Abstract
Pulsed-field gel electrophoresis and serotyping were performed for 544 isolates of Listeria monocytogenes, including 502 isolates recovered from contaminated samples from 31,705 retail ready-to-eat (RTE) food products and 42 isolates recovered from human cases of listeriosis. The isolates were from Maryland (294 isolates) and California (250 isolates) and were collected in 2000 and 2001. The isolates were placed into 16 AscI pulsogroups (level of relatedness within each group, ≥66%), 139 AscI pulsotypes (levels of relatedness, ≥25% to 100%), and eight serotypes (serotypes 1/2a, 1/2b, 1/2c, 3a, 3b, 4b, 4c, and 4d). The most frequently found pulsotypes belonged to either pulsogroup A (150 food isolates plus 4 clinical isolates) or pulsogroup B (104 food isolates plus 5 clinical isolates). The majority of the 502 food isolates were either serotype 1/2a (298 isolates) or serotype 1/2b (133 isolates), whereas the majority of the 42 clinical isolates were either serotype 1/2a (19 isolates) or serotype 4b (15 isolates). Additionally, 13 clinical isolates displayed pulsotypes also found in food isolates, whereas the remaining 29 clinical isolates displayed 24 unique pulsotypes. These data indicate that most (86%) of the L. monocytogenes subtypes found in the RTE foods sampled belonged to only two serotypes and that 90% of the isolates displayed 73 pulsotypes, with 107 isolates displaying pulsotype 1. These data should help define the distribution and relatedness of isolates found in RTE foods in comparison with isolates that cause listeriosis.
Listeria monocytogenes is a ubiquitous gram-positive facultative intracellular food-borne pathogen that causes listeriosis, particularly in high-risk groups, including the elderly and immunocompromised individuals, as well as pregnant women and their neonates. There are an estimated 2,500 cases of listeriosis that result in 500 deaths in the United States each year based on data from 1996 and 1997; the mortality rate approaches 28% (23). A more recent report of listeriosis cases from the Centers for Disease Control and Prevention (CDC) FoodNet active surveillance program indicated that there were 3 cases per million people in 2000 and 2001 (6, 7). Of the 13 known serotypes of L. monocytogenes, serotype 4b strains cause >50% of the listeriosis cases worldwide (29), whereas most of the L. monocytogenes strains found in food are serotype 1/2a and 1/2b (1, 11, 34). Contaminated refrigerated ready-to-eat (RTE) foods, some of which can support growth of L. monocytogenes, are of particular concern since such products typically are not heated prior to consumption. There is currently a “zero tolerance” policy for RTE foods in the United States, and under this policy any food contaminated with detectable levels of L. monocytogenes is deemed adulterated (30). As a result, in the United States between 1999 and 2004 there were five major recalls that alone were associated with 127 million pounds of RTE food due to contamination with L. monocytogenes (http://www.fsis.usda.gov/Fsis_Recalls/index.asp).
Highly discriminatory typing methods are essential for tracing contamination routes and the epidemiology of L. monocytogenes. Among the many methods used to type bacteria, pulsed-field gel electrophoresis (PFGE), amplified fragment length polymorphism analysis, random amplified polymorphic analysis, and ribotyping are the most useful for subtyping L. monocytogenes (32). Serotyping and phage typing have limited value as typing tools for L. monocytogenes since neither of these methods is discriminatory enough and since many strains are not typeable by phage typing and only three serotypes (serotypes 1/2a, 1/2b, and 4b) are responsible for 90% of the clinical cases of listeriosis (27, 29). For the DNA-based typing methods, random amplified polymorphic analysis and amplified fragment length polymorphism analysis results are both difficult to reproduce and difficult to standardize among laboratories. Although ribotyping is reproducible and can be automated, PFGE is more discriminatory, has very good reproducibility, and can be standardized (1, 4, 24, 37). Based on the excellent resolving power for epidemiological typing, PFGE is used by the CDC for PulseNet to track isolates of L. monocytogenes and other pathogens responsible for food-borne illness.
The strains used in this study were obtained through a large-scale market basket survey of a variety of retail RTE foods in Maryland and northern California in 2000 and 2001 (13). The eight categories of food surveyed were luncheon meats, deli salads, fresh soft “Hispanic-style” cheese, bagged salads, blue-veined cheese, soft mold-ripened cheese, smoked seafood, and seafood salads. Surveillance and monitoring activities of the FDA and U.S. Department of Agriculture in the mid-1990s indicated that up to 5% of some RTE foods, such as prepared deli salads and luncheon meats, may contain L. monocytogenes (17, 19). The other six food categories were chosen by Gombas et al. (13) to provide additional data for risk assessment. A total of 31,705 food samples were analyzed, and 577 of these samples tested positive for L. monocytogenes, for an overall prevalence of 1.82%. Of the 577 food samples that tested positive for L. monocytogenes using DNA-based screening methods, 505 yielded a viable isolate of L. monocytogenes, and a single isolate was retained from each sample (13). An additional 42 human clinical isolates from the same geographic regions and time periods were obtained for assessing the risk of L. monocytogenes from RTE foods in the United States and for a comparative analysis with the 505 food isolates (8, 13). The goal of the present study was to identify the “types” of the isolates mentioned above and to determine if certain subtypes were more often associated with certain foods and/or more often involved in cases of listeriosis.
