Abstract
We investigated the adaptation to milk of Streptococcus thermophilus LMG18311 using a proteomic approach. Two-dimensional electrophoresis of cytosolic proteins were performed after growth in M17 medium or in milk. A major modification of the proteome concerned proteins involved in the supply of amino acids, like the peptidase PepX, and several enzymes involved in amino acid biosynthesis. In parallel, we observed the upregulation of the synthesis of seven enzymes directly involved in the synthesis of purines, as well as formyl-tetrahydrofolate (THF) synthetase and serine hydroxy-methyl transferase, two enzymes responsible for the synthesis of compounds (THF and glycine, respectively) feeding the purine biosynthetic pathway. The analysis also revealed a massive increase in the synthesis of pyruvate formate-lyase (PFL), the enzyme which converts pyruvate into acetyl coenzyme A and formate. PFL has been essentially studied for its role in mixed-acid product formation in lactic acid bacteria during anaerobic fermentation. However, formate is an important methyl group donor for anabolic pathway through the formation of folate derivates. We hypothesized that PFL was involved in purine biosynthesis during growth in milk. We showed that PFL expression was regulated at the transcriptional level and that pfl transcription occurred during the exponential growth phase in milk. The complementation of milk with formate or purine bases was shown to reduce pfl expression, to suppress PFL synthesis, and to stimulate growth of S. thermophilus. These results show a novel regulatory mechanism controlling the synthesis of PFL and suggest an unrecognized physiological role for PFL as a formate supplier for anabolic purposes.
The thermophilic bacteria Streptococcus thermophilus is one of the most widely used lactic acid bacteria (LAB) in the dairy fermentation industry for yoghurt and cheese production. In the industrial implementation of S. thermophilus, fast-growing capacity is crucial to enable rapid and intense acidification of milk. Identification of functions activated during growth in milk should help to understand the molecular adaptation of this bacterium to milk and provide the basis for targeted strain selection.
In contrast with other LAB, the only environment from which S. thermophilus has been isolated is milk (38). In line with this restricted ecological niche, lactose, not glucose, is the preferred carbon source for S. thermophilus (31). The capacity to ferment lactose, the main sugar of milk, into lactic acid is essential for growth in milk and depends on a non-phosphotransferase system lactose permease (LacS) and a beta-galactosidase (LacZ) (40). Concerning the capacity of S. thermophilus to fulfill its amino acid needs during growth in milk, the species displays only a few amino acid auxotrophies compared to the model LAB, L. lactis (29, 33). The growth of Lactococcus lactis in milk depends largely on the activity of a cell-wall-bound proteinase (PrtP) that is responsible for casein hydrolysis, the main source of amino acids in milk. The presence of such a cell-wall-bound proteinase is, however, exceptional among S. thermophilus strains (37). This situation is likely explained by the amino acid biosynthetic capacities of S. thermophilus and its frequent association with Lactobacillus delbrueckii subsp. bulgaricus. This latter species possesses a very efficient cell-wall-bound proteinase whose activity is beneficial to both species (14). Several genetic and biochemical studies have focused on the characterization of S. thermophilus cytoplasmic peptidases (for examples, see references 1, 13, 28, 29, 34, 35, and 39), and the genome sequences revealed the presence of more than 20 potential cytoplasmic peptidases in the two strains analyzed (12). However, the role of these enzymes in nitrogen metabolism during growth in milk has not yet been established. More recently, transposon mutagenesis was used to isolate mutants of S. thermophilus deficient for growth in milk (17, 18). The genes inactivated in the mutants were associated with the peptide transport, branched-chain amino acid, and purine base biosynthetic capacities.
In this work, we investigated the adaptation of S. thermophilus to milk by comparing its proteomic profile after growth in synthetic medium or in milk. The analysis of the main variations of the proteome has shown that the physiology S. thermophilus is subjected to important changes during growth in milk that concern both nitrogen and carbon metabolism. A surprising finding was the strong upregulation of the pyruvate formate-lyase (PFL). The key role of this enzyme in the routing of the fermentation pathway prompted us to further study the regulation of its expression. We observed that pfl expression was upregulated in milk and found that formate or purine nucleotide starvation was responsible for pfl transcriptional activation, leading to the increase in the PFL level. This result established a novel link between a key fermentative enzyme and the nutritional resources of the environment.
MATERIALS AND METHODS
Bacterial strains, cultures, and media.
S. thermophilus LMG18311 (BCCM collection) was isolated from yogurt. Stock cultures were prepared in reconstituted low-heat 10% (wt/vol) Nilac skim milk (Nilac low-heat milk powder, NIZO, Ede, The Netherlands) that was autoclaved at 110°C for 12 min. At a pH value of 5.4 to 5.6, milk cultures were shock frozen in liquid nitrogen and maintained at −80°C. For M17-Lac cultures, precultures were inoculated with a colony isolated on M17 agar. Four other strains of S. thermophilus, LMD9, CNRZ385, CNRZ1585, and ATCC 19258, were used to generalize some of the results found with LMG18311. Like the majority of S. thermophilus strains, strain LM18311, chosen for the proteomic analysis, does not express a cell wall proteinase (PrtS−). For proteomic and transcript analysis, S. thermophilus was grown at 42°C either in M17 broth (Difco, Sparks, MD) supplemented with 1% (wt/vol) lactose (M17-Lac) or in skim Marguerite milk (La Laiterie, Villefranche sur Saône, France). Marguerite milk is a commercial milk sterilized by microfiltration. Before use, it was skimmed by centrifugation at 4°C at 5,000 × g for 30 min. The sterilization of milk by microfiltration avoids the chemical modifications and proteolysis induced by classical heat treatment. All cultures were performed by inoculating M17-Lac or milk with approximately 5 × 106 CFU/ml of stock cultures and incubating it without shaking. The pH of the culture was monitored. After disruption of cell chains by a 40-s treatment with a mechanical blender (Turax X620; Labo-Moderne, France) bacterial CFU were determined by plating dilutions on M17-Lac agar with an automatic spiral plater (AES Laboratoires, Combourg, France). The cell number was measured with the EC1 colony counter software (AES Laboratoires) after overnight incubation in anaerobic jars (Anaerocult A; Merck, Darmstadt, Germany). To study the effects of formate and purine on growth, milk was supplemented with 10 mM sodium-formate or 50 μg/ml of adenine and guanine.
Cytoplasmic protein extract preparation for proteome analysis.
