Abstract
Microbial communities in coastal subsurface sediments are scarcely investigated and have escaped attention so far. But since they are likely to play an important role in biogeochemical cycles, knowledge of their composition and ecological adaptations is important. Microbial communities in tidal sediments were investigated along the geochemical gradients from the surface down to a depth of 5.5 m. Most-probable-number (MPN) series were prepared with a variety of different carbon substrates, each at a low concentration, in combination with different electron acceptors such as iron and manganese oxides. These achieved remarkably high cultivation efficiencies (up to 23% of the total cell counts) along the upper 200 cm. In the deeper sediment layers, MPN counts dropped significantly. Parallel to the liquid enrichment cultures in the MPN series, gradient cultures with embedded sediment subcores were prepared as an additional enrichment approach. In total, 112 pure cultures were isolated; they could be grouped into 53 different operational taxonomic units (OTU). The isolates belonged to the Proteobacteria, “Bacteroidetes,” “Fusobacteria,” Actinobacteria, and “Firmicutes.” Each cultivation approach yielded a specific set of isolates that in general were restricted to this single isolation procedure. Analysis of the enrichment cultures by PCR and denaturing gradient gel electrophoresis revealed an even higher diversity in the primary enrichments that was only partially reflected by the culture collection. The majority of the isolates grew well under anoxic conditions, by fermentation, or by anaerobic respiration with nitrate, sulfate, ferrihydrite, or manganese oxides as electron acceptors.
The Wadden Sea, located on the southern shores of the North Sea, is a highly productive tidal flat ecosystem. It is characterized by a high nutrient input from the land as well as from the open sea. This nutrient supply stimulates intense benthic primary production, fuelling microbial activities in the upper sediment layers. As a result, oxygen is depleted within the uppermost few millimeters of the sediment. While nitrate, iron oxihydroxides, and manganese oxides are important electron acceptors for the degradation of organic matter within the upper anoxic horizons of the sediment, sulfate reduction becomes the predominant anaerobic degradation process in deeper layers.
To date, microbiological investigations dealing with intertidal sediments have generally been restricted to the uppermost 10 to 40 cm (20, 27, 28, 31, 55), the zone of the highest microbial activities. However, in the last decade, the discovery of the deep biosphere within marine sediments and its extension to several hundreds of meters below the surface has indicated that a major part of the microbial biosphere might be present in subsurface sediments (35, 51). Yet only a few investigations have dealt with coastal subsurface sediments, and these have focused on geochemical parameters and biogeochemical activities and have demonstrated microbial activities even at a depth of some meters (7, 23).
Although in most cases cultivation-based methods failed to detect the most abundant members of microbial communities in situ (2, 18, 49), we have chosen a cultivation-based approach, because it offers the opportunity to isolate indigenous microorganisms in pure culture. In turn, microorganisms that are active in situ can be identified using molecular biological methods such as microautoradiography in combination with fluorescence in situ hybridization (12, 30) or stable isotope probing (5, 37). However, these methods cannot reveal the whole spectrum of physiological capacities that are necessary to understand the ecology of a single bacterial species. Hence, for the investigation of microbial adaptations to environmental conditions, pure cultures remain crucial (22).
Intertidal flats on the German North Sea coast were studied down to a depth of 5.5 m below surface in a combined and detailed microbiological (this study) and molecular biological (R. Wilms et al., submitted for publication) approach that was complemented by geochemical characterization of the sediments (E. Freese et al., submitted for publication). In the present study, two different cultivation approaches were chosen for their practicability. A most-probable-number (MPN) approach was used for the quantitative assessment of microbial communities using a defined medium with many different carbon compounds, each at a low concentration, in combination with different electron acceptors targeting different physiological groups. As an alternative enrichment technique, undisturbed sediment sections were placed in substrate gradients and were used for enrichment and isolation of microorganisms as well. This approach should help to avoid the potential substrate shock (46) and minimize disturbance during sampling.
MATERIALS AND METHODS
Study area and sampling.
Sediment samples were taken from tidal flats located close to the island of Spiekeroog in the East Frisian Wadden Sea on the German North Sea coast (19). Two sites were analyzed: Neuharlingersieler Nacken (NSN) (53°43.270′N, 7°43.718′E) and Gröninger Plate (GP) (53°43.638′N, 07°45.960′E). At low tide, 6-m-long aluminum tubes were driven into the sediment using a vibrocorer provided by the Senckenberg Institute (Department for Marine Research, Wilhelmshaven, Germany). After retrieval, the aluminum liners were cut longitudinally, leaving behind a potentially contaminated surface. By use of a sterile spatula, the uppermost few millimeters of the freshly exposed sediment surface were removed. Sediment samples were taken aseptically from the undisturbed sediment underneath using sterile plastic syringes with cut-off tips.
Physical and chemical parameters.
Temperature was measured at the sediment surface and at the bottom of the core immediately after sampling. Concentrations of oxygen in pore water were determined by means of needle electrodes (Microscale Measurements, The Hague, The Netherlands) (38). Sediment density and porosity were determined according to Bak (3). Pore water chloride and sulfate concentrations were measured by ion chromatography with conductivity detection (Sykam, Gilching, Germany) (40).
Total cell counts.
Sediment samples were fixed by the addition of sterile glutaric dialdehyde (filtered with a 0.2-μm-pore-size filter; final concentration, 2%) and were stored at 4°C in the dark. Prior to analysis, sterile-filtered Tween 80 (final concentration, 0.01%) was added to the fixed sediment slurries and the samples were ultrasonicated (3 times, for 10 s each time). An aliquot of the fixed sediment slurry (5 to 10 μl) was diluted 1,000-fold in particle-free sterile PBS buffer (0.9 g NaCl, 15 mM sodium phosphate buffer, pH 7.4), thoroughly shaken, and filtered through a white polycarbonate membrane (pore size, 0.2 μm; diameter, 25 mm; Anodisc 25; Whatman, Maidstone, United Kingdom). Staining with 4′,6′-diamidino-2-phenylindole (DAPI) and counting were performed according to Süß et al. (48).
Growth media.
For preparation of sediment slurries, MPN series, and gradient tubes and for the isolation of pure cultures, an artificial seawater medium was used. The anoxic medium used contained the following components: NaCl (24.3 g · liter−1), MgCl2 · 6H2O (10.0 g · liter−1), CaCl2 · 2H2O (1.5 g · liter−1), KCl (0.66 g · liter−1), Na2SO4 (4.0 g · liter−1), KBr (0.1 g · liter−1), H3BO3 (0.0025 g · liter−1), SrCl2 · 6H2O (0.04 g · liter−1), NH4Cl (0.021 g · liter−1), KH2PO4 (0.0054 g · liter−1), and NaF (0.003 g · liter−1). The medium was supplemented with 1 ml · liter−1 of the trace element solution SL 10 (54) and 0.2 ml · liter−1 of a selenite and tungstate solution (53). After autoclaving, the medium was cooled under an atmosphere of N2-CO2 (80:20, vol/vol). Per liter of medium, 10 ml of a vitamin solution (4) and 30 ml of a CO2-saturated sodium bicarbonate solution (1 mol · liter−1) were added from sterile stocks. Anoxic medium was reduced by addition of a sterile sodium sulfide solution (final concentration, 1.2 mmol · liter−1) and a sterile acid ferrous chloride solution (final concentration, 0.5 mmol · liter−1). If necessary, the pH was adjusted to 7.2 to 7.4 by addition of sterile HCl or Na2CO3.
For oxic incubations, a slightly modified medium was used. Instead of bicarbonate and CO2, the medium was buffered with HEPES (2.38 g · liter−1) and the pH of the oxic medium was adjusted to 7.2 to 7.4 with NaOH prior to autoclaving. After autoclaving, the oxic medium was cooled down under air and supplemented with vitamins and sodium bicarbonate (0.2 g · liter−1) as described above.
For tests on aerobic growth and for the maintenance of pure cultures, a dilute yeast extract-peptone-glucose (YPG) medium was used. It consisted of the HEPES-buffered oxic seawater described above amended with yeast extract (0.03 g · liter−1), peptone (0.06 g · liter−1), sodium dl-lactate (1 mmol · liter−1), glucose (1 mmol · liter−1), vitamins, and sodium bicarbonate (0.2 g · liter−1).
Substrates for MPN series.
As an electron donor and a carbon source for all MPN series, a mixture of monomeric compounds was used. This mixture contained the 20 common l-amino acids, the short-chain fatty acids formate, acetate, propionate, butyrate, valerate, and caproate, the alcohols methanol, ethanol, n-propanol, and n-butanol, and dl-malate, fumarate, succinate, dl-lactate, glycerol, and glucose (0.1 mmol liter−1 each).
For the assessment of different physiological groups, the monomer mix was combined with different electron acceptors. (i) For aerobic microorganisms, the oxic medium was used. (ii) For anaerobic bacteria, including sulfate reducers, the anoxic sulfate-containing standard medium was used. (iii) For fermenting microorganisms, an anoxic but sulfate-free medium was used. (iv) For manganese-reducing bacteria, manganese oxide (5 mM) was added to the anoxic medium. (v) For iron-reducing bacteria, amorphous ferric hydroxide (5 mM) was added to the anoxic medium.