MATERIALS AND METHODS
Bacterial strains.
Of the 505 samples that yielded viable isolates of L. monocytogenes in the study mentioned above (13), 3 were subsequently confirmed to contain Listeria innocua (one isolate each of serotypes 3b and 1/2b) and Listeria seeligeri (one isolate of serotype 4b) using ribotyping (Riboprinter microbial characterization system; Qualicon/DuPont, Wilmington, DE) and API 10 300 Listeria test strips (BioMérieux, St. Louis, MO). After these three strains were eliminated, 502 L. monocytogenes strains obtained from the following six categories of food remained: luncheon meats, bagged salads, deli salads, cheese, seafood salads, and smoked seafood. For this study, fresh soft “Hispanic-style” cheese, blue-veined cheese, and soft mold-ripened cheese were grouped as a single food category, referred to as cheese. Also included in the present study were 42 human clinical isolates that represented all reported sporadic human cases of listeriosis from Maryland and northern California in 2000 and 2001 (8). The clinical isolates were kindly provided by the CDC (Atlanta, GA).
Pulsed-field gel electrophoresis.
Listeriae were embedded in agarose, lysed, washed, and digested with the restriction enzyme AscI (New England Biolabs, Beverly, MA) overnight (12 to 16 h) at 37°C essentially as described previously (14). Electrophoresis was performed in a 1% agarose gel using 0.5× Tris-borate-EDTA buffer with a Chef Mapper XA (Bio-Rad Laboratories, Hercules, CA) at 6 V/cm for 21 h with switch times of 4 to 40 s. Gels were stained for 30 min at room temperature with a 10-mg/ml ethidium bromide (Invitrogen, Carlsbad, CA) solution and photographed. The AscI-digested DNA from a laboratory control strain of L. monocytogenes (MFS1435; pulsotype 84, serotype 1/2a) was included as a reference.
Data analyses.
Pattern images were acquired using a Bio-Rad Gel Doc system with the Multi Analyst software program (version 1.1; Bio-Rad) and were compared using the Applied Maths BioNumerics software package (version 4.0; Applied Maths, Saint-Martins-Latem, Belgium). Pattern clustering was performed using algorithms within BioNumerics, specifically the unweighted pair group method using arithmetic averages (UPGMA) and the Dice correlation coefficient with a position tolerance of 1.0%.
Statistical analyses.
The frequencies of pulsotypes and serotypes were examined by using a log linear model using PROC CATMOD of the SAS statistical system (version 8.0; SAS Institute Inc., Cary, N.C.) to investigate if there were any changes in the distribution of strain types due to state, food category, lineage, and/or serotype. Associations between categorical variables (food category, serotype, season, geography) were examined by chi-square analysis (SAS), and a P value of <0.05 was considered significant. Additionally, the “fusion levels” (i.e., the levels at which two clusters join or fuse to become a larger cluster) from the cluster analysis dendrogram were examined.
Serotyping.
Serotyping was performed by using the FDA-BAM procedure, with minor modifications (3). Briefly, isolates were grown in brain heart infusion (BHI) motility agar (BHI broth with 0.3% agar) (Difco Laboratories Inc., Sparks, MD) for 48 h at 25°C, inoculated into 8 ml BHI broth, and incubated overnight at 25°C. Next, 0.5% formaldehyde (Sigma, St. Louis, MO) was added, and the cells were incubated for an additional 4 h at 25°C and then tested for reaction to the flagellar (H) antigen (Denka Seiken Listeria Rabbit set; Accurate Chemical, Westbury, NY) by incubating the cells and sera at 50°C for 2 h and examining the tubes visually for agglutination. For somatic (O) antigen testing, cells from the motility agar described above were inoculated into 8 ml of BHI broth, incubated overnight at 37°C, and then steamed for 30 min, centrifuged, resuspended in 1% formal saline, and tested by the slide method for agglutination with the O antigens (Accurate Chemical). A set of four strains (serotype 4b strain MFS53 [=CDC F2365], serotype 4b strain MFS96 [=CDC H7858], serotype 4c strain MFS108 [=ATCC 19116], and serotype 1/2a strain MFS110 [=F6854]) from the Microbial Food Safety Research Unit culture collection and an additional strain, ATCC 19114 (serotype 4a), were used as controls. The serotypes for 20 of the 42 clinical isolates provided by the CDC matched the serotype data derived in our laboratory, except in two cases. We identified strain MFS1090 as a serotype 4d strain, whereas the CDC identified this isolate as a serotype 4b strain. Likewise, we identified strain MFS 1097 as a serotype 4b strain, whereas the CDC identified this isolate as a serotype 1/2b strain. These isolates were tested three different times with two different batches of sera to confirm the serotypes. Additionally, these two isolates were tested using a multiplex PCR described by Doumith et al. (10), which identifies serotypes 1/2a, 1/2b, 1/2c, and 4b. By both methods, MFS1097 was confirmed to be a serotype 4b strain; however, the reaction does not discern between serotypes 4b and 4d, and therefore MFS1090 was identified as a serovar 4 strain by this method.