The cellular protein extracts were prepared from 400-ml cultures. Before centrifugation, a 0.33 volume of 1 M trisodium citrate and a 0.13 volume of buffered saline solution (0.145 M sodium chloride, 0.016 M sodium β-glycerophosphate, 0.1% Tween 80, pH 7) were added to cultures. This procedure, described by Guimont et al. (23), avoids the precipitation of caseins at the time of centrifugation. Cells were harvested by centrifugation (8,000 × g for 10 min at 4°C), and the cell pellet was washed twice with ice-cold extraction buffer, pH 7.0 (5 mM sodium phosphate, 1 mM EDTA, 2 mM β-mercaptoethanol), and resuspended in 20 mM Na phosphate buffer, pH 6.4. Protease Inhibitor Cocktail diluted 20 times (Sigma-Aldrich, St Louis, Mo.), 40 U/ml catalase (Sigma), and 10 mM tributylphosphine (Applied Biosystems) were added. The cells were mechanically disrupted with an FP120 FastPrep cell disruptor (Bio 101 Systems, Qbiogen, Inc., Irvine, CA) by 2 30-s cycles of homogenization at maximum speed with 1-min intervals in ice. The suspension was centrifuged at 5,000 × g for 15 min at 4°C to remove unbroken cells and large cellular debris. The supernatant was next collected and ultracentrifuged at 50,000 × g for 30 min at 4°C to remove cell envelope components. The total protein concentration was determined by the Coomassie protein assay reagent (Pierce, Rockford, Ill.) using bovine serum albumin as a standard.
Analytical 2-DE.
The procedures were essentially performed as described previously (22). Benzonase (1% [vol/vol], 10,000 U/ml; Merck, Darmstadt, Germany) and 1% (vol/vol) 1 M MgSO4 were added to a volume of cytosolic fraction corresponding to 50 μg (7-cm two-dimensional gel electrophoresis [2-DE]) or 350 μg (24-cm 2-DE) of protein. The reaction mixture was incubated at 37°C for 30 min and precipitated with 3 volumes of ice-cold methanol. The protein pellet was solubilized in 155 or 500 μl of isoelectric focusing buffer {7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 100 mM dithiothreitol, 0.5% pH 4 to 7 immobilized pH gradient (IPG) buffer (Amersham-Pharmacia Biotech, Uppsala, Sweden)} and loaded onto a 7-cm or 24-cm pH 4 to 7 IPG strip (Bio-Rad, Hercules, CA) using the in-gel rehydration technique (32).
Isoelectric focusing (IEF) of the 7-cm IPG strip was carried out at 15°C using the Protean II IEF cell (Bio-Rad). For 7-cm strips, IEF was performed for 30 min at 50- to 175-V ramp, 45 min at 175- to 2,000-V ramp, and 30 min at 2,000 V (total = 1.868 kVh). Second-dimension electrophoresis was performed on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis using precast 4 to 12% Bis-Tris ZOOM gel (Invitrogen, Carlsbad, CA). For the 24-cm strip, IEF was performed with the Ettan IPGphor system (Amersham Pharmacia Biotech, Uppsala, Sweden) with the following conditions: 11 h at 50 V, 50 to 300 V in 1 h, 300 to 1,500 V in 1 h, 1,500 to 8,000 V in 5 h, 5 h at 8,000 V (total = 68 kVh). After IEF was completed, the IPG strip was positioned onto SDS-polyacrylamide gel electrophoresis gels using 1% low-melting-point agarose in 150 mM Tris-HCl, pH 8.8. The second dimension was performed on 12% polyacrylamide gels (24 by 20 by 0.1 cm) in 25 mM Tris, 192 mM glycine, 0.1% SDS, pH 8.3, using the Ettan-Dalt II apparatus. Electrophoresis was run at 1 W/gel for 16 h at 4°C. The gels were stained with BioSafe colloidal Coomassie blue (Bio-Rad) for 1 h and destained with three successive washes in deionized water.
Gels were digitized using an Epson Expression 1640XL scanner controlled by the Silver Fast software. Image files were recorded at 256 gray levels. Images of 2-DE were edited and matched using the Image Master 4.01 software package (Amersham-Pharmacia).
The comparative analysis was performed by analyzing images from three independent cultures. The normalized intensity (NI) of each spot was calculated as the ratio between its intensity and the sum of the intensities of all spots in the gel. The mean intensity values of each spot were calculated on at least three gels and used to calculate the ratio (relative [n-fold] change) of expression level of the corresponding protein between the two experimental conditions (M17 versus milk). A protein was included in the list of up- or downregulated proteins with the following criteria: (i) it was present in the three duplicate biological experiments, (ii) a minimum of a twofold change in NI was observed between the two conditions. The average NI for the three independent experiments was used to calculate the ratios.
MALDI-TOF (mass spectrometry) and database searching.
Mass spectrometry analyses were performed using a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) Voyager-DE-STR mass spectrometer (Applied Biosystems), operated in positive-ion reflector mode. Protein spots of interest were cut out and digested with porcine trypsin (Promega, Madison, Wis.) as described previously (22). Peptide mixtures were analyzed using an α-cyano-4-hydroxycinnamic acid matrix prepared at 4 mg/ml in 50% acetonitrile containing 0.1% trifluoroacetic acid. The trypsin autolytic peptides (842.5 and 2,211.1 Da) were used as internal calibrants. Peptides were selected in the mass range of 840 to 4,000 Da. A local copy of the MS-FIT program, developed by the University of California at San Francisco, was used to search the S. thermophilus proteome database deduced from the genome sequence (12). Search parameters were as follows: monoisotopic masses, maximum allowed peptide mass error of 30 ppm, consideration of one incomplete cleavage per peptide, and carbamidomethylation of cysteines and oxidation of methionines. To assign an identification we used the following criteria: (i) a minimum of four matching peptides, (ii) a sequence coverage of at least 20%, (iii) conformity between theoretical and experimental PM and pI. The 2-DE images and information on the protein spots can be obtained by downloading the PARIS software http://www.inra.fr/bia/J/imaste/paris/ (41).
Northern blot analysis of pfl mRNA.
S. thermophilus total RNA was prepared from 200-ml cultures. Before harvesting the cells, a 0.33 volume of 1 M trisodium citrate and a 0.13 volume of buffered saline solution (0.145 M sodium chloride, 0.016 M sodium β-glycerophosphate, 0.1% Tween 80, pH 7) were added to the media (23). Cells were collected by centrifugation for 5 min at 4°C at 10,000 × g, washed with 100 ml ice-cold extraction buffer, pH 7.0 (5 mM sodium phosphate, 1 mM EDTA, 2 mM β-mercaptoethanol), shock frozen, and stored at −80°C. Bacterial pellets were resuspended in 500 μl of 25 mM Tris-HCl (pH 7.6) with 60 mM EDTA and transferred into tubes containing 500 μl of acid phenol (pH 4.5) and 0.5 g of 0.1-mm-diameter glass beads (Sigma). Cells were mechanically broken with a Fastprep apparatus (2 30-s cycles of homogenization at maximum speed with 1-min intervals on ice). RNA was then extracted and purified with the TRIzol reagent kit (Gibco-BRL) as previously described (15). RNA samples were resuspended in 100 μl of TE (10 mM Tris, 1 mM EDTA, pH 7.6) and quantified by measuring A260 and A280. Both RNA purity and integrity were controlled by separating a sample on agarose gel to ensure that mRNA, tRNA, and rRNA precursors could be seen. For Northern blot analysis, 20 μg of glyoxylated RNA samples were separated by electrophoresis in 1% (wt/vol) agarose gel containing 10 mM sodium iodoacetic acid in 10 mM sodium phosphate buffer (pH 7) (15). RNA was transferred to Hybond N nylon membranes (Amersham) and fixed by UV treatment. Blots were stained with bromophenol blue to ensure that equivalent amounts of total RNA were loaded in each sample. RNA extraction and hybridization experiments were carried out on two independent blots with similar results. A 690-bp PCR-amplified pfl gene probe obtained with primers pfl1 (5′-GAACGTGACCTTGCTCGTGG-3′) and pfl3 (5′-GCCATTTGAACNGCTTCRTAG-3′) was used for hybridization. The probe was labeled with the ECL Direct Nucleic Acid Labeling and Detection systems (Amersham Biosciences, Little Chalfont, United Kingdom) according to the manufacturer's procedures.