For the preparation of amorphous manganese oxides, 2 g H2O2 (35%) was added to 50 ml of a 1 M MnCl2 solution and the reaction was started by the dropwise addition of 10 ml of 4 M NaOH. The assay mixture was centrifuged (20,000 × g) for 10 min. The supernatant was collected separately and again treated with H2O2 and NaOH. Finally, the particulate manganese oxides were washed twice, resuspended in distilled water to a final volume of 50 ml, and autoclaved. The resulting manganese oxides represented a mixture of MnOOH and MnO2.
Amorphous ferrihydrite was prepared by careful titration of 8.0 g FeCl3 · 6H2O dissolved in 70 ml of water with 4 M NaOH to a final pH of 7.0. The precipitate was washed twice with distilled water and resuspended in distilled water to a final concentration of 400 mmol · liter−1.
Substrates for gradient tubes.
For gradient cultures, two different substrate combinations were used: (i) TCA, a mixture of 20 common l-amino acids (final concentration, 0.25 mM each) and dl-malate, fumarate, succinate, and dl-lactate (final concentration, 2 mM each) and (ii) ALC, a mixture of the short-chain fatty acids formate, acetate, propionate, butyrate, valerate, and caproate (final concentration, 2 mM each) as well as the alcohols methanol, ethanol, n-propanol, and n-butanol (final concentration, 2 mM each). All concentrations were calculated by considering the total end volume of the gradients.
Preparation and incubation of MPN series.
Viable counts were determined by the MPN method. Sediment slurries were prepared immediately after sampling by diluting in anoxic seawater and were used as an inoculum. Subsamples of these slurries were pasteurized (70°C, 15 min) and were used for the determination of viable counts of spores.
MPN series for anaerobic microorganisms were prepared in an anaerobic hood using polypropylene deep-well plates (Beckman, Fullerton, CA) containing 900 μl of medium per well. For the deeper layers, MPN series with three parallels and six dilution steps were prepared (48). For the surface layers, where higher numbers were expected, MPN series with five parallels and eight dilutions were made. As an inoculum, 100 μl of sediment slurry was added to each well of the first dilution and diluted in 10-fold steps. After inoculation, the plates were covered with sterile lids (CAPMAT; Beckman, Fullerton, CA) sealing each well separately. The MPN plates were put into gas-tight plastic bags equipped with a gas-generating and catalyst system for anoxic conditions (Anaerocult C mini; Merck, Darmstadt, Germany).
MPN series for aerobic microorganisms were prepared in microtiter plates (Corning, New York, NY) in a microbiological cabinet. Every well contained 180 μl of medium. MPN series for aerobes were inoculated with 20 μl of sediment slurry. After inoculation, the plates were covered with sterile lids (Corner Notch Lid; Corning, New York, NY) and wrapped with Parafilm to avoid water loss by evaporation.
On each plate four dilution series were left without inoculum as a control. All MPN series were incubated for 12 weeks at 20°C. Growth was checked by epifluorescence microscopy after staining with DAPI. MPN counts were calculated according to De Man (14). Ninety-five percent confidence levels obtained in the present study were in the range of three to five times and one-third to one-fifth of the MPN values.
Preparation of gradient cultures.
We have established enrichment cultures using undisturbed sediment samples embedded in substrate gradients. These offer two main advantages. The microorganisms remain in their habitual surroundings, and due to the diffusion from the bottom of the tube, substrate concentrations increase only slowly. This helps to avoid a substrate shock (46) but still supplies enough substrate to sustain visible growth.
Substrate gradients were prepared as follows. The substrate was placed at the bottom of a glass tube, and 1 ml of 4% agar was added. After cooling and solidifying, this substrate reservoir was overlaid with 6 ml of anoxic substrate-free mineral medium mixed with 3 ml of 4% agar. This agar-solidified medium overlay served as a spacer between the substrate reservoir and the embedded sediment. The spacer was in turn overlaid with 5 ml anoxic substrate-free mineral medium. To this medium 2 ml of 4% agar was added, and the tubes were kept at 50°C. After a sample of sediment (1 cm3, taken by use of syringes with cut-off tips) was added, the tube was immediately placed in ice-cold water in order to minimize a potential heat shock. The headspace of the tubes was flushed with N2-CO2 (80:20, vol/vol), and the tubes were sealed with butyl rubber stoppers.
Isolation of pure cultures.
Pure cultures were isolated from the highest positive dilutions of the MPN series. All subculturing and isolation were performed using the same media. Agar plates were used for aerobes, and deep agar dilution series were used for anaerobes. Subculturing and isolation from gradient cultures were also done in gradient tubes; the agar-solidified top layer was replaced by liquid culture or deep agar dilution series, respectively. The purity of cultures was tested by microscopy and denaturing gradient gel electrophoresis (DGGE) as described by Süß et al. (48).
Physiological characterization.
At least one representative strain of each operational taxonomic unit (OTU) was characterized physiologically. For tests on anaerobic growth, a slightly modified sulfate-free anoxic seawater medium containing resazurin (0.25 mg · liter−1) was used. After autoclaving, the medium was reduced by the addition of sterile sodium dithionite until the redox indicator turned colorless. Fermentative growth was tested with glucose (5 mmol · liter−1), an amino acid mixture (final concentration, 2 mmol · liter−1), and dl-lactate (10 mmol · liter−1) as single substrates, but also with anoxic YPG and a sulfate-free anoxic monomer medium. Tests for anaerobic respiration were performed with either sodium acetate or lactate (10 mmol · liter−1) as an electron donor in combination with one of the following electron acceptors: sulfate (28 mmol · liter−1), nitrate (10 mmol · liter−1), Fe(OH)3 (20 mmol · liter−1), or MnO2 (10 mmol · liter−1). Sulfide formation was checked as described by Widdel (52). Nitrite and ammonium were analyzed photometrically (24). Tests were prepared in an anaerobic cabinet as described for the MPN series. All assays were prepared in duplicate and were incubated at 20°C for at least 2 weeks in the dark. In some cases when the results were inconsistent, experiments were repeated in completely filled screw-cap tubes. Growth was checked by fluorimetry (33a). From each well 200 μl was transferred to a black microplate (Nunc 237108; VWR International, Darmstadt, Germany), and 50 μl of a Sybr Green I (Molecular Probes, Leiden, The Netherlands) solution (2,000-fold dilution in TE buffer [200 mM Tris and 50 mM EDTA, pH 8]) was added. Fluorescence was measured after a 12-h incubation in the dark at 4°C using a fluorescence microplate reader (BMG Labtechnologies, Offenburg, Germany). Growth was scored as positive if at least fivefold higher fluorescence than in substrate-free controls was detected.
Isolation of nucleic acids from pure cultures, PCR, and sequencing.
For DNA extraction, cultures were centrifuged and resuspended in 50 μl sterile distilled water, and 100 μl Tris-HCl (10 mM, pH 8.0) and 100 μl lysozyme (0.8 mg · ml−1) were added. The assay mixtures were incubated at room temperature for 10 min, and after addition of 40 μl of a sodium dodecyl sulfate (SDS) solution (9.6 ml of 20% SDS added to 2.4 ml of 0.5 M sodium acetate [pH 7.5] and 66.4 ml of distilled water) and 60 μl of 3 M sodium acetate, the suspension was incubated for 1 h on ice, followed by three freeze-thaw cycles. During each cycle the suspension was frozen at −80°C for 3 min and immediately boiled for 3 min. DNA was purified from the lysates using phenol-chloroform extraction followed by ethanol precipitation.
Almost-complete 16S rRNA gene fragments were amplified by PCR using oligonucleotide primers 8f (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492r (5′-GGT TAC CTT GTT ACG ACT T-3′). PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany). Sequencing was performed using the DYEnamic Direct Cycle Sequencing kit (Amersham Biosciences, Freiburg, Germany) as described previously (48).
DGGE analysis of sediment and enrichment cultures.
DNA for DGGE analysis was extracted from original sediments, from sediment embedded in gradient tubes, and from liquid MPN cultures. For extraction, 500 mg sediment or 100 μl liquid culture plus 600 μl TE-buffer (10 mM Tris and 1 mM EDTA, pH 7.5) was transferred into 1.7-ml tubes (Sorenson Bioscience Inc., West Salt Lake City, UT) containing 500 mg of 0.1-mm zirconia-silica beads (Biospec Products, Bartlesville, OK) and 500 μl of an SDS mixture. The samples were homogenized twice, for 1 min each time, on a bead beater (Biospec Products, Bartlesville, OK) followed by centrifugation at 15,000 rpm for 15 min at 4°C in a microcentrifuge (Eppendorf, Hamburg, Germany). DNA was purified from the extracts using phenol-chloroform extraction followed by ethanol precipitation. DNA fragments were amplified using primers GC357f and 907r. DGGE was carried out as described by Süß et al. (48) using an INGENYphorU-2 system (Ingeny, Leiden, The Netherlands) and a 6% (wt/vol) polyacrylamide gel containing denaturant gradients of 50 to 70% for separation of PCR products. The gels were stained for 2 h with 1× SybrGold (Molecular Probes, Leiden, The Netherlands) in 1× Tris-acetate-EDTA buffer and washed for 20 min in distilled water prior to UV transillumination. Individual DNA bands were excised from the gel with sterile scalpels, and the DNA was eluted into 50 μl molecular-grade water (Eppendorf, Hamburg, Germany) by incubation at 4°C. For subsequent sequence analysis, PCR products of DGGE bands were purified using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and sequenced as described above.