RESULTS
Serotyping.
Eight different serotypes were found among the 502 food and 42 clinical L. monocytogenes isolates (Table 1). As anticipated, the majority of the 502 food isolates recovered (298 isolates or 60%) were serotype 1/2a. Most of the remaining food isolates were either serotype 1/2b (133 isolates or 26%) or serotype 4b (44 isolates or 9%). The majority of the clinical isolates were either serotype 1/2a (19 isolates or 45%), serotype 4b (15 isolates or 36%), or serotype 1/2b (6 isolates or 14%). More clinical isolates were obtained from Maryland (27 of the 42 isolates) than from California (15 of the 42 isolates) (Table 1). When all 544 isolates were considered, 250 isolates (46%) were obtained from California and 294 isolates (54%) were obtained from Maryland.
TABLE 1.
Distribution of isolates, pulsotypes, and pulsogroups among serotypes and food categories
| Serotype | No. of isolatesa
|
Total no. of pulsotypesa,b | Total no. of pulsogroupsb | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lunch meats
|
Bagged salads
|
Deli salads
|
Cheese
|
Seafood salads
|
Smoked seafood
|
Total food
|
Clinical
|
|||||||||||
| CA | MD | CA | MD | CA | MD | CA | MD | CA | MD | CA | MD | CA | MD | CA | MD | |||
| 1/2a | 5 | 14 | 10 | 4 | 37 | 60 | 16 | 3 | 10 | 63 | 61 | 12 | 140 | 158 | 5 | 14 | 71 | 12 |
| 1/2b | 7 | 24 | 0 | 0 | 37 | 20 | 5 | 6 | 3 | 13 | 4 | 16 | 55 | 78 | 4 | 2 | 35 | 9 |
| 1/2c | 3 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 5 | 3 | 0 | 1 | 3 | 1 |
| 3a | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 1 | 0 | 0 | 1 | 1 |
| 3b | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 5 | 2 | 0 | 0 | 3 | 2 |
| 4b | 1 | 0 | 4 | 1 | 9 | 7 | 2 | 0 | 8 | 5 | 0 | 7 | 24 | 19 | 5 | 10 | 29 | 7 |
| 4c | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 1 |
| 4d | 0 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 1 | 0 | 4 | 3 |
| Total | 25 | 45 | 14 | 7 | 85 | 87 | 23 | 9 | 22 | 82 | 66 | 37 | 235 | 267 | 15 | 27 | ||
Twenty-five pulsotypes were displayed by the isolates from lunch meats, 10 pulsotypes were displayed by the isolates from bagged salads, 52 pulsotypes were displayed by the isolates from deli salads, 10 pulsotypes were displayed by the isolates from cheese, 33 pulsotypes were displayed by the isolates from seafood salads, 34 pulsotypes were displayed by the isolates from smoked seafood, 115 pulsotypes were displayed by the isolates from all types of food, and 35 pulsotypes were displayed by the clinical isolates.
Altogether, there were 139 pulsotypes and 16 pulsogroups.
When the food categories were examined according to serotype and geography, there were some apparent differences that were statistically significant (Table 1). For example, 61 serotype 1/2a isolates were found in smoked seafood from California, while in Maryland only 12 serotype 1/2a isolates were found in smoked seafood. However, in the seafood salad category, 63 serotype 1/2a isolates were found in Maryland, while only 10 serotype 1/2a isolates were found in California. When deli salads were examined, 37 serotype 1/2a isolates and 37 serotype 1/2b isolates were found in California; however, in Maryland 60 serotype 1/2a isolates and only 20 serotype 1/2b isolates were found. Beyond these three categories, there were either too few samples and isolates and/or there were no statistically significant differences (P > 0.05) for the remaining food categories in relation to serotype. When rare serotypes were examined, two serotype 4c isolates were found in this study, and both were recovered from bagged salads from Maryland in 2000. Of the six serotype 4d isolates, one was recovered from deli salad (California), one was a clinical isolate (California), and four were found in luncheon meats; however, all four luncheon meat isolates were from Maryland and were recovered from products purchased on the same day. Four of the nine serotype 1/2c isolates were found in deli salad, seafood salad, smoked seafood, and a clinical setting (one isolate each), while the other five isolates were found in luncheon meats. Of the seven serotype 3b isolates, five were from California, and four of these were found in luncheon meats (Table 1); two of the latter isolates were recovered from products purchased on the same day, and two were recovered from products purchased on different days. Additionally, all six serotype 3a isolates were found in luncheon meat samples, and two of these were isolated from products purchased on the same day. The luncheon meat category is interesting because a large portion (19 of 70 isolates) of the less common serotypes (serotypes 1/2c, 3a, 3b, and 4d) were found in luncheon meats and most of these isolates were collected in California. Only one isolate recovered from luncheon meat was a serotype 4b isolate (Table 1). However, the bagged salads contained five serotype 4b isolates and no serotype 1/2b isolates.