Measurements of formic acid.
Skim milk cultures of S. thermophilus LMG18311 were performed as described above. Cells were harvested at different times after inoculation, and deproteinization of cultures supernatants was performed with trichloroacetic acid at a final concentration of 1.2% (vol/vol). Samples were allowed to stand for 10 min on ice and centrifuged at 5,000 × g for 10 min at 4°C. Supernatants were then filtrated on Centriplus filter devices (YM-30,000 MWCO) (Millipore Corporation) according to the manufacturer's recommendations. Immediately prior to formate dosage, neutralization of filtrates to a final pH of 7 was carried out with 1 M NaOH and samples were briefly centrifuged. Formate was measured enzymatically by using formate dehydrogenase with the Test combination kit for determination of formic acid (r-Biopharm, Saint Didier au Mont d'Or, France) according to the manufacturer's protocol and using a known formic acid dilution as the standard.
RESULTS
Proteome profiles of S. thermophilus LMG18311 grown in M17 and milk.
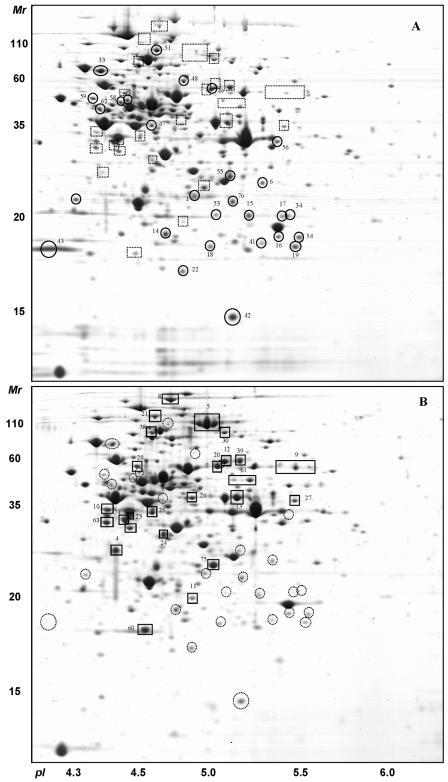
The proteome of S. thermophilus has been studied on protein extracts from cells cultivated in M17-Lac and microfiltrated skim milk. For the proteome analysis, bacteria were harvested during the exponential growth phase at a cell density corresponding to 5.0 × 107 CFU/ml in milk (pH 6.35) and 7.5 × 107 CFU/ml (pH 6.7) for M17-Lac. Cytoplasmic proteins were separated by 2-DE in the pH range 4 to 7 (Fig. 1). More than 350 protein spots were detected on this gel system, and 150 unique proteins were identified by peptide mass fingerprinting using MALDI-TOF mass spectrometry. We found that the glycolytic enzymes represent almost 20% of the total spot volume detected on pH 4 to 7 2-DE. This property of the cytoplasmic acidic proteome of S. thermophilus is reminiscent of that found for L. lactis (22); it thus seems that this is a common property of the proteome of the lactic acid bacteria, for which the central metabolism is based on homolactic fermentation. All modifications observed in protein level between the two media are summarized in Table 1.
FIG. 1.
2-DE (pH range, 4 to 7) of intracellular extracts of S. thermophilus LMG18311 after growth in M17-Lac (A) or milk (B). Proteins (350 μg) were loaded on an IPG strip and stained with colloidal Coomassie blue. The proteins differentially expressed are indicated by circles (higher level in M17) or squares (higher level in milk).
TABLE 1.
List of identified proteins of S. thermophilus LMG18311 varying between M17 and milka
| Level in milk and spot no. | Gene | pIth | Mrth (103) | pIexp | Mr exp (103) | Sequence coverage (%) | Accession no. | Fold change | Protein identification |
|---|---|---|---|---|---|---|---|---|---|
| Higher level in milk | |||||||||
| Carbohydrate metabolism | |||||||||
| 4 | tpi | 4.7 | 26.6 | 4.7 | 32.2 | 71 | YP_139013.1 | 2.3 | Triose phosphate isomerase |
| 5 | pfl | 5.2 | 87 | 5.1 | 78.5 | 53 | YP_140075 | >100 | Pyruvate formate-lyase |
| Nucleotide biosynthesis | |||||||||
| 13 | purB | 5.2 | 49.5 | 5.2 | 45.6 | 77 | YP_138590.1 | 4.5 | Adenylosuccinate lyase |
| 11 | purC | 5.0 | 27 | 5.0 | 24.7 | 69 | YP_138575.1 | 4.4 | SAICAR synthetase |
| 10 | purD | 4.7 | 45.1 | 4.7 | 41.9 | 30 | YP_138585.1 | 5.9 | GAR synthetase |
| 9 | purF | 5.4 | 56.5 | 5.5 | 52.6 | 49 | YP_138577.1 | 10 | PRPP amidotransferase |
| 12 | purH | 5.4 | 58.5 | 5.2 | 58.8 | 48 | YP_138580.1 | 5.5 | IMP cyclohydrolase |
| 8 | purL | 5.0 | 136 | 4.9 | 94.8 | 50 | YP_138576 | 5.6 | FGAM synthetase |
| 74 | purM | 4.7 | 36.3 | 4.7 | 39.1 | 31 | YP_138578.1 | >30 | AIR synthetase |
| Amino acids biosynthesis | |||||||||
| 25 | aspC3 | 4.9 | 42.8 | 4.9 | 41.4 | 43 | YP_138988.1 | 3.5 | Aspartate aminotransferase |
| 24 | bcaT | 4.9 | 37.5 | 4.9 | 35.7 | 27 | YP_139111.1 | 3.7 | Branched-chain amino acid aminotransferase |
| 26 | cysD | 5.0 | 47.5 | 5.0 | 45.4 | 70 | YP_139462.1 | 3.2 | O-acetylhomoserine sulfhydrylase |
| 75 | cysM1 | 5.2 | 32.3 | 5.1 | 30.1 | 25 | YP_138901.1 | 2.7 | Cysteine synthase serine |
| 27 | glyA | 5.4 | 44.6 | 5.5 | 45.1 | 55 | YP_139261.1 | 2 | Hydroxymethyltransferase |
| 23 | ilvC | 4.7 | 37.4 | 4.7 | 40 | 49 | YP_140285.1 | >10 | Ketol-acid reductoisomerase |
| 28 | thrC | 4.8 | 53.9 | 4.8 | 56.9 | 30 | YP_140292.1 | 2 | Threonine synthase |
| Translation | |||||||||
| 39 | lysS | 5.3 | 58.2 | 5.2 | 59.2 | 37 | YP_139201.1 | 2.7 | Lysyl-tRNA synthetase |
| 38 | metS | 4.8 | 76.2 | 4.8 | 73.2 | 51 | YP_138976.1 | 4.1 | Methionyl-tRNA synthetase |
| Miscellaneous | |||||||||
| 20 | fhs | 5.2 | 59.8 | 5.1 | 57 | 47 | YP_139285.1 | 8.3 | Formate-tetrahydrofolate ligase |