Phylogenetic analysis.
The partial 16S rRNA sequences of the isolates and the DGGE bands were compared to those in GenBank using the BLAST function (1) and were aligned with each other and the closest relatives using ClustalX (50). Distance matrices were calculated using the dnadist program within the PHYLIP 3.6 package (J. Felsenstein, Department of Genome Science, University of Washington [http://evolution.genetics.washington.edu/phylip.html]) by applying the algorithm of Jukes and Cantor. Isolates with sequence similarities of at least 97% (44) were grouped into a single OTU.
Nucleotide sequence accession numbers.
The sequences obtained in this study are available from EMBL under accession numbers AJ786027 to AJ786110 for the isolates and AJ889123 to AJ889184 for the DGGE bands.
RESULTS
Chemical and physical parameters.
Sediment samples were taken from the two sampling sites NSN and GP in June 2002. Cores were 450 cm (NSN) and 550 cm (GP) long. Sediments at the two sampling sites differed with respect to their bulk parameters. At the NSN site, sand dominated along the upper 160 cm, followed by a porous layer of sand and remains of a buried mussel (Mytilus edulis Linné) bed between depths of 160 and 220 cm (Fig. 1). Downwards, the sediment consisted mainly of mud and clay (grain size, <63 μm). At the GP site, sediments were more homogenously composed with depth and were mainly dominated by a mixture of sand and mud (Freese et al., submitted). On the sampling date, temperatures were 20°C at the sediment surface and around 12°C at the bottom of the core at both sites.
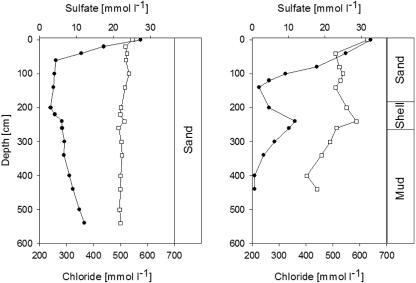
FIG. 1.
Depth profiles of sulfate (•) and chloride (□) and major lithological facies in sediments from the Gröninger Plate (left) and Neuharlingersieler Nacken (right) sites.
Oxygen within the sediments was detected only within the uppermost 3 mm (data not shown). Pore water sulfate concentrations at the Neuharlingersieler Nacken and Gröninger Plate sites ranged between 27 and 32 mM at the sediment surface (Fig. 1) and decreased with depth. Minimum values were detected at 100 (GP) to 140 (NSN) cm below surface. At the NSN site, sulfate concentrations increased again below that level and reached a local maximum within the mussel shell-rich layers at a depth interval from 240 cm to 260 cm (Fig. 1). As indicated by the pore water chloride profiles, low sulfate concentrations were not the result of an influx of groundwater.
Total- and viable-cell counts.
Total cell counts showed only minor differences between the two sites. At the sediment surface, around 1 × 109 cells · g−1 were detected (Table 1). An approximately 10-fold decrease was observed within the uppermost 50 cm of the sediments. Below that level, cell counts decreased less with depth and still reached values of around 1 × 107 to 4 × 107 cells · g−1 at the bottom of the core at both sites.
TABLE 1.
MPN counts obtained with oxic and anoxic media supplemented with different electron acceptors of sediments from the Neuharlingersieler Nacken and Gröninger Plate sites
| Site and sediment depth (cm) | Total cell count (g−1)a | MPN count (g of sediment−1)a obtained by:
|
||||
|---|---|---|---|---|---|---|
| Oxic incubation | Anoxic incubation
|
|||||
| Sulfate free | Sulfate | Iron hydroxide | Manganese oxide | |||
| NSN | ||||||
| 0.5 | 8.06 × 108 | 3.96 × 105 | 3.60 × 106 | 3.60 × 105 | 7.21 × 107 | 7.21 × 107 |
| 5 | 6.52 × 108 | 3.27 × 104 | 4.16 × 104 | 2.97 × 104 | 1.49 × 108 | 5.95 × 107 |
| 50 | 1.93 × 108 | 1.38 × 105 | 1.60 × 104 | 6.07 × 104 | 3.04 × 107 | 3.04 × 107 |
| 100 | 2.50 × 108 | 6.37 × 102 | 3.19 × 105 | 1.27 × 104 | 3.51 × 104 | 6.69 × 104 |
| 200 | ND | 1.23 × 102 | ND | ND | 2.71 × 106 | 1.23 × 105 |
| 300 | 1.53 × 107 | 1.37 × 102 | ND | ND | ND | ND |
| 400 | ND | 8.76 × 101 | ND | ND | 2.63 × 103 | 4.38 × 102 |
| 450 | 6.46 × 107 | 9.19 × 101 | ND | ND | ND | ND |
| GP | ||||||
| 0.5 | 1.39 × 109 | 1.25 × 106 | 5.49 × 104 | 5.49 × 106 | 2.75 × 108 | 4.99 × 106 |
| 5 | 5.85 × 108 | 3.14 × 105 | 2.23 × 102 | 1.71 × 105 | 2.00 × 107 | 2.00 × 105 |
| 50 | 8.27 × 107 | 4.00 × 105 | 1.53 × 104 | 3.64 × 105 | 1.82 × 105 | 7.27 × 105 |
| 100 | 1.27 × 108 | 6.10 × 104 | 3.05 × 104 | 1.04 × 103 | 4.57 × 104 | 4.57 × 105 |
| 200 | ND | 3.32 × 104 | 4.22 × 102 | 8.44 × 103 | 9.04 × 101 | 1.21 × 102 |
| 300 | 6.13 × 107 | 2.29 × 102 | 1.48 × 104 | 8.65 × 102 | ND | ND |
| 400 | ND | 2.33 × 103 | 3.62 × 103 | 5.69 × 103 | ND | ND |
| 500 | 6.46 × 107 | 1.22 × 103 | 3.04 × 104 | 1.03 × 103 | 5.70 × 102 | 1.14 × 103 |
| 550 | 4.65 × 107 | 4.27 × 102 | 2.85 × 103 | 9.69 × 102 | ND | ND |
ND, not determined.
Viable counts were determined by the MPN method. At both sites, the highest counts were achieved at the surface or within a 5-cm depth (Table 1) with medium supplemented with Fe(OH)3 as an electron acceptor. At the NSN site, 1.5 × 108 cells · g−1 were detected within a 5-cm depth, corresponding to 26% of the total cell count. At the GP site, the maximum cultivation success with Fe(OH)3 as an electron acceptor was in a similar range (2.8 × 108 cells · g−1, corresponding to 24% of the total cell count). Except for the layers near the surface at the GP site, MPN counts with manganese oxide as an electron acceptor were in a range similar to those with Fe(OH)3.
Oxic and anoxic media without metal oxides yielded similar MPN counts in the uppermost two sediment layers at the two sites, which were 100- to 1,000-fold lower than those found when metal oxides were used as electron acceptors. Surprisingly, at both sites, MPN counts of aerobes were as high as or higher than those obtained with metal oxide-free anoxic medium.
At the NSN site, the MPN counts showed only minor variations within the upper 50 cm. Beneath that level, the numbers of aerobes declined, while with metal oxides high numbers were still achieved even for the mussel shell-rich layer. However, MPN counts decreased strongly in the mud-and-clay-dominated bottom layers irrespective of the medium used. At the GP site, MPN counts with Fe(OH)3, sulfate, and oxygen showed a steady decrease with depth, while for the sulfate-free anoxic medium, almost no decrease with depth was found. The sulfate-free medium delivered the highest counts for the deepest layers from 300 cm downwards. At a depth of 500 cm, 30,400 cells per g of sediment were still detected.
Contribution of endospores to the viable community.
In order to estimate the potential contribution of inactive endospores to the viable counts, parallel MPN series were inoculated with pasteurized sediment slurries. For surface sediments at the Neuharlingersieler Nacken site, the MPN counts obtained with pasteurized samples were approximately 1% of those obtained with untreated samples (Table 2). Absolute numbers of spores that could be stimulated to grow increased with depth and reached a maximum at a depth of 50 cm, where 1,500 to 5,500 per gram of sediment were retrieved. Beneath that level, MPN counts with untreated and pasteurized slurries were in a similar range (Table 2), but absolute numbers of spores that were stimulated to germinate decreased.
TABLE 2.