Statistical analysis was done to determine if there was any seasonality associated with the isolation of L. monocytogenes from food. The recovery of L. monocytogenes was evenly distributed over the spring, winter, and autumn in both states. However, in the summer the recovery of L. monocytogenes from food was significantly different between the two FoodNet sites (P < 0.05); the prevalence of the organism was twice as high in California as it was in Maryland.
PFGE.
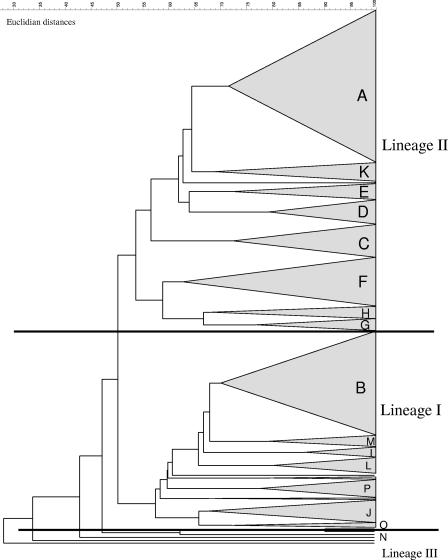
L. monocytogenes genomic DNA was digested using AscI, which generated 9 to 11 bands (range, 7 to 12 bands). The 544 isolates were placed into 16 AscI pulsogroups (Fig. 1) that were comprised of 139 distinct AscI pulsotypes. Pulsogroups were established both empirically based on the number and arrangement of fragments and computationally based on the levels of relatedness using the Dice similarity coefficient and UPGMA. Isolates were considered to have the same pulsotype when the numbers and locations of the bands were indistinguishable. A unique pulsotype was defined as a fingerprint displayed by only one isolate in our data set. Each of the 16 pulsogroups contained 1 to 21 distinct pulsotypes, with an overall similarity value of ≥66% using the Dice correlation coefficient and UPGMA. The largest pulsogroup, pulsogroup A, contained 150 food isolates and four clinical isolates; these isolates were serotype 1/2a (148 isolates) or 3a (6 isolates) (Table 2). The second largest pulsogroup, pulsogroup B, contained 104 food isolates and five clinical isolates; these isolates were serotype 1/2b (102 isolates), 3b (6 isolates), or 4b (1 isolate) (Table 2). Isolates that were distinct and that did not associate with any other pulsogroups were placed into pulsogroup N (17 isolates). Therefore, pulsogroup N contained most of the “outliers” with low levels of relatedness (25 to 60%) to the vast majority of the isolates in the entire set. Of the 17 isolates in pulsogroup N, 8 were clinical isolates, and five of these clinical isolates were serotype 4b. The fusions levels (the clustering of smaller groups into a larger group) were examined. The first significant break occurred when lineage I and lineage II isolates were joined; thus, there was significant grouping into these two lineages.
FIG. 1.
Dendrogram for 16 pulsogroups, pulsogroups A to P.
TABLE 2.
Pulsogroups, pulsotypes, and serotypes of L. monocytogenes isolates recovered from retail RTE foods and clinical cases in California and Maryland in 2000 and 2001
| Pulsogroup | No. of pulsotypes | No. of isolates | Serotype(s) (no. of isolates) | No. of food categories represented |
|---|---|---|---|---|
| A | 15 | 154 | 1/2a (148), 3a (6) | 6 |
| B | 21 | 109 | 1/2b (102), 3b (6), 4b (1) | 5 |
| C | 11 | 34 | 1/2a (9), 1/2b (2) | 4 |
| D | 7 | 26 | 1/2a (26) | 3 |
| E | 12 | 23 | 1/2a (14), 1/2c (9) | 6 |
| F | 5 | 50 | 1/2a (50) | 5 |
| G | 5 | 13 | 1/2a (13) | 3 |
| H | 6 | 12 | 1/2a (10), 1/2b (2) | 4 |
| Ia | 3 | 12 | 4b (8), 4d (4) | 4 |
| J | 10 | 20 | 4d (1), 1/2a (1), 1/2b (1), 4b (17) | 5 |
| Ka | 8 | 20 | 1/2a (17), 1/2b (2), 4b (1) | 3 |
| L | 5 | 16 | 1/2b (15), 3b (1) | 3 |
| Ma | 3 | 11 | 1/2b (11) | 2 |
| N | 15 | 17 | 4b (7), 1/2b (3), 1/2a (5), 4c (2) | 5 |
| O | 5 | 9 | 4b (9) | 3 |
| P | 8 | 18 | 4d (1), 1/2a (1), 1/2b (1), 4b (15) | 2 |
| Total | 139 | 544 |
No clinical isolates were in pulsogroups I, K and M.