| 30 | pepXP | 5.2 | 85.5 | 5.2 | 73.3 | 46 | YP_140089.1 | 2.5 | X-prolyl dipeptidyl peptidase |
| 63 | ppaC | 4.6 | 33.8 | 4.7 | 38.6 | 66 | YP_138906.1 | 2.3 | Manganese-dependent inorganic pyrophosphatase |
| 60 | sodA | 5.1 | 24.7 | 4.8 | 21.1 | 52 | YP_139228.1 | 3.7 | Superoxide dismutase (Mn) |
| Lower level in milk | |||||||||
| Nucleotide biosynthesis and salvage | |||||||||
| 14 | deoD | 4.9 | 26.2 | 4.9 | 23.4 | 32 | YP_139567.1 | 0.35 | Purine nucleoside phosphorylase |
| 15 | punA | 5.3 | 25.7 | 5.3 | 28.9 | 45 | YP_139571.1 | 0.2 | Purine nucleoside phosphorylase |
| 19 | pyrE | 5.6 | 25.7 | 5.5 | 21.9 | 38 | YP_139445.1 | 0.14 | Orotate phosphoribosyltransferase |
| 18 | pyrF | 5.2 | 25.2 | 5.1 | 22.1 | 40 | YP_139444.1 | 0.23 | Orotidine-5-phosphate decarboxylase |
| 17 | pyrH | 5.6 | 26.3 | 5.4 | 25.6 | 52 | YP_138966.1 | 0.05 | Uridylate kinase |
| 16 | upp | 5.4 | 23 | 5.4 | 23.1 | 53 | YP_138890.1 | 0.38 | Uracil phosphoribosyltransferase |
| Translation | |||||||||
| 49 | argS | 5.1 | 62.9 | 5.1 | 58.1 | 59 | YP_138592.1 | 0.33 | Arginyl-tRNA synthetase |
| 41 | defB | 5.4 | 22.7 | 5.3 | 22.4 | 72 | YP_138691.1 | 0.15 | Polypeptide deformylase |
| 47 | prfA | 4.9 | 40.6 | 4.9 | 44.7 | 51 | YP_139258.1 | 0.46 | Peptide chain release factor 1 |
| 48 | prfC | 5.0 | 58.4 | 5.0 | 61.7 | 34 | YP_139992.1 | 0.04 | Peptide chain release factor 3 |
| 42 | rplJ | 8.6 | 20.8 | 5.2 | 16.8 | 60 | YP_139058.1 | 0.16 | 50S ribosomal protein L10 |
| 43 | rpsB | 5.2 | 28.3 | 4.3 | 31.8 | 57 | YP_138617.1 | 0.01 | 30S ribosomal protein S2 |
| 51 | typA | 4.9 | 68.7 | 4.9 | 78.2 | 38 | YP_139237.1 | 0.09 | GTP-binding protein TypA/BipA (tyrosine phosphorylated protein A) |
| Fatty acid biosynthesis | |||||||||
| 56 | cfa | 5.4 | 45.1 | 5.4 | 40 | 45 | YP_138668.1 | 0.22 | Cyclopropane-fatty-acyl-phospholipid synthase |
| 54 | fabG | 5.6 | 25.7 | 5.5 | 23.1 | 44 | YP_138920.1 | 0.13 | Beta-ketoacyl-ACP reductase |
| 55 | fabK | 5.2 | 33.7 | 5.2 | 32.2 | 48 | YP_138918.1 | 0.12 | trans-2-enoyl-ACP reductase II |
| Cell division protein | |||||||||
| 58 | ftsA | 4.8 | 49.9 | 4.8 | 52.6 | 55 | YP_139242.1 | 0.24 | Cell division protein |
| 59 | ftsZ | 4.6 | 46.5 | 4.7 | 54.2 | 46 | YP_139243 | 0.16 | Cell division protein |
| Miscellaneous | |||||||||
| 62 | nox | 4.7 | 50.1 | 4.7 | 50.1 | 30 | YP_139728.1 | 0.32 | NADH oxidase (H2O forming) |
| 33 | ptsl | 4.7 | 62.9 | 4.7 | 66.6 | 49 | YP_139711.1 | 0.4 | Phosphoenolpyruvate-sugar phosphotransferase system enzyme I |
| 53 | rr01 | 5.0 | 26.6 | 5.2 | 25.8 | 32 | YP_138854.1 | 0.35 | Response regulator (homolog to csrR/covR Spy) |
| 1 | scrK | 5.1 | 32.3 | 5.1 | 28.6 | 24 | YP_140147.1 | 0.31 | Fructokinase |
| 22 | ssbB | 5.0 | 18.7 | 5.0 | 19.8 | 50 | YP_140167.1 | 0.19 | Single-stranded binding protein |
pIth, theoretical isoelectric point; pIexp, experimental isoelectric point.
Among the 23 proteins downregulated in milk, 16 belong to three functional classes: purine and pyrimidine salvage, translation, and fatty acid biosynthesis. While the levels of two peptide chain release factors (PrfA and PrfC) and two acidic ribosomal proteins (L10 and S2) decreased in milk, the amount of the elongation factors EF-Tu, EF-Ts, and EF-G, which are abundant proteins, stayed at similar levels in the two media. The fatty acid biosynthesis group contains two enzymes involved in the elongation of the fatty acid chain (FabG at FabK) and the enzyme cyclopropane fatty acid synthase, which has been shown to be responsible for the in situ synthesis of cyclopropane fatty acids (20).
On the other hand, most of the upregulated proteins in milk were associated with amino acid and purine base metabolism. A major modification of the proteome concerned enzymes involved in the synthesis or supply of amino acids. The amounts of two enzymes, CysD (O-acetylhomoserine sulfhydrylase) and CysM1 (cysteine synthase), were found to increase in milk. These proteins are potentially responsible for cysteine synthesis from acetyl serine. Most of the dairy strains of S. thermophilus have the ability to synthesize branched-chain amino acids (BCAA) (Leu, Ile, Val), and mutants in which ilvB or ilvC genes are inactivated present a strong growth deficit in milk (18, 19). The present proteomic analysis showed that BcaT (BCAA aminotransferase) and IlvC (ketol-acid reductoisomerase) were produced at higher levels during growth in milk. We identified several peptidases by 2-DE (PepC, PepN, PepQ, PepXP), and only one them, the X-prolyl dipeptidyl peptidase PepXP was found to be upregulated during growth in milk. This is in contrast with the situation found for L. lactis, where pepX was shown to belong to the same regulon as pepC and pepN (see Discussion).
