of the supernatants at 600 nm.
(ii) Comparison of the adsorption property of cells onto CIS and control resins. (a) Cell adsorption ability of CIS, amino silica, or pure silica.
We conducted tests to examine the adsorption of microbial cells onto resins. Gram-negative E. coli, A. hydrophila, and P. aeruginosa; gram-positive B. subtilis, M. luteus, S. aureus, S. epidermidis, L. casei, S. mutans, S. sobrinus, and S. salivarius; and the yeasts S. cerevisiae, S. ludwigii, and S. pombe were chosen for testing with CIS, amino silica, or pure silica. The adsorption of these strains onto three resins in the presence of 50 mM cysteine is shown in Fig. 1. Most of the species adsorbed well onto CIS except for P. aeruginosa and L. casei. These organisms did not adsorb onto CIS at all. Pure silica showed low adsorption for all strains. CIS showed no adsorption for P. aeruginosa and L. casei, but amino silica and pure silica showed low adsorption (Fig. 1). CIS showed negative selected adsorption for P. aeruginosa and L. casei. CIS adsorbed more E. coli and S. cerevisiae cells than amino silica (Fig. 1). Almost all bacteria adsorbed well onto CIS except for P. aeruginosa and L. casei, so CIS was determined to be a good carrier for adsorption of bacterial cells. This result provides the basis for the application of a new concentration method for bacteria.
In the experiments described below, the use of CIS as a carrier of microbial cells was studied in detail.
(b) Viability of cells adsorbed onto CIS and amino silica.
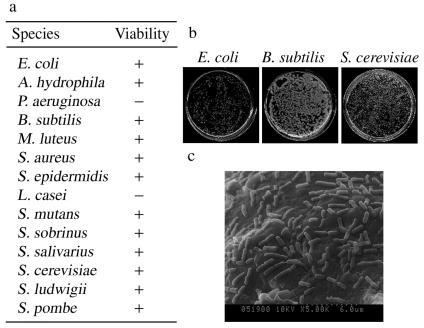
After adsorption, cell-bound CIS was spread onto agar plates in order to quantitatively determine the viability of the cells. All species formed colonies well, except for those that did not adsorb (P. aeruginosa and L. casei). These results demonstrated that the antibacterial ability of chitosan was suppressed by immobilization (Fig. 2). Figure 2b shows colony formation of three species after adsorption onto CIS. The effect of immobilization with chitosan was that microbial cells contacted the CIS on only one side of the cell, and the number of amino groups on the surface of CIS was low compared to that with soluble chitosan (20). For these reasons, the antibacterial effect was suppressed. A SEM observation of the adsorption state of B. subtilis on CIS is shown in Fig. 2c. Many cells were adsorbed on the CIS surface dispersedly.
FIG. 2.
Cell adsorption and viability. (a) Viability of microbial cells adsorbed onto CIS. After adsorption of cells onto CIS, CIS was washed five times with 10 mM MOPS (pH 7). Two microliters of CIS was spread onto agar plates prepared with nutrient media. +, colony formation; −, no colony formation. (b) The photos show colonies of E. coli, B. subtilis, and S. cerevisiae. (c) SEM of B. subtilis absorbed onto CIS (magnification, ×5,000).
(c) Zeta potential of cells.
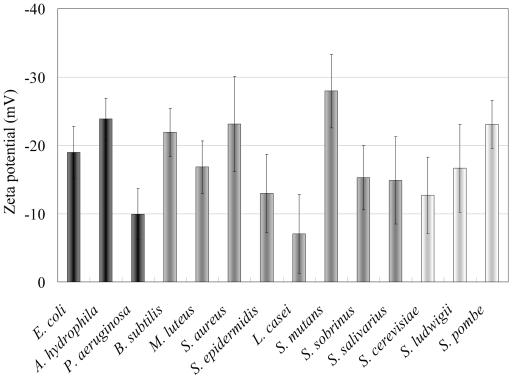
Since the surface charge of cells should be one of the important factors for adsorption, the electrostatic surface charges of 14 strains were examined. The negative surface charges ranged from −7.0 mV for L. casei to −27.9 mV for S. mutans (Fig. 3). The surfaces of CIS and amino silica both have many amino groups; therefore, their surfaces were positively charged. The initial attraction of adsorption could have resulted from a mutual electrostatic force, because P. aeruginosa and L. casei, both weak negatively charged strains (Fig. 3), did not adsorb onto silica at all (Fig. 1). The mechanism of cell adsorption may depend on cell size and shape, the presence of flagella, and other factors. However, it seems that adsorption of cells onto silica was mainly due to electrostatic forces, because P. aeruginosa and L. casei, which did not adsorb onto CIS, had weak negative charges (Fig. 3).