Among the 42 clinical isolates, there were five sets that displayed the same pulsotype (four sets of two isolates and one set of four isolates); therefore, 35 pulsotypes were displayed by the clinical isolates (Table 1). Additionally, 13 clinical isolates displayed pulsotypes that were also found among the food isolates, and these 13 isolates were serotype 1/2a (6 isolates), serotype 1/2b (4 isolates), serotype 1/2c (1 isolate), and serotype 4b (2 isolates). The one clinical isolate that was serotype 1/2c displayed a pulsotype that was also displayed by two isolates recovered from luncheon meats, and all three isolates were from Maryland and were recovered in 2000; however, the clinical strain was recovered in January 2000, while both luncheon meat isolates were recovered in August 2000. Other than this serotype 1/2c isolate, five additional clinical isolates were recovered in the same year and state as a food isolate which displayed an identical pulsotype (one isolate from bagged salad, one isolate from seafood salad, one isolate from deli salad, and two isolates from luncheon meats). The deli salad isolate was from California, and the remaining four isolates were from Maryland. After clinical isolates that displayed the same pulsotype as a food isolate were accounted for, the clinical isolates contributed 24 unique pulsotypes not found among the food isolates. Of these 24 unique clinical pulsotypes, 1 was displayed by a serotype 4d isolate, 9 were displayed by serotype 4b isolates, 13 were displayed by serotype 1/2a isolates, and 1 was displayed by a serotype 1/2b isolate. Of note, there were 43 serotype 4b food isolates and 15 serotype 4b clinical isolates, for a total of 58 serotype 4b strains that displayed a total of 29 pulsotypes (Table 1); nine of these pulsotypes were displayed by clinical isolates, and 20 were displayed by food isolates (data not shown).
At least one isolate with a unique pulsotype was found in each food category sampled. The number of pulsotypes per category of food varied from 52 pulsotypes found in deli salads to 10 pulsotypes each found in bagged salads and cheese (Table 1). Of note, bagged salads, from which the fewest number of isolates were recovered, displayed considerable heterogeneity based on the number of pulsotypes (10) per number of isolates (21) (Table 1). Isolates from the seafood salad (104 isolates), smoked seafood (103 isolates), and luncheon meat (70 isolates) categories displayed 33, 34, and 25 pulsotypes, respectively (Table 1). Many pulsotypes, such as pulsotypes 1, 12, and 25, were found in isolates recovered from more than one food category. However, pulsotype 11 was displayed only by 18 isolates recovered from deli salads in California collected in 2000 and 2001. Additionally, some pulsotypes were not associated exclusively with one food category but were recovered from one state. There were 18 pulsotypes exclusively from California and 19 pulsotypes exclusively from Maryland. For example, pulsotype 78 (serotype 1/2a) was displayed by 22 isolates recovered from smoked seafood and one isolate recovered from bagged salads, all from California.
The most common pulsotype, pulsotype 1 (Fig. 2), which was in pulsogroup A, was displayed by 107 of the 544 isolates (Table 3). Pulsogroup A, the largest group, contained 154 isolates that were 71% related and displayed 15 different pulsotypes (Table 2). The second most common pulsotype, pulsotype 78 (Fig. 2), was displayed by 23 isolates (Table 3). These 23 isolates plus an additional 27 isolates were all serotype 1/2a, were 71% related, and formed pulsogroup F, the third largest group (Table 2). The third most common pulsotype, pulsotype 10 (Fig. 2), was in pulsogroup B, the second largest group (109 isolates), which was comprised of 80 isolates that were 82% related. An additional 29 isolates that were 70% related to the isolates mentioned above (Table 2) completed pulsogroup B, which contained 21 different pulsotypes, making this the most diverse pulsogroup in terms of pulsotypes. The remaining pulsogroups were comprised of 9 to 34 isolates, and each pulsogroup contained from 3 to 15 pulsotypes, with levels of relatedness ranging from 66 to 86% (Table 2).
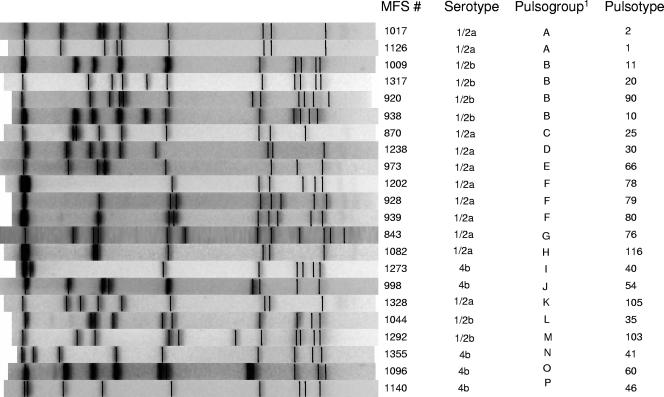
FIG. 2.
Representative pulsotypes for each of the 16 pulsogroups.
TABLE 3.