The proteome of Streptococcus thermophilus LMG18311 cultivated in milk was also characterized by an increase in the level of seven enzymes involved in the de novo purine biosynthetic pathway. These proteins participate in the biosynthesis of GMP and AMP. In parallel, we observed an increase of Fhs (formyl-tetrahydrofolate synthetase), which is involved in the formation of folic acid derivatives, and of GlyA (serine hydroxymethyl transferase) responsible for the conversion of serine into glycine. Both enzymes are implied in the supply of the methyl group in various anabolic pathways, including purine biosynthesis.
One of the most impressive changes in the proteome pattern concerned PFL. While the enzyme was barely detectable in M17-grown cells, PFL was one of the 10 most abundant proteins in cells cultivated in milk. This intriguing result prompted us to further analyze the role and control of the synthesis of this protein responsible for the conversion of pyruvate into formate and acetyl-coenzyme A.
Transcriptional activation of S. thermophilus pfl gene in milk.
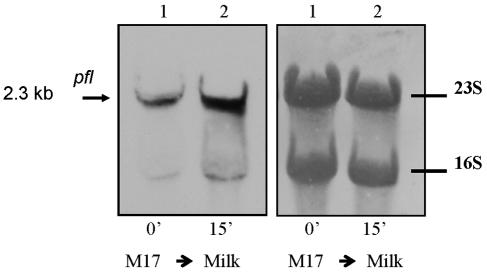
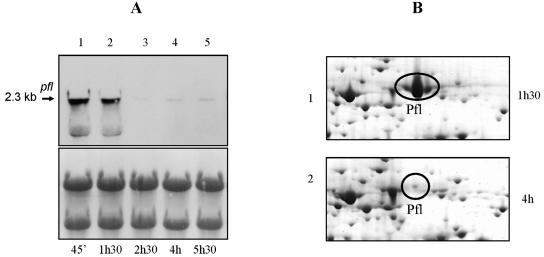
Previous studies performed on the LAB S. bovis and L. lactis have shown that the expression of PFL is controlled at the transcriptional level (3, 5). The expression of the S. thermophilus pfl gene was studied in cells grown in M17-Lac and after transfer to milk. Northern blot analysis with an internal fragment of the pfl gene as a probe allowed us to detect a single transcript of 2.3 kb under both conditions (Fig. 2). The size of the transcript agreed with the length of the transcriptional unit of pfl deduced from the nucleotide sequence (2,309 bp) and shows that pfl gene from S. thermophilus is monocistronic. The presence of a potential stem-loop structure (12.2 Kcal mol−1) in the downstream region of the pfl gene is indicative of a potential rho-independent transcription terminator. Similar organization of the pfl gene has been demonstrated in S. bovis (7) and L. lactis (3). When S. thermophilus cultures grown exponentially in M17-Lac medium were shifted to milk for 15 min, a strong increase in the abundance of the pfl mRNA was observed (Fig. 2). This result indicated that the transcriptional activation of pfl in milk took place rapidly, suggesting that a yet unknown intrinsic property of the medium has triggered this response. We then measured the levels of pfl mRNA during a complete growth cycle in milk by Northern blot analysis (Fig. 3A). The pfl transcript abundance was maximal during the exponential phase (time points, 45 and 90 min) but drastically decreased at the very start of stationary phase (time, 2 h, 30 min and later). The proteome analysis performed in parallel showed that the same trend was observed at the protein level (Fig. 3B).
FIG. 2.
Northern blot of RNA extracted after exponential growth in M17 (1) and after 15-min transfer in milk (2) and hybridized with a pfl probe. The size of the transcript (in kilobases) is shown on the left. On the right, the same membrane is shown stained with bromophenol blue.
FIG. 3.
mRNA and proteomic analyses of pfl of S. thermophilus LMG18311 after growth in milk. (A) Northern blot of RNA extracted during exponential growth (lanes 1 and 2) or stationary phase (lanes 3 to 5) in milk and hybridized with a pfl probe. The size of the transcript (in kilobases) is shown on the left. On the right, the same membrane is shown stained with bromophenol blue. (B) 2-DE of the intracellular proteins of S. thermophilus LMG18311 extracted during exponential growth (1 h, 30 min; lane 1) or stationary phase (4 h; lane 2) in milk. The picture represented a zoom of the Pfl region of a pH 4 to 7 gel.
Formate and purine bases as repressors of S. thermophilus pfl gene expression.
The proteome and transcriptional data suggested that PFL was important for the development of S. thermophilus in milk. Previous studies on S. bovis and L. lactis have shown that activation of PFL lead to excretion of formate in the medium (5, 7, 9, 25, 26). To determine if the increase in S. thermophilus PFL was followed by secretion of formate in the medium, formate concentration was measured in the supernatant of milk cultures at several growth times (see Materials and Methods). The concentration was quite constant during the growth and did not significantly differ from that of noninoculated milk (about 4.3 mg/liter) (see Discussion). This result indicated either that the increase in PFL was not associated with a higher formate production or that the formate produced was not excreted by the cells.
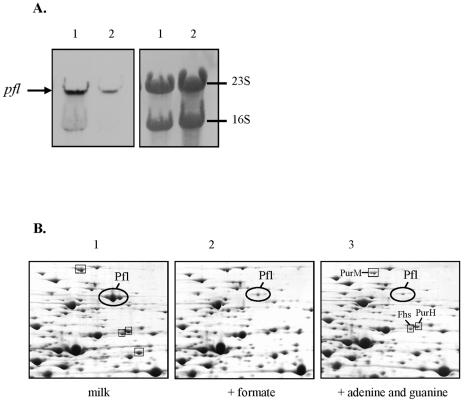
The proteomic results showed the activation of de novo purine biosynthesis in milk. Because this pathway requires formate as a 1-carbon unit, we hypothesized that the upregulation of PFL synthesis was driven by the cell formate needs. In agreement with this hypothesis, we next showed that formate could act as a repressor of PFL synthesis. Transcriptional and proteome analysis of S. thermophilus LMG18311 grown in milk supplemented with formate (5 mM) or adenine plus guanine (50 μM each) were performed (Fig. 4). When formate was supplied to milk, the pfl mRNA abundance sharply decreased (Fig. 4A) and the amount of PFL became almost undetectable (Fig. 4B). When purines, but not formate, were added to milk, a similar strong reduction in the level of PFL was observed (Fig. 4B). In addition, the amounts of PurM, PurH, and Fhs proteins were also strongly diminished in the presence of purines but not formate. This result strongly suggested that PFL synthesis and the purine biosynthetic pathway are linked in S. thermophilus.