FIG. 3.
Zeta potentials (millivolts) of 14 species under the adsorption condition of 50 mM cysteine, pH 5.0. Results for gram-negative bacteria are indicated in black, results for gram-positive bacteria are shown in gray, and results for yeasts are shown in white.
Moreover, by comparing the results of desorption (Table 2 and Fig. 4a), we could not find any correlations. For example, a more negatively charged species, S. mutans, was efficiently recovered (Fig. 3, Table 2, and Fig. 4a). It was thought that the adsorption of cells was caused by mainly electrostatic force at first, but other forces (for example, van der Waals and hydrophobic interactions between cells and CIS) were worked after the attraction of cells onto CIS (19). The main driving force would be changed in the course of the adsorption process. For that reason, adsorption and desorption mechanism were thought to be complicated.
TABLE 2.
Desorption of microbial cells by various solutions
| Solutionb | Recovery (%)a
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-negative bacteria
|
Gram-positive bacteria
|
Yeasts
|
||||||||||
| E. coli | A. hydrophila | B. subtilis | M. luteus | S. aureus | S. epidermidis | S. mutans | S. sobrinus | S. salivarius | S. cerevisiae | S. ludwigii | S. pombe | |
| 1 M NaCl | + (63) | + (60) | − | + (56) | − | − | + (71) | − | − | + (81) | − | − |
| 1% CHAPS | − | − | − | − | − | − | − | − | − | − | − | − |
| 1% Tween 20 | − | − | − | − | − | − | − | − | − | − | − | − |
| 30 mM SDS | − | + (53) | + (87) | + (61) | − | − | − | − | − | + (80) | − | − |
| 1% sodium polyphosphate | − | − | − | + (64) | − | − | + (68) | − | − | + (63) | − | − |
| 0.1 M arginine | − | − | − | + (71) | + (69) | − | + (95) | − | − | + (72) | − | − |
| 0.1 M Tris | − | − | − | + (57) | + (59) | − | + (82) | − | − | + (69) | − | − |
| 0.1 M histidine | − | − | − | − | − | − | − | − | − | + (70) | − | − |
| 0.1 M imidazole | − | − | − | − | − | + (52) | + (53) | − | − | + (68) | − | − |
| 0.1 M citric acid | − | + (82) | − | − | − | − | − | − | − | + (63) | − | − |
| 0.1 M acetic acid | − | + (51) | − | − | − | − | − | − | − | + (60) | − | − |
| 0.1 M arginine-1% CHAPS | − | − | + (86) | + (64) | − | + (52) | + (100) | + (61) | − | + (87) | − | − |
| 0.1 M Tris-1% CHAPS | − | − | + (62) | + (94) | − | − | + (94) | − | − | + (82) | − | − |
| 0.1 M histidine-1% CHAPS | − | − | − | − | − | − | + (89) | − | − | + (98) | − | − |
| 0.1 M imidazole-1% CHAPS | − | − | − | − | + (69) | − | + (75) | − | − | + (74) | − | − |
| 0.1 M citric acid-1% CHAPS | + (76) | + (84) | − | − | − | − | − | + (54) | − | − | − | − |
| 0.1 M acetic acid-1% CHAPS | − | + (91) | − | − | − | − | − | − | − | − | − | − |
−, 0 to 50% recovery; +, 50 to 100% recovery. Values in parentheses are actual percent recoveries.
For details about the 1 M NaCl, 0.1 M arginine—1% CHAPS, and 0.1 M citric acid—1% CHAPS conditions, see Fig. 4.
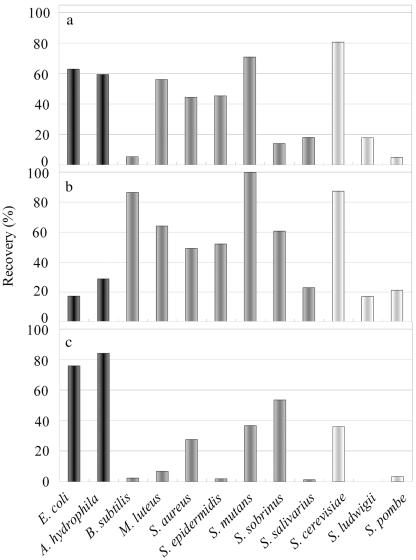
FIG. 4.