MPN counts of spores obtained after inoculation with pasteurized sediment slurry from the Neuharlingersieler Nacken site
| Sediment depth (cm) | MPN count of spores g of sediment−1 (% of MPN counts of spores in untreated samples) after:
|
|
|---|---|---|
| Oxic incubationa | Anoxic incubationa,b | |
| 0.5 | 3,170 (0.8) | 1,080 (0.3) |
| 5 | 1,190 (3.6) | 1,200 (4.0) |
| 50 | 5,520 (4.0) | 1,530 (2.6) |
| 100 | 130 (20.0) | 250 (2.0) |
| 200 | 120 (100.0) | ND |
| 300 | 100 (75.0) | ND |
| 400 | 90 (100.0) | ND |
| 450 | 90 (100.0) | ND |
The sulfate-containing medium was used.
ND, not determined.
Isolation of pure cultures.
To identify the most abundant microorganisms growing in the various media, pure cultures were isolated from the highest positive MPN dilutions or from sediment embedded in gradient tubes. In total, 112 pure cultures were obtained, 64 from the Gröninger Plate site and 48 from the Neuharlingersieler Nacken site (Table 3). About one-third of these isolates were obtained from gradient cultures. The different isolates were affiliated with the major bacterial phyla on the basis of partial 16S rRNA gene sequences and could be grouped into 53 different OTUs within the Alphaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, “Bacteroidetes,” “Fusobacteria,” Actinobacteria, and “Firmicutes.” Of the different OTUs, 28 were represented only by a single strain, while 8 OTUs comprised two strains and 17 comprised three to eight strains (Table 3). This remarkably high diversity of different phylotypes was in part due to the investigation of two separate sampling sites obviously harboring different bacterial communities. For example, Actinobacteria and “Fusobacteria” were retrieved exclusively from the Neuharlingersieler Nacken site, while in turn some of the Bacillus-related strains were obtained only from the Gröninger Plate site.
TABLE 3.
Phylogenetic grouping of pure cultures according to their partial 16S rRNA gene sequences
| Closest relative (% sequence similarity) | No. of isolates obtained from the following:
|
Metabolic capacitiesc
|
Spore formation in stationary-phase cultures | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Origin deptha (cm)
|
Source of isolation
|
||||||||||||||
| MPN seriesb
|
Gradient
|
Aerobic growth | Fermentation | Growth by anaerobic respiration
|
|||||||||||
| 0-5 | 50-100 | 200-550 | Oxic | Anox. | -Sulf. | TCA | ALC | NO3− | Mn4+ | Fe3+ | SO42− | ||||
| Alphaproteobacteria | |||||||||||||||
| Rhizobium rubiT (97) | G1 | 1 | ++ | − | − | − | − | − | − | ||||||
| Paracoccus carotinifaciensT (99) | G1 | 1 | + | Gl | − | − | − | − | − | ||||||
| Roseobacter gallaeciensisT (96) | N1 | 1 | + | − | − | − | − | − | − | ||||||
| Ruegeria atlanticaT (99) | N2 | 2 | + | − | − | − | − | − | − | ||||||
| Roseovarius nubinhibensT (96) | G1 | 1 | + | − | − | − | − | − | − | ||||||
| Sphingobium yanoikuyaeT (98) | G1 | 1 | − | +d | ND | ND | ND | ND | − | ||||||
| Gammaproteobacteria | |||||||||||||||
| Glaciecola pallidulaT (96) | N1 | 1 | ++ | − | − | − | − | − | − | ||||||
| Marinobacter lipolyticusT (98) | N1 | 1 | + | Gl, A | +e | (+) | (+) | − | − | ||||||
| Shewanella affinisT (98) | G3 | 3 | + | Gl, A | − | + | − | − | − | ||||||
| Shewanella violaceaT (97) | N2 | 2 | + | A | +e | + | − | − | − | ||||||
| Halomonas ventosaeT (99) | G1 | 1 | + | Gl, A | +f | − | − | − | − | ||||||
| Psychromonas profundaT (96) | 1 | + | Gl, A | − | − | (+) | − | − | |||||||
| Photobacterium profundumT (97) | G2 | N1 | 1 | 2 | + | Gl, A | +e | − | − | − | − | ||||
| Vibrio splendidusT (98) | G3, N2 | N2 | 7 | + | Gl, A | − | − | − | − | − | |||||
| Deltaproteobacteria | |||||||||||||||
| Desulfuromonas palmitatisT (94) | G1 | 1 | − | ND | ND | ND | ND | ND | − | ||||||
| Malonomonas rubraT (97) | N1 | 1 | − | Gl | +e | + | − | − | − | ||||||
| Desulfovibrio acrylicusT (92) | G1 | 1 | − | − | ND | ND | ND | + | − | ||||||
| Desulfovibrio acrylicusT (99) | G1 | 1 | − | − | +g | + | − | + | − | ||||||
| Desulfovibrio aespoeensisT (94) | G3 | G4 | 2 | 6 | − | − | − | + | + | + | − | ||||
| Desulfovibrio aespoeensisT (92) | N1 | 1 | − | A, L | +g | + | − | + | − | ||||||
| Desulfovibrio zosteraeT (97) | G2, N1 | 2 | 1 | − | A | − | + | − | + | − | |||||
| Desulfovibrio seneziiT (91) | G1 | 1 | − | − | +g | − | + | + | − | ||||||
| Desulfobacterium catecholicumT (96) | G1 | 1 | − | − | − | + | − | + | − | ||||||
| “Fusobacteria,” Ilyobacter tartaricusT (93) | N2 | N1 | 3 | − | A | − | − | − | − | − | |||||
| “Bacteroidetes” | |||||||||||||||
| Salegentibacter salegensT (93) | G1 | 1 | + | Gl, A | +g | (+) | − | − | − | ||||||
| Tenacibaculum mesophilumT (92) | G2 | G1 | 1 | 2 | + | Gl | − | − | − | − | − | ||||
| Cytophaga fermentansT (87) | N2 | 2 | − | Gl | − | − | − | − | − | ||||||
| Actinobacteria | |||||||||||||||
| Cellulomonas biazoteaT (96) | N2 | 2 | + | − | − | − | − | − | − | ||||||
| Citricoccus muralisT (98) | N2 | N1 | 3 | + | − | +f | + | − | − | − | |||||
| Micrococcus luteusT (99) | N1 | 1 | + | − | − | − | − | − | − | ||||||
| “Firmicutes” | |||||||||||||||
| Bacillus simplexT (96) | G4 | 4 | + | − | +e | − | − | − | − | ||||||
| Bacillus hwajinpoensisT (99) | G1 | G3 | G2 | 6 | + | Gl, A | − | − | − | − | + | ||||
| Bacillus aquimarisT (99) | G2 | G1 | 3 | ++ | − | − | − | − | − | + | |||||
| Bacillus cohniiT (98) | N2 | 2 | + | Gl | − | − | − | − | + | ||||||
| Bacillus firmusT (98) | G1 | 1 | + | + | +e | − | − | − | + | ||||||
| Bacillus novalisT (96) | G1 | 1 | ++ | − | − | − | − | − | + | ||||||
| Bacillus infernusT (97) | G1, N1 | 1 | 1 | + | Gl, A | − | − | − | − | + | |||||
| Bacillus simplexT (99) | N1 | G1 | 2 | ++ | − | − | − | − | − | + | |||||
| Halobacillus salinusT (99) | G1 | 1 | ++ | − | − | − | − | − | + | ||||||
| Bacillus haloduransT (94) | G2 | N3 | 5 | ++ | − | − | − | − | − | − | |||||
| Paenibacillus glucanilyticusT (98) | N1 | 1 | + | Gl | ND | ND | ND | ND | + | ||||||
| Clostridium lentocellumT (95) | N2 | 2 | − | + | ND | − | − | − | − | ||||||
| Tepidibacter thalassicusT (96) | G3 | 2 | 1 | − | + | − | − | − | − | + | |||||
| Tepidibacter formicigenesT (95) | G1 | 1 | − | + | − | − | − | − | + | ||||||
| Clostridium novyiT (95) | N1 | 1 | − | + | ND | ND | ND | ND | − | ||||||
| Alkaliphilus transvaalensisT (95) | G1 | 1 | − | + | ND | ND | ND | ND | − | ||||||
| Alkaliphilus crotonatoxidansT (92) | N1 | 1 | − | + | ND | ND | ND | ND | − | ||||||
| Eubacterium angustumT (91) | N1 | N1 | G2 | 1 | 3 | − | Gl, A | − | + | − | − | − | |||
| Desulfosporosinus auripigmentiT (97) | G1, N3 | 2 | 2 | − | L | (+)g | + | + | + | + | |||||
| Sedimentibacter hydroxybenzoicusT (94) | G1 | 1 | + | Gl | +e | (+) | (+) | − | + | ||||||
| Parasporobacterium paucivoransT (93) | G1 | 1 | + | + | ND | ND | ND | ND | + | ||||||
| Staphylococcus equorumT (99) | G1 | G1, N1 | 3 | ++ | − | − | − | − | − | − | |||||
| Staphylococcus succinusT (99) | G1 | N2 | 3 | ++ | − | − | − | − | − | − | |||||
The prefix G before the number of isolates stands for the GP site; the prefix N stands for the NSN site.