FIG. 4.
mRNA and proteomic analyses of pfl of S. thermophilus LMG18311 after growth in milk supplemented or not with formate or adenine plus guanine. (A) Northern blot of RNA extracted from S. thermophilus LMG18311 after exponential growth in milk supplemented (lane 2) or not (lane 1) with 5 mM formate. The arrow indicates the 2.3-kb pfl transcript. On the right, the corresponding blots of extracted RNA after bromophenol blue coloration are presented. (B) 2-DE of the intracellular proteins of S. thermophilus LMG18311 extracted during exponential growth in milk in the presence (lane 2) or not (lane 1) of formate (5 mM) or adenine plus guanine (50 μM) (lane 3). The picture represented a zoom of the Pfl region of a pH 4 to 7 gel.
Formate supply improves S. thermophilus growth in milk.
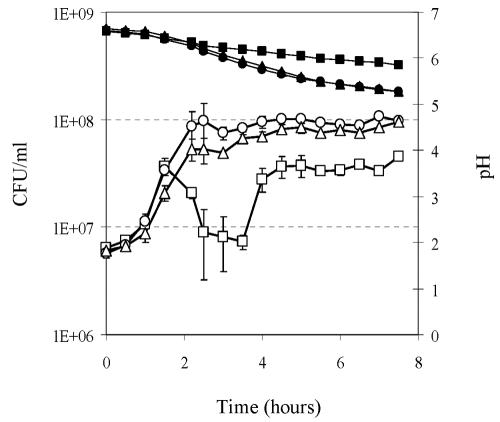
Interestingly, it was also noticed during formate and purine supplementation experiments that the addition of both compounds to milk enhanced the growth of S. thermophilus and acidification of the medium (Fig. 5). While similar exponential growth rates were monitored for all cultures, the final biomass reached by the LMG18311 strain was about threefold higher in supplemented media than in milk. This growth-stimulating effect of formate in milk, which was also observed for other S. thermophilus strains (see Materials and Methods; data not shown), suggested that the supply of formate might be a limiting step for the metabolism of S. thermophilus growing in milk. Curiously, we observed for strain LMG18311 a transient cell susceptibility to the mechanical treatments used for cell chain disruption before plating. This was observed at the end of the exponential growth in milk (lasting between 2 and 4 h after milk inoculation) and was abolished when milk was supplemented with formate or purines (Fig. 5). This peculiar growth characteristic of the strain studied was not further investigated.
FIG. 5.
Growth and acidification curves of S. thermophilus LMG18311 in milk supplemented or not with formate or adenine plus guanine. The evolution of CFU (open symbols) and pH (filled symbols) in milk (squares) and milk supplemented with either 5 mM formate (triangles) or 50 μM adenine plus guanine (circles) is shown.
DISCUSSION
To ensure basic cellular functions and rapid multiplication, bacteria must take into account the physicochemical characteristics of their environment. Milk is known to be poor in purine bases and free amino acids (19, 30). The proteomic data indicate that S. thermophilus adapts to these nutritional limitations by the induction of enzymes implicated in the corresponding biosynthetic and conversion pathways.
First, several proteins involved in the supply of amino acids were more abundant in milk-cultivated cells. Caseins that are the main source of amino acid in milk are rich in prolyl residues (5 to 17%). Therefore, the intracellular degradation of peptides issued from the caseins requires enzymes specifically devoted to the hydrolysis of the prolyl peptidic bond. In milk, we observed an increased level of the peptidase PepX (X-prolyl dipeptidyl aminopeptidase), which is responsible for the production of X-Pro dipeptides from the N terminus of peptides. In LAB, PepX is the only peptidase that efficiently cleaves these sequences (27). In L. lactis, pepX belongs to the CodY regulon, which is activated by a low intracellular level of BCAA (21). In the course of the present study, we identified by 2-DE two other peptidases (PepC, PepN) that also belong to the CodY regulon in Lactococcus lactis. However, and in contrast to L. lactis (M.-Y. Mistou, unpublished), we did not find evidence for a higher synthesis of these peptidases in S. thermophilus grown in milk. As noted above, the expression of these genes is dependent upon the intracellular BCAA concentration. Since L. lactis and S. thermophilus do not have similar requirements for these amino acids, the synthesis of these enzymes is not subjected to the same constraints in the two dairy bacteria cultivated in milk. While L. lactis is auxotrophic, S. thermophilus possesses a functional BCAA biosynthetic pathway (see below). In addition, two enzymes involved in BCAA biosynthesis, IlvC and BcaT, are synthesized at a higher level in milk in S. thermophilus. These results are in good agreement with genetic experiments having shown that inactivation of ilvC led to a reduced-growth phenotype of S. thermophilus in milk (18). Induction of the BCAA biosynthetic pathway in S. thermophilus is thus likely associated with a modification of the transcriptional regulatory mechanisms dependent on the BCAA intracellular concentration. Other enzymes involved in Cys, Met, Asp, and Ser biosynthesis were also more abundant in milk. In this respect, it must be mentioned that the lactococcal AspC, homologous to the streptococcal AspC3 enzymes, was found to have an essential role in the growth of L. lactis in milk (16). This result suggests that aspartate aminotransferase, responsible for the synthesis of aspartate from oxaloacetate also plays a key role in the development of S. thermophilus in milk.
A second hallmark of the proteome profile of S. thermophilus grown in milk is the synthesis of enzymes directly involved in the de novo purine biosynthetic pathway (Table 1). In parallel, glycine synthase (GlyA) and formate-tetrahydrofolate ligase (Fhs), two proteins that are involved in the intracellular production of the methyl group donors glycine and 10-formyltetrahydrofolate, respectively, were also upregulated. Both substrates are feeding the purine biosynthetic pathway. In L. lactis, glyA and fhs have been shown to belong to the PurR regulon that includes proteins upregulated under purine starvation conditions (11). Genes controlled by the transcriptional repressor PurR display a 13-mer sequence called the PurBox located upstream of their promoter. Similar potential Pur boxes (consensus sequence: AWWWCCGAACWWT) were identified upstream of the transcriptional units of purCLFMNH, purDEKB1, and fhs in the strain LMG18311.
Finally, PFL was the strongest overexpressed protein of S. thermophilus LMG18311 during exponential growth in milk. On mini 2-DE, we observed a similar increase of PFL for 4 strains (see Materials and Methods) of S. thermophilus isolated from various dairy products coming from different geographical locations (data not shown). Upregulation of PFL in milk is thus likely common to the S. thermophilus species.
The enzyme catalyzes the nonoxidative transformation of pyruvate into acetyl-coenzyme A and formate. In general, a posttranslational process catalyzed by a PFL-activating enzyme is required to obtain the radical, active form of PFL. In Streptococcus bovis, the enzyme has been characterized previously (8); it is encoded by the act gene, which is also present under the name pflA in the genome of S. thermophilus. It is thus likely that a posttranslational activation mechanism of PFL occurs in S. thermophilus. Further studies will be required to understand how the system functions and is regulated during growth in milk.
The role of PFL in the modulation of the final fermentation products prompted us to further study the control of its expression. Northern blot analysis indicated (i) that PFL synthesis is regulated at the transcriptional level in S. thermophilus, (ii) that the expression of pfl is rapidly induced during exponential growth in milk but restricted to this growth phase, and (iii) that it is strongly reduced in the presence of formate, one of the products of the reaction it catalyzes. The amount of cellular PFL analyzed by 2-DE was in line with the observed changes in mRNA levels, as already observed for S. bovis (4).