Recovery of cells from CIS with three solutions shown Table 1. Shown are (a) 1 M NaCl, (b) 0.1 M arginine-1% CHAPS, and (c) 0.1 M citric acid-1% CHAPS. Results for gram-negative bacteria are indicated in black, results for gram-positive bacteria are indicated in gray, and results for yeast are indicated in white.
(iii) Desorption and fractionation of cells. (a) Desorption of cells from CIS.
Many desorption conditions were tested for E. coli and A. hydrophila; gram-positive B. subtilis, M. luteus, S. aureus, S. epidermidis, S. mutans, S. sobrinus, and S. salivarius; and the yeasts S. cerevisiae, S. ludwigii, and S. pombe, and 17 conditions are reported in Table 2. Several interesting tendencies were observed (Table 2).
As shown in Table 2, 0.1 M citric acid with an ampholytic detergent (1% CHAPS) was good for desorption of gram-negative E. coli and A. hydrophila, and 1 M NaCl was also moderately effective.
Gram-positive B. subtilis was efficiently desorbed from CIS by 30 mM SDS or 0.1 M l-arginine with 1% CHAPS (Table 2). M. luteus, S. epidermidis, S. mutans, and S. sobrinus were also recovered well with 0.1 M l-arginine in 1% CHAPS. On the other hand, for gram-positive M. luteus, the best condition for desorption was 0.1 M Tris-1% CHAPS (Table 2). In particular, S. mutans was desorbed well by several different desorption agents in Table 2.
In the case of the eukaryotic S. cerevisiae, S. ludwigii, and S. pombe, the desorption patterns were dramatic (Table 2). Although S. cerevisiae was easily washed off of CIS by most desorption agents, the others were never released (Table 2).
Although none of the organisms was desorbed by CHAPS alone, the detergent worked favorably as a support for the other desorption agents (Table 2). Tween 20 was also found to work like CHAPS (data not shown). S. ludwigii and S. pombe were not desorbed even in their presence. Each of the agents was quite selective, and some species were not released from CIS at all (Table 2), suggesting that chromatography can be achieved by selecting specific kinds and concentrations of “eluting” agents.
To illustrate the differences among species based on the screening results (Table 2), Fig. 4 shows the three most remarkable conditions (1 M NaCl, 0.1 M Arg-1% CHAPS, and 0.1 M citric acid-1% CHAPS). In the presence of 1 M NaCl, recovery of cells from CIS differed significantly depending on the species. E. coli, A. hydrophila, M. luteus, S. aureus, S. epidermidis, S. mutans, and S. cerevisiae were efficiently recovered under these conditions (Fig. 4a). Moreover, a tendency for gram-positive bacteria like B. subtilis and S. mutans to be desorbed relatively well by 0.1 M Arg-1% CHAPS (Fig. 4b) and for gram-negative bacteria like E. coli and A. hydrophila to be desorbed well by 0.1 M citric acid-1% CHAPS (Fig. 4c) was seen.
Desorption by 1 M NaCl seemed to be caused by an ion-exchange effect (Fig. 4a). The tendency of gram-positive bacteria to be relatively well desorbed by 0.1 M Arg-1% CHAPS (Fig. 4b) and of gram-negative bacteria to be well desorbed by 0.1 M citric acid-1% CHAPS (Fig. 4c) indicated that some affinity recognition of the gram-positive or -negative surface may be occurring.
In comparison, citric acid (pKa1 = 3.13; 25°C) is a stronger acid than acetic acid (pKa1 = 4.76; 25°C). For E. coli and A. hydrophila, the use of citric acid resulted in higher cell recovery than with acetic acid. This result suggested that citric acid was attracted to the amino groups of chitosan and that these specific interactions (such as electrostatic and hydrophobic forces) for CIS were stronger than cell adsorption (18). Similarly, when the basic amino acids were compared, arginine (pKa = 12.5; 25°C) has a more basic character than histidine (pKa = 6.0; 25°C). Table 2 shows that arginine use resulted in better recovery of gram-positive bacteria than histidine. It seemed that the specific interaction between the negatively charged bacterial cells and arginine was stronger than that with histidine. Moreover, a detergent would help to detach the weakened bonds caused by the use of agents such as citric acid and arginine.