Anox., anoxic; -Sulf., anoxic sulfate-free MPN series.
++, strictly aerobic; +, facultatively anaerobic; (+), only weak growth, or growth not present in all strains tested; −, no growth; ND, not determined. Gl, glucose; A, amino acid mix; L, lactate.
Substrate mixture.
Reduction to nitrite.
Reduction to dinitrogen.
Reduction to ammonium.
In general, different phylotypes were isolated from gradient tubes and MPN series. Of the 17 OTUs comprising three strains or more, only 2 (those related to Tenacibaculum mesophilum and to Eubacterium angustum) were retrieved from MPN series as well as from gradient tubes. Sulfate-reducing bacteria were isolated almost exclusively from the gradient tubes.
Even more than by the kind of cultivation approach, the results of the isolation attempts were determined by the redox conditions of the enrichment procedure. Oxic and anoxic MPN series or anoxic gradient cultures never yielded strains belonging to the same OTU. However, this finding is surprising, since a large fraction of strains obtained under anoxic conditions turned out to be only facultatively anaerobic, e.g., Shewanella, Vibrio, or Bacillus strains. On the other hand, two-thirds of the isolates obtained from the oxic MPN series did not grow in any of the anaerobic growth tests at all and are therefore considered to be strictly aerobic (Table 3).
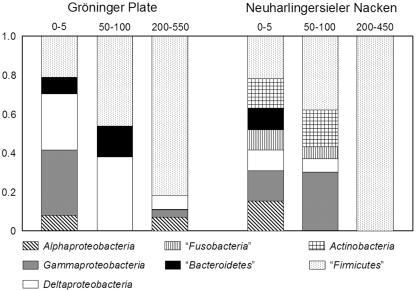
By the culture approach chosen here, a shift of phylogenetic groups along the sediment column was found. A higher number of different phylogenetic groups was retrieved from the surface layers (0.5 to 5 cm) than from the bottom layers below a depth of 100 cm. While predominantly Proteobacteria were obtained from the upper layers, “Firmicutes” dominated the strain collection for the deeper layers (Fig. 2). Only a few OTUs, comprising strains related to the genera Desulfovibrio, Bacillus, Eubacterium, and Staphylococcus, had representatives isolated from the surface as well as from the subsurface layers.
FIG. 2.
Relative contributions of the different phylogenetic groups to the numbers of isolates present in the culture collection obtained from different depths at the Neuharlingersieler Nacken and Gröninger Plate sites. The numbers above the columns indicate depth in centimeters, and the numbers on the left indicate the relative contributions to the culture collection from the respective depths.
Physiological characterization of the pure cultures.
To obtain information on the potential ecological niches of the isolates, physiological experiments were performed. The majority of the OTUs (20 out of 53) were found to comprise strictly anaerobic isolates, in particular those affiliated with the Deltaproteobacteria and the “Fusobacteria.” But among the “Firmicutes” also, several strict anaerobes were found. Facultative anaerobes represented a similar number of phylotypes (17 of 53 OTUs). While most of the strictly aerobic bacteria were isolated from the sediment surface (e.g., some members of the Alphaproteobacteria and Actinobacteria), some were isolated even from the deepest sediment layers. The latter, however, were mostly Bacillus-related strains, most likely present as inactive endospores in situ.
Most of the anaerobic bacteria grew in mineral medium by fermentation of a single substrate (glucose) or a mixture of substrates (amino acids). However, some anaerobes (for example, the Tepidibacter-related strains) did require a more complex medium (monomer or YPG) to grow fermentatively. Nitrate was the electron acceptor most frequently used for anaerobic respiration. In most cases nitrate was reduced to nitrite, less frequently to N2 or ammonium. Some fermenters also grew by iron or manganese reduction. However, strong differences among the strains were observed. While the Shewanella-related strains grew relatively fast on metal oxides, other strains needed at least 2 weeks to achieve slight turbidity and detectable metal reduction (e.g., the Marinobacter- and Psychromonas-related strains). On metal oxides, the sulfate-reducing bacteria generally grew very fast and to high cell densities. However, sulfate-reducing bacteria were scored positive for iron or manganese reduction only if they grew as fast as with sulfate as electron acceptor, in order to exclude growth by sulfur cycling (47).
Molecular comparison of the different enrichment methods.
Molecular analysis of the original sediment samples, the gradient cultures, and the MPN series was performed using PCR and DGGE. The different bacterial types within the different samples were identified by excision and sequencing of the DGGE bands and compared to the pure cultures and the original sediment. The enrichment cultures obtained by the different cultivation approaches revealed a high diversity of microorganisms in total, but also within the single enrichments. In almost all samples, at least five but sometimes as many as nine different bands were detected (Fig. 3). Most of them could be assigned to the Alpha-, Gamma-, or Deltaproteobacteria, the “Firmicutes,” the “Bacteroidetes,” or the Spirochaetes (Table 4). However, several of these sequences (e.g., bands IE2, IIC2, and IIIA7) were only very distantly related to species already known (less than 10% sequence similarity).
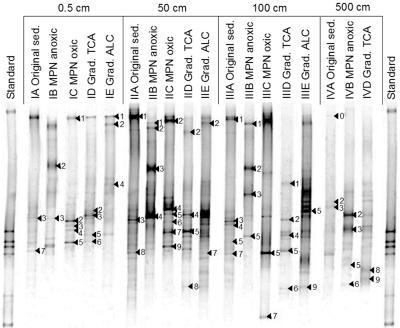
FIG. 3.
Comparison of the different enrichment cultures and the microbial community in situ (original sediments [sed.]) by PCR and DGGE using primers specific to Eubacteria. Enrichment cultures in gradient tubes with carboxylic and amino acids are labeled “Grad. TCA”; those with alcohols and fatty acids are labeled “Grad. ALC.” Bands that were excised for sequence analysis are numbered consecutively, beginning at the upper edge of the gel. Arrowheads indicate only those DGGE bands that gave reliable results after excision and sequence analysis.
TABLE 4.
Phylogenetic affiliations of genotypes detected in the different enrichments inoculated with sediments from the Gröninger Plate site by PCR-DGGE and sequence analysis of DGGE bands and comparison with the original sediment samples
| Depth (cm) | Samplea | Total no. of bands detected/seq.b | Bandc | Closest relative sequence or species in GenBank (% sequence similarity) | Phylogenetic affiliation | Correspondence to other bands |
|---|---|---|---|---|---|---|
| 0.5 | Original | 7/2 | IA3 | Methylomicrobium buryatenseT (90) | Gammaproteobacteria | |
| IA7 | Anaeromyxobacter dehalogenansT (90) | Deltaproteobacteria | ||||
| M anox | 6/2 | IB2 | Alkalibacterium olivapovliticusT (92) | Firmicutes | IIB3 | |
| IB3 | Desulfovibrio aespoeensisT (94) | Deltaproteobacteria | ||||
| M ox | 5/5 | IC1 | Psychroserpens burtonensisT (95) | Bacteroidetes | ||
| IC2 | Ruegeria atlanticaT (96) | Alphaproteobacteria | IC3 | |||
| IC3 | Ruegeria atlanticaT (98) | Alphaproteobacteria | IC2 | |||
| IC4 | Roseovarius toleransT (96) | Alphaproteobacteria | IID5 | |||
| IC5 | Roseovarius toleransT (98) | Alphaproteobacteria | ||||
| GR TCA | 7/4 | ID2 | Sulfitobacter brevisT (97) | Alphaproteobacteria | ||
| ID3 | Alkaliphilus crotonatoxidansT (95) | Firmicutes | ||||
| ID5 | Dethiosulfovibrio acidaminovoransT (92) | Firmicutes | ||||
| ID6 | Uncultured marine alphaproteobacterium BY-95 (97) | Alphaproteobacteria | ||||
| GR ALC | 5/3 | IE1 | Uncultured Cytophagales bacterium, clone Hyd24-40 (97) | Bacteroidetes | ||
| IE2 | Riemerella columbinaT (87) | Bacteroidetes | ||||
| IE4 | Uncultured spirochete clone CD5C5 (97) | Spirochaeta | ||||
| 50 | Original | 9/4 | IIA1 | Chloroplast Thalassiosira eccentrica (99) | ||
| IIA3 | Uncultured gammaproteobacterial isolate, DGGE band GWS-SE-4 (97) | Gammaproteobacteria | ||||
| IIA6 | Desulfonema magnumT (90) | Deltaproteobacteria | ||||
| IIA8 | Anaeromyxobacter dehalogenansT (90) | Deltaproteobacteria | ||||
| M anox | 6/4 | IIB1 | Cytophaga fermentansT (89) | Bacteroidetes | ||
| IIB2 | Alkalibacterium olivapovliticusT (90) | Firmicutes | ||||
| IIB3 | Alkalibacterium olivapovliticusT (92) | Firmicutes | IB2 | |||
| IIB4 | Desulfovibrio aespoeensisT (94) | Deltaproteobacteria | ||||
| M ox | 9/6 | IIC2 | Cytophaga sp. 0sq516 (84) | Bacteroidetes | ||
| IIC4 | Roseobacter gallaeciensisT (96) | Alphaproteobacteria | ||||
| IIC5 | Sulfitobacter mediterraneusT (97) | Alphaproteobacteria | ||||
| IIC6 | Sulfitobacter dubiusT (96) | Alphaproteobacteria | ||||
| IIC7 | Bacillus aquimarisT (96) | Firmicutes | ||||
| IIC9 | Bacillus indicusT (97) | Firmicutes | ||||
| GR TCA | 8/4 | IID2 | Cytophaga fermentansT (89) | Bacteroidetes | ||