Predominant pulsotypes and pulsogroups of L. monocytogenes isolates recovered from retail RTE foods and clinical cases in California and Maryland in 2000 and 2001
| Pulsotypea | No. of isolatesb | Pulsogroupc |
|---|---|---|
| 1 | 107 | A |
| 78 | 23 | F |
| 10 | 22 | B |
| 11 | 18 | B |
| 20 | 18 | B |
| 2 | 16 | A |
| 25 | 14 | C |
| 30 | 13 | D |
| 80 | 13 | F |
| 17 | 12 | B |
| 79 | 11 | F |
| 105 | 11 | K |
| 35 | 11 | L |
The 13 pulsotypes listed account for 50% of all isolates. The remaining 126 pulsotypes not listed were displayed by 10 or fewer isolates.
A total of 289 of the 544 isolates examined displayed the pulsotypes listed.
The pulsotypes listed were in 7 of the 16 pulsogroups identified.
Two serotype 4c isolates, one serotype 1/2a isolate, one serotype 1/2b isolate, and one serotype 4b isolate formed the group of “outliers” in pulsogroup N that, when included in the analysis, reduced the overall relatedness of the isolates. One of the two serotype 4c isolates was only 25% related and the other was only 31% related to the rest of the isolates on the dendrogram. The overall level of similarity of all of the isolates on the dendrogram with these outliers included was around 25%; however, when the outliers were removed, the overall level of similarity of the isolates increased to 47%, as calculated using the Dice correlation coefficient and UPGMA.
DISCUSSION
In this study we determined the PFGE profiles and serotypes of 502 food isolates and 42 clinical isolates of L. monocytogenes and demonstrated that the clinical isolates were a heterogeneous group that were often distinct from the food isolates; only 13 of 42 (31%) pulsotypes displayed by the clinical isolates were found in the food isolates tested. A companion study (15) that used ribotyping to subtype the food isolates mentioned above along with an additional 492 clinical isolates (including the 42 isolates used in this study) also found that L. monocytogenes populations associated with RTE foods differed significantly from human clinical isolates. Although ribotyping is a useful subtyping method, it is not as discriminatory as PFGE; therefore, PFGE is more useful for discerning closely related strains, for examining subtypes, and/or for tracing contamination routes. More specifically, Gray et al. (15) identified a total of 36 ribotypes, while we identified a total of 139 pulsotypes among the 502 food isolates. Additionally, Gray and colleagues (15) examined a much larger set of clinical isolates and identified 54 ribotypes among the 492 clinical isolates, while we identified a total of 35 pulsotypes among our 42 clinical isolates. Both studies indicated that there is heterogeneity among clinical isolates and that there are distinct populations of food and clinical isolates of L. monocytogenes.
One of the goals of this study was to examine whether certain subtypes of L. monocytogenes are more commonly associated with specific categories of food. For example, in the study of Gray et al. (15) mentioned above, isolates displaying ribotype DUP-1062D were recovered 26 times from smoked seafood, and isolates displaying ribotypes DUP-1042B and DUP-1062A were isolated more frequently from seafood salads collected in Maryland (15). Sauders et al. (28) examined 125 retail food isolates and 40 environmental isolates from retail establishments in New York state from 1997 to 2002 and found 29 ribotypes; two of these ribotypes were significantly more common among food isolates and had been reported as being associated with smoked fish processing plants in previous studies (28). Other studies have also reported that a specific ribotype, pulsotype, and/or serotype is more frequently associated with seafood (2, 22, 24, 28). In this study, within each food category there were too few isolates to examine the association of pulsotypes with food types statistically; however, we did find some pulsotypes that were specific to certain food types by visual examination. For example, pulsotype 11 (serotype 1/2b) was confined to 18 isolates recovered from deli salads in California, and pulsotype 90 (serotype 1/2b) was confined to nine isolates recovered from smoked seafood in Maryland. However, previous studies and our work indicate that many pulsotypes are found in more than one food type, which indicates that there is no clear correlation between certain subtypes and specific food products (1, 2, 31).
A number of studies have examined the presence of L. monocytogenes subtypes in food processing environments. For instance, a study done in pork slaughtering and cutting plants examined 287 isolates that yielded 17 ApaI PFGE types and four serotypes (serotypes 1/2a, 3a, 1/2c, and 3c) (12). This study found that there was overrepresentation of one L. monocytogenes PFGE type that accounted for 90% of the isolates; all of these isolates were serotype 1/2a (12). A study that examined the prevalence of L. monocytogenes in frankfurter packages from seven plants demonstrated that 90% had the same ribotype profile and serotype (34). Other investigators have also reported that 75 to 90% of isolates from food and food processing environments tested belonged to serogroup 1/2 (1, 9, 11, 27, 31). About 50% of our food isolates displayed only 13 pulsotypes, and 88% of the isolates belonged to serogroup 1/2 (60% belonged to serotype 1/2a, 27% belonged to serotype 1/2b, and 1.6% belonged to serotype 1/2c). One explanation for this is that specific subtypes of L. monocytogenes in food products may have originated as persistent strains in the food processing environment that subsequently contaminated the finished products; this possibility is supported by the results of a number of studies (9, 16, 18, 20, 21, 22, 24, 28). Collectively, our results and those of other investigators indicate that serotype 1/2a isolates displaying a few pulsotypes represent the majority of isolates found in food and food processing environments.