However, bacterial cell surfaces contain a variety of charged polymers such as lipopolysaccharides and glycoconjugates, which contain an array of ionizable groups such as carboxyl, phosphate, sulfate, and amino moieties. Therefore, the mechanisms responsible for desorption (Table 2) can be exceedingly complex, providing the variation necessary for differential separation.
Fractionation of microbial cells by selective desorption.
Figure 5 depicts the separation of mixtures of two different species of microbial cells. In accordance with the data shown in Table 2 and Fig. 4, suitable conditions for each species were selected. Fractionation of the mixture of E. coli and B. subtilis is shown in Fig. 5a, fractionation of the mixture of S. cerevisiae and B. subtilis is shown in Fig. 5b, and fractionation of the mixture of E. coli and S. cerevisiae is shown in Fig. 5c. Solutions A and B contained citric acid-cysteine-Tween 20 and l-arginine-cysteine-Tween 20, respectively. Solution A was effective for desorbing E. coli from CIS, and then when solution B was used, B. subtilis was eluted (graph in Fig. 5a). Citric acid appeared to be specific for E. coli; other acids, such as HCl, acetic acid, and oxalic acid, were not effective (Table 2). Solution B efficiently desorbed B. subtilis from CIS. Solutions C (50 mM cysteine-1 M NaCl) and D (50 mM cysteine-10 mM NaCl) desorbed S. cerevisiae from CIS well, with good maintenance of viability. In Fig. 5b, solution C was first used and then solution B was used. Similarly, solutions D and A were employed for Fig. 5c. The cells at the peak points of release from CIS were examined by microscopy (photographs in Fig. 5a to c), and clear separation was observed. Most of the desorbed cells from CIS were motile (photographs in Fig. 5a to c). In these experiments, it was noteworthy that in order to gain high recovery from CIS, E. coli and B. subtilis needed to be adsorbed in the presence of cysteine. A weak bond between CIS and cysteine might be present, and subsequently, cysteine might disturb the adsorption force between CIS and the cells, exerting a weakening effect.
In conclusion, this study showed that intact cells with different surface structures could be selectively fractionated with chitosan immobilized on silica (Fig. 5a to c). Fractionation of cells and selective desorption of cells by the very familiar saccharide chitosan superficially appears to be very surprising. We aim to clarify details in the future, but here we could confirm the feasibility of a new method for cell separation with a variety of target cells. Moreover, CIS might be utilized for bacterial adsorption in the fields of medicine and food. Examples include the concentration of bacteria from large-scale solutions in preparation for more sensitive PCR detection or antibody detection. CIS could broadly separate bacterial genus or species, so the loss of primer or antibody should be minimized. Clearly, combinations of various methods may offer the best and most sensitive approach.
Acknowledgments
We thank Kazuo Fukushima, Department of Microbiology, School of Dentistry at Matsudo, Nihon University, for providing bacterial cultures. We thank the Laboratory of Masahiko Abe, Department of Pure and Applied Chemistry, Tokyo University of Science, for use of the scanning electron microscope and NICOMP 380 ZLS. We acknowledge Tatsuo Tsunoda, Research Center for Compact Chemical Process, National Institute of Advanced Industrial Science and Technology, for help with the scanning electron microscope observation.
REFERENCES
- 1.Arvidsson, P., F. M. Plieva, I. N. Savina, V. I. Lozinsky, S. Fexby, L. Bülow, I. Y. Galaev, and B. Mattiasson. 2002. Chromatography of microbial cells using continuous supermacroporous affinity and ion-exchange columns. J. Chromatogr. A 977:27-38. [DOI] [PubMed] [Google Scholar]
- 2.Benesch, J., and P. Tengvall. 2002. Blood protein adsorption onto chitosan. Biomaterials 23:2561-2568. [DOI] [PubMed] [Google Scholar]
- 3.Black, J. G. 1996. Microbiology principles and applications, 3rd ed. Prentice-Hall, Upper Saddle River, N.J.