| IID4 | Clostridium populetiT (90) | Firmicutes | IIID2 | |||
| IID5 | Roseovarius toleransT (95) | Alphaproteobacteria | IC4 | |||
| IID8 | Thermanaerovibrio acidaminovoransT (89) | Firmicutes | IIID6, IIIE9 | |||
| GR ALC | 9/3 | IIE2 | Desulfovibrio aespoeensisT (94) | Deltaproteobacteria | ||
| IIE7 | Lamprocystis purpureaT (90) | Gammaproteobacteria | ||||
| 100 | Original | 7/5 | IIIA1 | Tenacibaculum mesophilumT (89) | Bacteroidetes | |
| IIIA3 | Thiorhodovibrio winogradskyiT (91) | Gammaproteobacteria | ||||
| IIIA4 | Uncultured gammaproteobacterium GWS-SE-5 (95) | Gammaproteobacteria | ||||
| IIIA5 | Uncultured deltaproteobacterium GWS-SE-6 (87) | Deltaproteobacteria | ||||
| IIIA7 | Anaeromyxobacter dehalogenansT (88) | Deltaproteobacteria | ||||
| M anox | 5/4 | IIIB1 | Cytophaga fermentansT (90) | Bacteroidetes | ||
| IIIB2 | Alkalibacterium olivapovliticusT (92) | Firmicutes | ||||
| IIIB3 | Uncultured bacterium BURTON-31 (95) | Bacteroidetes | ||||
| IIIB5 | Desulfovibrio gabonensisT (95) | Deltaproteobacteria | ||||
| M ox | 7/3 | IIIC5 | Bacillus hwajinpoensisT (97) | Firmicutes | ||
| IIIC7 | Cellulomonas fimiT (95) | Firmicutes | ||||
| GR TCA | 6/5 | IIID1 | Cytophaga fermentansT (89) | Bacteroidetes | ||
| IIID2 | Clostridium populetiT (92) | Firmicutes | IID4 | |||
| IIID4 | Aminobacterium colombienseT (91) | Firmicutes | ||||
| IIID5 | Dethiosulfovibrio acidaminovoransT (89) | Firmicutes | ||||
| IIID6 | Thermanaerovibrio acidaminovoransT (89) | Firmicutes | IID8, IIIE9 | |||
| GR ALC | 9/2 | IIIE5 | Cytophaga fermentansT (92) | Bacteroidetes | ||
| IIIE9 | Thermanaerovibrio acidaminovoransT (89) | Firmicutes | IID8, IIID6 | |||
| 500 | Original | 7/4 | IVA0 | Thalassiosira eccentrica chloroplast (99) | ||
| IVA2 | Uncultured deltaproteobacterium clone PI_6RA04 (82) | Deltaproteobacteria | ||||
| IVA3 | Uncultured bacterium Seq13(S5) (88) | Chloroflexi | ||||
| IVA6 | Thermobacillus xylanilyticusT (94) | Firmicutes | ||||
| M anox | 6/5 | IVB2 | Clostridium homopropionicumT (86) | Firmicutes | ||
| IVB3 | Tepidibacter formicigenesT (94) | Firmicutes | ||||
| IVB5 | Desulfofaba hanseniiT (85) | Deltaproteobacteria | ||||
| IVB6 | Desulfotomaculum sp. strain 175 (89) | Firmicutes | ||||
| M ox | 0/0 | |||||
| GR TCA | 9/2 | IVD8 | Sporotomaculum syntrophicumT (96) | Firmicutes | ||
| IVD9 | Desulfotomaculum gibsoniaeT (95) | Firmicutes | ||||
| GR ALC | 0/0 |
Original, original sediment sample. The following enrichments were investigated: MPN series under anoxic (M anox) or oxic (M ox) conditions as well as gradient tubes supplemented with carboxylic and amino acids (GR TCA) or with n-alcohols and fatty acids (GR ALC).
seq., number of bands that could be unambiguously identified by sequence analysis.
Individual bands are labeled for the origin depth (I to IV), for the sample (A to E), and with a consecutive number (1 to 9) within the lane in the DGGE gel (see Fig. 3).
At the sediment surface, the Alphaproteobacteria were the most frequently detected group in the enrichments. At a depth of 50 cm, they were still present in high numbers, but they were not detected in enrichments from depths of 100 and 500 cm. “Firmicutes” were found at all depths but were absolutely dominant at depths of 100 cm and 500 cm. These results seemed to confirm the composition of the culture collection. However, direct comparison of the sequences derived from the DGGE bands with those of the isolates originating from the same enrichments revealed that only three of the phylotypes identified from the DGGE bands (IIE2, IIC7, IIC5) were obtained in pure culture. These DGGE bands matched isolates that were related to Desulfovibrio aespoeensis, Bacillus aquimaris, and Bacillus hwajinpoensis. In addition, one DGGE band (IIB4) obtained from anoxic MPN series corresponded to Desulfovibrio aespoeensis-related isolates, though none of the isolates was obtained from this particular MPN series. While 7 isolates were detected in the enrichment cultures, the remaining 105 strains were not retrieved in the respective enrichment cultures by PCR and DGGE. At least in some cases, this might be due to the fact that not all DGGE bands could be sequenced. Several bands, in particular from the original sediment sample, gave ambiguous sequences, in most cases due to the fact that several phylotypes were present in a single band and the sequences obtained revealed many ambiguities. However, the results indicate that subculturing during the isolation procedure led to a population shift from the types dominating the primary enrichment to less-dominant but presumably faster-growing types.
Most enrichment cultures were unique in their phylogenetic composition and showed little overlap with enrichments achieved by other methods or by use of different media. With a single exception (bands IIID6 and IIIE9), MPN series and gradient tubes from the same depth did not reveal matching DGGE bands, indicating that every enrichment approach was highly selective. This finding was supported by the fact that in some cases the same phylotype was enriched using the same approach but from two different depths (e.g., bands IID4 and IIID6 or bands IB2 and IIB3). Microscopic investigations of the enrichments revealed high morphological diversity and the presence of several unusually shaped bacteria, as, for example, large spirochetes or chains of vacuolated cells. In parallel to the shake procedure, an attempt was made to isolate some of these types directly by micromanipulation (21) and to transfer single cells into fresh medium. However, none of these mechanically separated cells grew in subculture.
DISCUSSION
This study presents the results of an analysis of microbial communities in intertidal sediments on the German North Sea coast from the surface down to the subsurface at a 550-cm depth. The use of improved media for MPN series resulted in high cultivation efficiency and, in combination with enrichment cultures in gradients, allowed the isolation of a highly diverse culture collection. Since the different media and enrichment strategies were selective for specific bacterial groups, the use of different approaches turned out to be a basic requirement for achieving high diversity.
Improving cultivation efficiency.
In most previous investigations, MPN counts in marine sediments rarely exceeded 0.25% of the total cell count (2, 20, 27, 31). Only in a few cases could larger fractions of microbial communities be cultured from sediments (16, 55) or sediment-like habitats such as rice paddies (8). However, in the present study, consistently high cultivation efficiencies, as high as 23% of the total cell count, were achieved throughout the upper 50 cm of the sediment. One reason might be the use of a medium containing a wide range of different carbon sources, stimulating the growth of different microorganisms. A very similar medium was recently shown to increase cultivation efficiency even in several-thousand-year-old sediments of the Eastern Mediterranean Sea (48).
The highest MPN counts were obtained with oxidized metal species as electron acceptors. It is well documented that in marine sediments manganese or iron reduction can be as important as sulfate reduction (6), whereas little information on the numerical abundance of metal-reducing bacteria in marine sediments is available. Their numbers, estimated as viable counts, can be in a range similar to those reported for sulfate-reducing bacteria (27, 28). However, since a large variety of (fermentable) substrates was present in the medium, not all organisms enriched by this method are necessarily iron or manganese reducers. Interestingly, high numbers of Fe- and Mn-reducing bacteria were detected even in strictly anoxic sediment layers where no Fe(III) and Mn(IV) should be available. However, it is known that even some sulfate reducers, like the isolates presented here, can couple carbon oxidation to iron reduction (9). Some of the isolates presented here grew slowly under anoxic conditions with metal oxides but showed hardly any growth without an external electron acceptor (e.g., the isolates related to Marinobacter and Salegentibacter). These organisms might be active in situ in a syntrophic relationship and can use metal oxides as an unspecific electron sink (32), which would also explain their slow growth. We therefore have to assume that MPN counts of metal-reducing bacteria in anoxic sediment layers comprise a large metabolic variety of strict and facultative anaerobes.