There are a number of reasons that could explain the predominance of serotype 1/2a isolates recovered from the RTE foods sampled in this study. A food sample could be contaminated with multiple types of L. monocytogenes, as has been reported elsewhere (2, 34), but only specific types may survive and replicate in food. In the present study only one isolate was retained from each food sample tested; therefore, it is probable that only the predominant L. monocytogenes isolates were recovered. This conclusion is supported by a study that used PFGE to monitor a five-strain cocktail consisting of serotype 4b and 1/2a isolates of L. monocytogenes in frankfurter packages for 90 days at 4°C, in which one strain accounted for the statistical majority (83%) of the isolates found in the package. The isolate was a serotype 1/2a strain that was initially recovered from a pork processing plant, indicating that specific subtypes may survive better in certain food products (25). However, it has also been noted that selective enrichment procedures can affect which types or lineages of L. monocytogenes are recovered from food (5). The existence of three distinct lineages of L. monocytogenes has been described previously (4, 26, 35, 36), and the majority of our isolates (312 isolates) belonged to lineage II (serotypes 1/2a, 1/2c, and 3a), while a smaller number (183 isolates) belonged to lineage I (serotypes 1/2b, 3b and 4b). Bruhn et al. (5) demonstrated that lineage II strains could outcompete lineage I strains in UVM I broth, which contains the selective compounds nalidixic acid and acriflavine (5). In the current study, the selective compounds were present in all of the enrichment broths used (13) and, therefore, could have contributed to overrepresentation of lineage II strains. Other contributions could be related to different susceptibilities to sublethal stress, differences in growth rates, and background flora that may mask the recovery of Listeria spp.
There have been several studies in which the workers compared clinical isolates with food and/or environmental isolates (15, 22, 27). Revazishvili et al. (27) examined 74 environmental isolates, 94 clinical isolates, and seven isolates with unknown origins by a variety of methods and found that the clinical isolates had distinct PFGE types which did not cluster together or with the PFGE types of the environmental isolates and were not closely related according to the PFGE-based dendrogram (27). In the present study, the 42 clinical isolates also did not cluster on the dendrogram and were not closely related compared to the food isolates. Ribotyping of 492 human isolates and 502 food isolates by Gray et al. (15) identified 20 ribotypes that occurred only once in the data set, 19 of which were clinical isolates. Additionally, these authors observed that one ribotype was displayed by 30% of the food isolates but by only 1.8% of the clinical isolates (15). These data establish that isolates with genetic profiles that match clinical subtypes are sporadically found in food, while other subtypes apparently not associated with listeriosis are more common in food.
Examination of the previously published data indicates that clinical isolates are more heterogeneous than food isolates and display more unique pulsotypes. For instance, in a study that included 41 clinical isolates obtained from sporadic cases of listeriosis in Austria between 1997 and 2000 the workers found 37 unique pulsotypes (33). In another study the workers examined 48 clinical isolates from sporadic listeriosis cases in Belgium in 2001 and found 34 different AscI profiles and 38 different ApaI profiles, providing additional support for the hypothesis that listeriosis involves a variety of genetically distinct strains (37). In the present study, our serotype 4b isolates, both food isolates and clinical isolates, were by far the most heterogeneous group, displaying 29 pulsotypes for 58 isolates.
One of the unique aspects of the present study was the availability and subsequent direct comparison of both food and clinical isolates obtained from the same geographic regions and during the same time frames. Overall, most (86%) of the L. monocytogenes subtypes found in the RTE foods sampled belonged to only two serotypes, and 90% of the isolates displayed 73 pulsotypes. However, the clinical isolates that were obtained from sporadic cases of listeriosis displayed unique pulsotypes that were displayed by only a few of the isolates recovered from RTE foods sampled in this study. The clinical isolates therefore contributed to the overall heterogeneity of the isolates examined. Future work needs to focus on the distinct pulsotypes found in clinical isolates and the source, frequency, and survival of these isolates in food. While there are overlapping populations of L. monocytogenes that cause illness and that are found in food, there is also a subset of isolates that are genetically very distinct and are not readily found in food but are capable of causing illness in humans.
Acknowledgments
We extend our appreciation to the following colleagues at the Eastern Regional Research Center in Wyndmoor, PA: Jennielyn Bumanlag and Brian Dirks for help in the lab, Darrell Bayles and James Smith for critical reviews of the manuscript, and John Phillips for statistical analyses of the data. We also extend our appreciation to Tim Barrett (CDC, Atlanta, GA) for providing the clinical isolates, Dave Gombas (International Fresh-cut Produce Association, Alexandria, VA) for expert advice and assistance, and Jim Lindsay (ARS, NPS, Beltsville, MD) for facilitating this collaboration.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Aarnisalo, K., T. Autio, A. Sjoberg, J. Lunden, H. Korkeala, and M. Suihko. 2003. Typing of Listeria monocytogenes isolates originating from the food processing industry with automated ribotyping and pulsed-field gel electrophoresis. J. Food Prot. 66:249-255. [DOI] [PubMed] [Google Scholar]
- 2.Autio, T., J. Lunden, M. Fredriksson-Ahomaa, J. Bjorkroth, and A.-M. Sjoberg. 2002. Similar Listeria monocytogenes pulsotypes detected in several foods originating from different sources. Int. J. Food Microbiol. 77:83-90. [DOI] [PubMed] [Google Scholar]
- 3.Bennet, R. W., and R. E. Weaver. 2001. Serodiagnosis of Listeria monocytogenes. FDA-BAM procedure. [Online.] www.cfsan.fda.gov/∼ebam/bam-11.html.