- 4.Caruso, P., M. T. Gorris, M. Cambra, J. L. Palomo, J. Collar, and M. M. López. 2002. Enrichment double-antibody sandwich indirect enzyme-linked immunosorbent assay that uses a specific monoclonal antibody for sensitive detection of Ralstonia solanacearum in asymptomatic potato tubers. Appl. Environ. Microbiol. 68:3634-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, D., P. J. Hanna, K. Altmann, A. Smith, P. Moon, and L. S. Hammond. 1992. Development of monoclonal antibodies that identify Vibrio species commonly isolated from infections of humans, fish, and shellfish. Appl. Environ. Microbiol. 58:3694-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, B., K. Kim, Y. Yoo, S. Oh, J. Choi, and C. Kim. 2001. In vitro antimicrobial activity of a chitooligosaccharide mixture against Actinobacillus actinomycetemcomitans and Streptococcus mutans. Int. J. Antimicrob. Agents 18:553-557. [DOI] [PubMed] [Google Scholar]
- 7.Ebersole, R. C., and R. M. McCormick. 1993. Separation and isolation of viable bacteria by capillary zone electrophoresis. Biotechnology 11:1278-1282. [DOI] [PubMed] [Google Scholar]
- 8.Helander, I. M., E.-L. Nurmiaho-Lassila, R. Ahvenainen, J. Rhoades, and S. Roller. 2001. Chitosan disrupts the barrier properties of the outer membrane of gram-negative bacteria. Int. J. Food Microbiol. 71:235-244. [DOI] [PubMed] [Google Scholar]
- 9.Inglis, G. D., and L. D. Kalischuk. 2003. Use of PCR for direct detection of Campylobacter species in bovine feces. Appl. Environ. Microbiol. 69:3435-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaspers, E., and J. Overmann. 1997. Separation of bacterial cells by isoelectric focusing, a new method for analysis of complex microbial communities. Appl. Environ. Microbiol. 63:3176-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambertz, S. T., R. Lindqvist, A. Ballagi-Pordány, and M.-L. Danielsson-Tham. 2000. A combined culture and PCR method for detection of pathogenic Yersinia enterocolitica in food. Int. J. Food Microbiol. 57:63-73. [Google Scholar]
- 12.Li, X., N. Boudjellab, and X. Zhao. 2000. Combined PCR and slot blot assay for detection of Salmonella and Listeria monocytogenes Int. J. Food Microbiol. 56:167-177. [DOI] [PubMed] [Google Scholar]
- 13.Liu, H., Y. Du, X. Wang, and L. Sun. 2004. Chitosan kills bacteria through cell membrane damage. Int. J. Food Microbiol. 95:147-155. [DOI] [PubMed] [Google Scholar]
- 14.Liu, X. D., S. Tokura, M. Haruki, N. Nishi, and N. Sakairi. 2002. Surface modification of nonporous glass beads with chitosan and their adsorption property for transition metal ions. Carbohydr. Polymers 49:103-108. [Google Scholar]
- 15.No, H. K., N. Y. Park, S. H. Lee, and S. P. Meyers. 2002. Antibacterial activity of chitosans and chitosan oligomers with different molecular weights. Int. J. Food Microbiol. 74:65-72. [DOI] [PubMed] [Google Scholar]
- 16.Strand, S. P., K. M. Vårum, and K. Østgaard. 2003. Interactions between chitosans and bacterial suspensions: adsorption and flocculation. Colloids Surf. B Biointerf. 27:71-81. [Google Scholar]
- 17.Strand, S. P., T. Nordengen, and K. Østgaard. 2002. Efficiency of chitosans applied for flocculation of different bacteria. Water Res. 36:4725-4752. [DOI] [PubMed] [Google Scholar]
- 18.Takatsuji, W., and H. Yoshida. 1998. Adsorption on polyaminated highly porous chitosan: equilibria. Ind. Eng. Chem. Res. 37:1300-1309. [Google Scholar]
- 19.Vacheethasanee, K., and R. E. Marchant. 2000. Nonspecific Staphylococcus epidermidis adhesion: contributions of biomaterial hydrophobicity and charge, p. 73-90. In Y. H. An and R. J. Friedman (ed.), Handbook of bacterial adhesion. Principles, methods and applications, 1st ed. Humana Press Inc., Totowa, N.J.
- 20.Yoshida, H., and T. Kataoka. 1989. Adsorption of BSA on cross-linked chitosan: equilibrium isotherm. Chem. Eng. J. 41:B11-B15. [Google Scholar]
- 21.Yu, H., and J. G. Bruno. 1996. Immunomagnetic-electrochemiluminescent detection of Escherichia coli O157 and Salmonella typhimurium in foods and environmental water samples. Appl. Environ. Microbiol. 62:587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]