Although oxic and anoxic isolation procedures resulted in completely different phylotypes, several of the anaerobic isolates turned out to be only facultatively anaerobic, e.g., the Shewanella isolates. Since these strains were obtained exclusively under anoxic conditions but were present in numbers similar to those of strains isolated under oxic conditions, as can be inferred from the MPN counts, it seems likely that the immediate exposure to air severely harms these cells. Similarly, a decreased oxygen partial pressure was shown to facilitate the enrichment and isolation of typical aerobic soil bacteria (45). It can be assumed that with the metal oxides a relatively oxidized electron acceptor was offered that circumvented the harmful effects of molecular oxygen (34). It might be advantageous for sediment bacteria to be able to cope with oxygen. Bioturbation can generate oxic microenvironments to depths of as much as 50 cm (29). Furthermore, the upper layers of tidal flats represent a rather unstable ecosystem. During storm events, large patches of sediment can be resuspended. Strict anaerobes, in turn, can be expected to be dependent on reducing conditions. Free pore water sulfide, however, was not detected along the upper 20 cm of the sediment (data not shown), indicating that several centimeters deep in the sediment, anoxic but still quite oxidized conditions prevail.
While high cultivation efficiencies were achieved along the uppermost 50 cm of the sediment, there was a decrease within deeper sediment layers. Oxidative stress as discussed above does not apply for the anoxic medium and therefore seems to be an unlikely cause. Small inoculum sizes, as in our study, have even been shown to increase viable counts (13). Substrate shock caused by high substrate concentrations (42, 46) has often been blamed for poor culturability. In fact, low substrate concentrations have been shown to improve cultivation success (10, 13, 25, 43, 45, 56). Although we used a substrate mixture containing different carbon sources, each of them was present in submillimolar concentrations, and a medium similar to that used in the present study achieved high MPN counts even in Mediterranean sediments up to 100,000 years old (48). Incubation time was shown to have an impact on viable counts as well, since some of the organisms that are abundant in situ might grow only slowly. However, 12 weeks, as in this study, should be long enough, as found for soil microbial communities (13). Molecular analysis of the same sediments (Wilms et al., submitted) revealed that bacteria belonging to a subgroup of the phylum Chloroflexi that is typically found in the marine subsurface (11, 36) were also dominant in the deep layers of the tidal flats investigated here. But since no representative of these presumably typical subsurface bacteria is available as a pure culture, we can only speculate about their nutritional demands.
Relatively low cultivation efficiencies could also be caused by overestimation of the number of potentially viable cells, particularly in deep sediments. DAPI stains living cells, irrespective of the metabolic state. However, it was estimated that the fraction of dead cells that can be stained might represent up to 70% of the total cell counts in marine sediments (33). On the other hand, the use of media with low substrate concentrations can possibly lead to problems with growth detection due to the low level of biomass formed. But even if a bacterium grows on only one of the substrates provided in the monomer mixture in the present study, e.g., lactate, and the growth yield reported for Acetobacterium woodii (41) is considered, theoretically enough biomass is formed for microscopic detection.
Microbial diversity detected by the different enrichment approaches.
In the present study a remarkable high diversity in the culture collection was achieved, outranging similar investigations on deep-sea pelagic environments (40) or sediments (15, 48). During the other studies almost all bacteria that could be cultured by the methods applied were obtained in pure culture as indicated by rarefaction analyses. Obviously, in the present study the number of OTUs detected is at least three times higher, although a similarly high number of isolates was obtained. One reason is likely to be the use of different cultivation and isolation procedures. As discussed above, almost no group was retrieved by more than a single method (oxic or anoxic, MPN or gradient culture). This result supports the view that no single method or medium is suitable to reveal the whole diversity within a single sample. It is likely that an even higher diversity could have been achieved if more medium variations had been used. On the other hand, tidal surface sediments are patchy, e.g., due to bioturbation, and offer a number of different microhabitats and redox horizons suitable for different physiological groups (29). Furthermore, the tidal sediments investigated in the present study revealed layers of different grain sizes such as sand and mud. At least for the deeper layers, the different sedimentological parameters of the two sampling sites might be one major reason for the different composition of the culture collection. As shown also by molecular methods, every sedimentological facies harbors its own specific microbial community (Wilms et al., submitted).
Phylogenetic analysis of the enrichment cultures.
DGGE analysis of the enrichment cultures revealed an even higher diversity than that present in the culture collection. Nevertheless, little correspondence between the enrichment cultures and the natural community was found. One of the main reasons was probably that only some of the bands yielded reliable sequences, in most cases due to the presence of several phylotypes in one band. It was expected that some of the types that are dominant in situ would be retrieved at least in the gradient tubes (26). Obviously, however, even the addition of low substrate concentrations or the application of substrate gradients seemed to stimulate less abundant types in particular. That community shifts can be achieved even by less thorough disturbance was shown by Eilers et al. (17), who found a dominance of easily culturable bacterial genera even after short-term incubation. On the other hand, we do not know which genotypes were present in the ambiguous bands. It can be assumed that at least some of our isolates remained undetected in the natural community or the primary enrichments.
Improved classical approaches such as agar plates have been applied successfully to soil microbial communities and led to the isolation of many yet uncultured groups of bacteria (25, 39). However, a basic requirement for the isolation of pure cultures, as performed in the present study, was the ability of the microorganisms to form colonies in deep agar or on agar plates. It can be imagined that several of the types observed in the enrichment cultures were unable to form colonies (43) and therefore were lost during isolation. The use of micromanipulation of single cells (21) as an alternative isolation strategy was unsuccessful. This might be due to cell damage during handling or to inability of these organisms to grow independently in the absence of syntrophic partners (26). For these organisms, alternative strategies such as dialysis cultures, dilution series, or growth on filters (43) will have to be used in the future.
Microbial communities in the subsurface of a tidal flat.
During the last decade the discovery of the “deep biosphere” has drawn much attention to deep-sea subsurface environments (15, 35, 36, 51), and it was suggested that the majority of prokaryotic life on earth is found in the subsurface (51). However, most of these investigations dealt with very deep layers, and so far it is not clear at which depth the deep biosphere starts. Whitman et al. (51) defined the subsurface in sediment as layers below a depth of 10 cm. This definition might fit sediments underlying oceans with low primary production and sedimentation. As an example, the 8,500-year-old sapropels in the Eastern Mediterranean Sea can be found at shallow depths of 17 to 35 cm below the sea floor (48). Coastal seas, however, show a much higher sedimentation rate. The estimated age of the buried mussel bed at a depth of approximately 200 cm at the Neuharlingersieler Nacken site was about 650 years (J. Rullkötter, personal communication). However, after this duration of burial, it is to be expected that only very recalcitrant organic material is present, and these layers can clearly be seen as a subsurface environment. This is also reflected by the composition of the culture collection. Almost no overlap between the surface layers (0 to 0.5 cm) and the deep layers (200 cm and below) was found (Table 3). This result is supported by a parallel analysis by PCR and DGGE using primers specific to Bacteria (Wilms et al., submitted). The finding that the deep layers (200 cm and below) are dominated by a subgroup of Chloroflexi, typically found in the subsurface, provides a link to the microbial communities within the “deep biosphere” that is present down to some hundred meters below the sea floor (36).
It is less clear whether the sediment layers at depths of 50 and 100 cm can be seen as part of the subsurface. However, these layers are already beneath the zone of bioturbation (29), are characterized by sulfidic conditions, and are unlikely to provide easily degradable organic matter to the microbial communities. Clear differences were found in the bacterial types isolated from these intermediate layers and those from the surface layers, for example, the lack of Alphaproteobacteria in the intermediate layers. However, these differences were less pronounced than the differences between the surface layers on the one hand and the shell and mud layers on the other. For this reason, it is suggested that these layers can be seen as subsurface habitats, but to clearly distinguish them from the deeper layers, the term “shallow subsurface” seems to be appropriate.
Acknowledgments
We thank the master and the crew of the RV Senckenberg, as well as Alexander Bartholomä, Maik Wilsenack, and Elke Tilch, for their help during sampling. We acknowledge Andrea Schlingloff and Jürgen Fröhlich for experimental help.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., W. Ludwig, and K. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bak, F. 1988. Ph.D. thesis. Sulfatreduzierende Bakterien und ihre Aktivität im Litoralsediment der Unteren Güll (Überlinger See). University of Constance, Constance, Germany.