The regulation of pfl expression and PFL activity has been intensively studied in S. bovis (4-10) and L. lactis (2, 3, 24-26). In anaerobic conditions and in the presence of slowly fermentable sugars, PFL is responsible for the shift from homolactic to mixed-acid fermentation in L. lactis (3, 25). In S. bovis, it was shown that pfl expression is also dependent on the nature of the fermentable carbon source via a CcpA-dependent mechanism (4, 9). In the strain LMG18311, a catabolite-responsive element (cre box) was detected upstream of pfl (sequence TGTAAGCGGTTACT, at −92 bp upstream of the putative ATG start codon), suggesting that the nature of the carbon source would also modulate pfl expression in S. thermophilus.
In Escherichia coli, the anaerobic regulation of pfl expression is under the control of ArcA and the global regulator FNR (fumarate and nitrate reduction) that controls transcription of genes involved in the adaptation to O2 limiting conditions. An FNR box (or anaerobox), which is present in the promoter of the FNR-regulated genes, has been characterized upstream of pfl in E. coli (36) and L. lactis (3). No obvious FNR regulator or FNR boxes located upstream of the pfl gene were found in the nucleotidic sequence of S. thermophilus LMG18311. Further work would nevertheless be required to conclude about the putative role of oxygen tension in the expression of pfl in S. thermophilus. However, neither the carbon source, the pH, nor aerobic conditions can be considered major determinants of the massive increase in the synthesis of PFL observed during cultivation in milk, since these parameters are similar in the M17-Lac cultures. In milk, the low level of purines suggests that the bacterial ability to synthesize the nucleic acid constituents plays an important role in their capacity to grow at high cell density. In this context, the supply of formate could be a limiting step in the metabolism of LAB growing in milk. Several experimental evidences support this hypothesis: (i) we did not observe an increased production of formate in milk culture supernatants, suggesting that the formate produced by S. thermophilus is metabolized by the bacteria rather than excreted, (ii) when purines were added to milk, not only were de novo purine biosynthesis proteins (PurM, PurH) repressed but also Fhs and PFL, indicating a link between these two proteins and purine biosynthetic pathway, (iii) the growth-stimulating effect observed when formate or purines was added to milk. In conclusion, our results indicated that the major factor controlling pfl expression in S. thermophilus in milk was the simultaneous starvation for formate and purines. Consequently, the regulation of pfl seems to involve a sensing mechanism of the product of the PFL activity. To our knowledge, the control of pfl expression by formate has not been reported previously and raises questions. Is the control of PFL expression by formate observed in S. thermophilus a specific adaptation to milk or does it also exist in nondairy bacteria? The molecular mechanism involved in the regulation of expression of pfl would also deserve attention. Finally, S. thermophilus is grown in association with L. delbrueckii subsp. bulgaricus for the production of yogurt. At the moment, the metabolic interactions that could exist between the two species cocultivated in milk are not well defined. A global approach of the proteome of S. thermophilus growing in milk provided us novel data on its adaptation to this complex medium. Our future studies will focus on dairy lactobacilli and on the interactions between these two species in milk.
Acknowledgments
We are grateful to Juhui Wang and Christophe Caron for the establishment of the PARIS database. We thank Christophe Gitton for technical advice for 2-DE and Véronique Monnet for critical reading of the manuscript.
S.D. was supported by the departement MICA (Microbiologie et Chaîne Alimentaire) of the Institut National de la Recherche Agronomique.
REFERENCES
- 1.Anastasiou, R., M. Papadelli, M. D. Georgalaki, G. Kalantzopoulos, and E. Tsakalidou. 2002. Cloning and sequencing of the gene encoding X-prolyl-dipeptidyl aminopeptidase (PepX) from Streptococcus thermophilus strain ACA-DC 4. J. Appl. Microbiol. 93:52-59. [DOI] [PubMed] [Google Scholar]
- 2.Arnau, J., F. Jorgensen, S. M. Madsen, A. Vrang, and H. Israelsen. 1998. Cloning of the Lactococcus lactis adhE gene, encoding a multifunctional alcohol dehydrogenase, by complementation of a fermentative mutant of Escherichia coli. J. Bacteriol. 180:3049-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnau, J., F. Jorgensen, S. M. Madsen, A. Vrang, and H. Israelsen. 1997. Cloning, expression, and characterization of the Lactococcus lactis pfl gene, encoding pyruvate formate-lyase. J. Bacteriol. 179:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asanuma, N., and T. Hino. 2000. Effects of pH and energy supply on activity and amount of pyruvate formate-lyase in Streptococcus bovis. Appl. Environ. Microbiol. 66:3773-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asanuma, N., and T. Hino. 2002. Molecular characterization and expression of pyruvate formate-lyase-activating enzyme in a ruminal bacterium, Streptococcus bovis. Appl. Environ. Microbiol. 68:3352-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asanuma, N., and T. Hino. 2001. Molecular characterization, enzyme properties and transcriptional regulation of phosphoenolpyruvate carboxykinase and pyruvate kinase in a ruminal bacterium, Selenomonas ruminantium. Microbiology 147:681-690. [DOI] [PubMed] [Google Scholar]
- 7.Asanuma, N., M. Iwamoto, and T. Hino. 1999. Structure and transcriptional regulation of the gene encoding pyruvate formate-lyase of a ruminal bacterium, Streptococcus bovis. Microbiology 145(Pt 1):151-157. [DOI] [PubMed] [Google Scholar]
- 8.Asanuma, N., T. Yoshii, and T. Hino. 2004. Molecular characteristics and transcription of the gene encoding a multifunctional alcohol dehydrogenase in relation to the deactivation of pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis. Arch. Microbiol. 181:122-128. [DOI] [PubMed] [Google Scholar]
- 9.Asanuma, N., T. Yoshii, and T. Hino. 2004. Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis. Appl. Environ. Microbiol. 70:5244-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asanuma, N., T. Yoshii, M. Kikuchi, and T. Hino. 2004. Effects of the overexpression of fructose-1,6-bisphosphate aldolase on fermentation pattern and transcription of the genes encoding lactate dehydrogenase and pyruvate formate-lyase in a ruminal bacterium, Streptococcus bovis. J. Gen. Appl. Microbiol. 50:71-78. [DOI] [PubMed] [Google Scholar]