- 4.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Rolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. de Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 6.Canfield, D. E., B. Thamdrup, and J. W. Hansen. 1993. The anaerobic degradation of organic matter in Danish coastal sediments: iron reduction, manganese reduction, and sulfate reduction. Geochim. Cosmochim. Acta 57:3867-3883. [DOI] [PubMed] [Google Scholar]
- 7.Chambers, R. M., J. T. Hollibaugh, C. S. Snively, and J. N. Plant. 2000. Iron, sulfur, and carbon diagenesis in sediments of Tomales Bay, California. Estuaries 23:1-9. [Google Scholar]
- 8.Chin, K. J., D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen. 1999. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl. Environ. Microbiol. 65:5042-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman, M. L., D. B. Hedrick, D. R. Lovley, D. C. White, and K. Pye. 1993. Reduction of Fe(III) in sediments by sulphate-reducing bacteria. Nature 361:436-438. [Google Scholar]
- 10.Connon, S. A., and J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coolen, M. J. L., H. Cypionka, A. M. Sass, H. Sass, and J. Overmann. 2002. Ongoing modification of Mediterranean Pleistocene sapropels mediated by prokaryotes. Science 296:2407-2410. [DOI] [PubMed] [Google Scholar]
- 12.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine Proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, E. R., S. J. Joseph, and P. H. Janssen. 2005. Effect of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Man, J. C. 1977. MPN tables for more than one test. Eur. J. Appl. Microbiol. 4:307-316. [Google Scholar]
- 15.D'Hondt, S., B. B. Jørgensen, D. J. Miller, A. Batzke, R. Blake, B. A. Cragg, H. Cypionka, G. R. Dickens, T. Ferdelman, K.-U. Hinrichs, N. G. Holm, R. Mitterer, A. Spivack, G. Wang, B. Benkins, B. Engelen, K. Ford, G. Gettemy, S. D. Rutherford, H. Sass, C. G. Skilbeck, I. W. Aiello, G. Guérin, C. H. House, F. Inagaki, P. Meiser, T. Naehr, S. Niitsuma, R. J. Parkes, A. Schippers, D. C. Smith, A. Teske, J. Wiegel, C. N. Padilla, and J. L. S. Acosta,. 2004. Distributions of microbial activities in deep subseafloor sediments. Science 206:2216-2221. [DOI] [PubMed] [Google Scholar]
- 16.Drake, H. L., N. G. Aumen, C. Kuhner, C. Wagner, A. Grießhammer, and M. Schmittroth. 1996. Anaerobic microflora of Everglades sediments: effects of nutrients on population profiles and activities. Appl. Environ. Microbiol. 62:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eilers, H., J. Pernthaler, and R. Amann. 2000. Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66:4634-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felske, A., A. Wolterink, R. Van Lis, W. M. de Vos, and A. D. L. Akkermans. 1999. Searching for predominant soil bacteria: 16S rDNA cloning versus strain cultivation. FEMS Microbiol. Ecol. 30:137-145. [DOI] [PubMed] [Google Scholar]
- 19.Flemming, B. W., and R. A. Davis, Jr. 1994. Holocene evolution, morphodynamics and sedimentology of the Spiekeroog Barrier island system (Southern North Sea). Senckenbergiana Maritima 24:117-155. [Google Scholar]
- 20.Freitag, T. E., T. Klenke, W. E. Krumbein, G. Gerdes, and J. I. Prosser. 2003. Effect of anoxia and high sulphide concentrations on heterotrophic microbial communities in reduced surface sediments (black spots) in sandy intertidal flats of the German Wadden Sea. FEMS Microbiol. Ecol. 44:291-301. [DOI] [PubMed] [Google Scholar]
- 21.Fröhlich, J., and H. König. 2000. New techniques for isolation of single prokaryotic cells. FEMS Microbiol. Rev. 24:567-572. [DOI] [PubMed] [Google Scholar]
- 22.Fry, J. C. 2000. Bacterial diversity and “unculturables”. Microbiol. Today 27:186-188. [Google Scholar]
- 23.Goldhaber, M. B., R. C. Aller, J. K. Cochran, J. K. Rosenfeld, C. S. Martens, and R. A. Berner. 1977. Sulfate reduction, diffusion, and bioturbation in Long Island Sound sediments: report of the foam group. Am. J. Sci. 277:193-237. [Google Scholar]
- 24.Grasshoff, K., K. Kremling, and M. Erhard. 1999. Methods of seawater analysis. Wiley-VCH, Weinheim, Germany.
- 25.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2000. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 27.Koretsky, C. M., C. M. Moore, K. L. Lowe, C. Meile, T. J. DiChristina, and P. van Cappellen. 2003. Seasonal oscillation of microbial iron and sulfate reduction in saltmarsh sediments (Sapelo Island, GA, USA). Biogeochemistry 64:179-203. [Google Scholar]
- 28.Kostka, J. E., B. Gribsholt, E. Petrie, D. Dalton, H. Skelton, and E. Kristensen. 2002. The rates and pathways of carbon oxidation in bioturbated saltmarsh sediments. Limnol. Oceanogr. 47:230-240. [Google Scholar]
- 29.Kristensen, E. 2000. Organic matter diagenesis at the oxic/anoxic interface in coastal marine sediments, with emphasis on the role of burrowing animals. Hydrobiologia 426:1-24. [Google Scholar]
- 30.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography: a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Llobet-Brossa, E., R. Rabus, M. E. Böttcher, M. Könneke, N. Finke, A. Schramm, R. L. Meer, S. Grötzschel, R. Roselló-Mora, and R. Amann. 2002. Community structure and activity of sulfate-reducing bacteria in an intertidal surface sediment: a multi-method approach. Aquat. Microb. Ecol. 29:211-226. [Google Scholar]
- 32.Lovley, D. R. 1991. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55:259-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luna, G. M., E. Manini, and R. Danovaro. 2002. Large fraction of dead and inactive bacteria in coastal marine sediments: comparison of protocols for determination and ecological significance. Appl. Environ. Microbiol. 68:3509-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Martens-Habbena, W., and H. Sass. Sensitive determination of microbial growth by nucleic acid staining in aqueous suspension. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 34.Nyström, T. 2002. Aging in bacteria. Curr. Opin. Microbiol. 5:596-601. [DOI] [PubMed] [Google Scholar]
- 35.Parkes, R. J., B. A. Cragg, S. J. Bale, J. M. Getliff, K. Goodman, P. A. Rochelle, J. C. Fry, A. J. Weightman, and S. M. Harvey. 1994. Deep bacterial biosphere in Pacific Ocean sediments. Nature 371:410-413. [Google Scholar]
- 36.Parkes, R. J., G. Webster, B. A. Cragg, A. J. Weightman, C. J. Newberry, T. G. Ferdelman, J. Kallmeyer, B. B. Jorgensen, I. W. Aiello, and J. C. Fry. 2005. Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature 436:390-394. [DOI] [PubMed] [Google Scholar]
- 37.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 38.Revsbech, N. P., and B. B. Jørgensen. 1986. Microelectrodes: their use in microbial ecology. Adv. Microb. Ecol. 9:293-352. [Google Scholar]
- 39.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 40.Sass, A. M., H. Sass, M. J. Coolen, H. Cypionka, and J. Overmann. 2001. Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania basin, Mediterranean Sea). Appl. Environ. Microbiol. 67:5392-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholten, J. C. M., and R. Conrad. 2000. Energetics of synthrophic propionate oxidation in defined batch and chemostat cocultures. Appl. Environ. Microbiol. 66:2934-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiba, T., R. T. Hill, W. Straube, and R. R. Colwell. 1995. Decrease in culturability of Vibrio cholerae caused by glucose. Appl. Environ. Microbiol. 61:2583-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simu, K., and Å. Hagström. 2004. Oligotrophic bacterioplankton with a novel single-cell life strategy. Appl. Environ. Microbiol. 70:2445-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kämpfer, M. C. J. Maiden, X. Nesme, R. Rosselló-Mora, J. Swings, H. G. Trüper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson, B. S., S. A. Eichhorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Straskrabova, V. 1983. The effect of substrate shock on populations of starving aquatic bacteria. J. Appl. Bacteriol. 54:217-224. [Google Scholar]
- 47.Straub, K. L., and B. Schink. 2004. Ferrihydrite-dependent growth of Sulfurospirillum deleyanum through electron transfer via sulfur cycling. Appl. Environ. Microbiol. 70:5744-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Süß, J., B. Engelen, H. Cypionka, and H. Sass. 2004. Quantitative analysis of bacterial communities from Mediterranean sapropels based on cultivation-dependent methods. FEMS Microbiol. Ecol. 51:109-121. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki, M. M. S. Rappé, Z. W. Haimberger, H. Winfield, N. Adair, J. Ströbl, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Widdel, F. 1980. Anaerober Abbau von Fettsäuren und Benzoesäure durch neu isolierte Arten sulfatreduzierender Bakterien. Ph.D. thesis. University of Göttingen, Göttingen, Germany.
- 53.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulphate-reducing bacteria, p. 3353-3378. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer, New York, N.Y.
- 54.Widdel, F., G. W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov. sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 143:286-294. [Google Scholar]
- 55.Wieringa, E. B. A., J. Overmann, and H. Cypionka. 2000. Detection of abundant sulphate-reducing bacteria in marine oxic sediment layers by a combined cultivation and molecular approach. Environ. Microbiol. 2:417-427. [DOI] [PubMed] [Google Scholar]
- 56.Zengler, K., G. Toledo, M. Rappé, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]