- 11.Beyer, N. H., P. Roepstorff, K. Hammer, and M. Kilstrup. 2003. Proteome analysis of the purine stimulon from Lactococcus lactis. Proteomics 3:786-797. [DOI] [PubMed] [Google Scholar]
- 12.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapot-Chartier, M. P., F. Rul, M. Nardi, and J. C. Gripon. 1994. Gene cloning and characterization of PepC, a cysteine aminopeptidase from Streptococcus thermophilus, with sequence similarity to the eucaryotic bleomycin hydrolase. Eur. J. Biochem. 224:497-506. [DOI] [PubMed] [Google Scholar]
- 14.Courtin, P., V. Monnet, and F. Rul. 2002. Cell-wall proteinases PrtS and PrtB have a different role in Streptococcus thermophilus/Lactobacillus bulgaricus mixed cultures in milk. Microbiology 148:3413-3421. [DOI] [PubMed] [Google Scholar]
- 15.Derzelle, S., E. Duchaud, F. Kunst, A. Danchin, and P. Bertin. 2002. Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl. Environ. Microbiol. 68:3780-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudley, E., and J. Steele. 2001. Lactococcus lactis LM0230 contains a single aminotransferase involved in aspartate biosynthesis, which is essential for growth in milk. Microbiology 147:215-224. [DOI] [PubMed] [Google Scholar]
- 17.Garault, P., D. Le Bars, C. Besset, and V. Monnet. 2002. Three oligopeptide-binding proteins are involved in the oligopeptide transport of Streptococcus thermophilus. J. Biol. Chem. 277:32-39. [DOI] [PubMed] [Google Scholar]
- 18.Garault, P., C. Letort, V. Juillard, and V. Monnet. 2000. Branched-chain amino acid biosynthesis is essential for optimal growth of Streptococcus thermophilus in milk. Appl. Environ. Microbiol. 66:5128-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garault, P., C. Letort, V. Juillard, and V. Monnet. 2001. La biosynthèse des acides aminés à chaîne branchée et des purines: deux voies essentielles pour une croissance optimale de Streptococcus thermophilus dans le lait. Lait 81:83-90. [Google Scholar]
- 20.Grogan, D. W., and J. E. Cronan, Jr. 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol Rev. 61:429-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227-1239. [DOI] [PubMed] [Google Scholar]
- 22.Guillot, A., C. Gitton, P. Anglade, and M. Y. Mistou. 2003. Proteomic analysis of Lactococcus lactis, a lactic acid bacterium. Proteomics 3:337-354. [DOI] [PubMed] [Google Scholar]
- 23.Guimont, C., M.-A. Chopard, J.-L. Gaillard, and J.-F. Chamba. 2002. Comparative study of the protein composition of three strains of Streptococcus thermophilus grown either in M17 or in milk. Lait 82:645-656. [Google Scholar]
- 24.Melchiorsen, C. R., N. B. Jensen, B. Christensen, K. Vaever Jokumsen, and J. Villadsen. 2001. Dynamics of pyruvate metabolism in Lactococcus lactis. Biotechnol. Bioeng. 74:271-279. [DOI] [PubMed] [Google Scholar]
- 25.Melchiorsen, C. R., K. V. Jokumsen, J. Villadsen, H. Israelsen, and J. Arnau. 2002. The level of pyruvate-formate lyase controls the shift from homolactic to mixed-acid product formation in Lactococcus lactis. Appl. Microbiol. Biotechnol. 58:338-344. [DOI] [PubMed] [Google Scholar]
- 26.Melchiorsen, C. R., K. V. Jokumsen, J. Villadsen, M. G. Johnsen, H. Israelsen, and J. Arnau. 2000. Synthesis and posttranslational regulation of pyruvate formate-lyase in Lactococcus lactis. J. Bacteriol. 182:4783-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mierau, I., E. R. Kunji, K. J. Leenhouts, M. A. Hellendoorn, A. J. Haandrikman, B. Poolman, W. N. Konings, G. Venema, and J. Kok. 1996. Multiple-peptidase mutants of Lactococcus lactis are severely impaired in their ability to grow in milk. J. Bacteriol. 178:2794-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motoshima, H., T. Shiraishi, F. Tsukasaki, and S. Kaminogawa. 2003. Purification, characterization, and gene cloning of lysyl aminoeptidase from Streptococcus thermophilus YRC001. Biosci. Biotechnol. Biochem. 67:772-782. [DOI] [PubMed] [Google Scholar]
- 29.Neviani, E., G. Giraffa, A. Brizzi, and D. Carminati. 1995. Amino acid requirements and peptidase activities of Streptococcus salivarius subsp. thermophilus. J. Appl. Bacteriol. 79:302-307. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson, D., and M. Kilstrup. 1998. Cloning and expression of the Lactococcus lactis purDEK genes, required for growth in milk. Appl. Environ. Microbiol. 64:4321-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poolman, B. 1993. Energy transduction in lactic acid bacteria. FEMS Microbiol. Rev. 12:125-147. [DOI] [PubMed] [Google Scholar]
- 32.Rabilloud, T., C. Valette, and J. J. Lawrence. 1994. Sample application by in-gel rehydration improves the resolution of two-dimensional electrophoresis with immobilized pH gradients in the first dimension. Electrophoresis 15:1552-1558. [DOI] [PubMed] [Google Scholar]
- 33.Reiter, B., and J. D. Oram. 1962. Nutritional studies on cheese starters. Dairy Res. 29:63-77. [Google Scholar]
- 34.Rul, F., and V. Monnet. 1997. Presence of additional peptidases in Streptococcus thermophilus CNRZ 302 compared to Lactococcus lactis. J. Appl. Microbiol. 82:695-704. [DOI] [PubMed] [Google Scholar]
- 35.Rul, F., V. Monnet, and J. C. Gripon. 1994. Purification and characterization of a general aminopeptidase (St-PepN) from Streptococcus salivarius ssp. thermophilus CNRZ 302. J. Dairy Sci. 77:2880-2889. [DOI] [PubMed] [Google Scholar]
- 36.Sawers, G. 1993. Specific transcriptional requirements for positive regulation of the anaerobically inducible pfl operon by ArcA and FNR. Mol. Microbiol. 10:737-747. [DOI] [PubMed] [Google Scholar]
- 37.Shahbal, S., D. Hemme, and M. Desmazeaud. 1991. High-cell wall-associated proteinase activity of some S. thermophilus (H-strain) correlated with a high acidification rate in milk. Lait 71:351-357. [Google Scholar]
- 38.Torriani, S., M. Vescovo, and L. Dicks. 1997. Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus: a review. Ann. Microbiol. Enzimol. 47:29-52. [Google Scholar]
- 39.Tsakalidou, E., R. Anastasiou, K. Papadimitriou, E. Manolopoulou, and G. Kalantzopoulos. 1997. Purification and characterisation of an intracellular X-prolyl-dipeptidyl aminopeptidase from Streptococcus thermophilus ACA-DC 4. J. Biotechnol. 59:203-211. [DOI] [PubMed] [Google Scholar]
- 40.van den Bogaard, P. T., M. Kleerebezem, O. P. Kuipers, and W. M. de Vos. 2000. Control of lactose transport, beta-galactosidase activity, and glycolysis by CcpA in Streptococcus thermophilus: evidence for carbon catabolite repression by a non-phosphoenolpyruvate-dependent phosphotransferase system sugar. J. Bacteriol. 182:5982-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, J., C. Caron, M. Y. Mistou, C. Gitton, and A. Trubuil. 2004. PARIS: a proteomic analysis and resources indexation system. Bioinformatics 20:133-135. [DOI] [PubMed] [Google Scholar]