- 4.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruhn, J. B., B. F. Vogel, and L. Gram. 2005. Bias in the Listeria monocytogenes enrichment procedure: lineage 2 strains outcompete lineage 1 strains in University of Vermont selective enrichments. Appl. Environ. Microbiol. 71:961-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2001. Preliminary FoodNet data on the incidence of foodborne illnesses-selected sites, United States, 2000. Morb. Mortal. Wkly. Rep. 50:241-246. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2002. Preliminary FoodNet data on the incidence of foodborne illnesses-selected sites, United States, 2001. Morb. Mortal. Wkly. Rep. 51:325-329. [PubMed] [Google Scholar]
- 8.Chen, Y., W. H. Ross, V. N. Scott, and D. E. Gombas. 2003. Listeria monocytogenes: low levels equal low risk. J. Food Prot. 66:570-577. [DOI] [PubMed] [Google Scholar]
- 9.Coillie, E. V., H. Werbrouck, M. Heyndrickx, L. Herman, and N. Rijpens. 2004. Prevalence and typing of Listeria monocytogenes in ready-to-eat food products on the Belgian market. J. Food Prot. 67:2480-2487. [DOI] [PubMed] [Google Scholar]
- 10.Doumith, M., C. Buchrieser, P. Glaser, C. Jacquet, and P. Martin. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilot, P., A. Genicot, and P. Andre. 1996. Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J. Clin. Microbiol. 34:1007-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giovannacci, I., C. Ragimbeau, S. Queguiner, G. Salvat, J.-L. Vendeuvre, V. Carlier, and G. Ermel. 1999. Listeria monocytogenes in pork slaughtering and cutting plants: use of RAPD, PFGE, and PCR-REA for tracing and molecular epidemiology. Int. J. Food Microbiol. 53:127-140. [DOI] [PubMed] [Google Scholar]
- 13.Gombas, D. E., Y. Chen, R. S. Clavero, and V. N. Scott. 2003. Survey of Listeria monocytogenes in ready-to-eat foods. J. Food Prot. 66:559-569. [DOI] [PubMed] [Google Scholar]
- 14.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 15.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heir, E., B. Lindstedt, O. Rotterud, T. Vardund, G. Kapperud, and T. Nesbakken. 2004. Molecular epidemiology and disinfectant susceptibility of Listeria monocytogenes from meat processing plants and human infections. Int. J. Food Microbiol. 96:85-96. [DOI] [PubMed] [Google Scholar]
- 17.Hitchins, A. D. 1996. Assessment of alimentary exposure of Listeria monocytogenes. Int. J. Food. Microbiol. 30:71-85. [DOI] [PubMed] [Google Scholar]
- 18.Lappi, V. R., J. Thimothe, K. K. Nightingale, K. Gall, V. N. Scott, and M. Wiedmann. 2004. Longitudinal studies on Listeria in smoked fish plants: impact of intervention strategies on contamination patterns. J. Food Prot. 67:2500-2514. [DOI] [PubMed] [Google Scholar]
- 19.Levine, P., B. Rose, S. Green, G. Ransom, and W. Hill. 2001. Pathogen testing of ready-to-eat meat and poultry products collected at federally inspected establishments in the United States, 1990 to 1999. J. Food Prot. 64:1188-1193. [DOI] [PubMed] [Google Scholar]
- 20.Lukinmaa, S., K Aarnisalo, M.-L. Suihko, and A. Siitonen. 2004. Diversity of Listeria monocytogenes isolates of human and food origin studied by serotyping, automated ribotyping and pulsed-field gel electrophoresis. Clin. Microbiol. Infect. 10:562-568. [DOI] [PubMed] [Google Scholar]
- 21.Lunden, J. M., T. J. Autio, A.-M. Sjoberg, and H. J. Korkeala. 2003. Persistent and nonpersistent Listeria monocytogenes contamination in meat and poultry processing plants. J. Food Prot. 66:2062-2069. [DOI] [PubMed] [Google Scholar]
- 22.Martinez, I., L. Rorvik, V. Brox, J. Lassen, M. Seppola, L. Gram, and B. Fonnesbech-Vogel. 2003. Genetic variability among isolates of Listeria monocytogenes from food products, clinical samples, and processing environments estimated by RAPD typing. Int. J. Food Microbiol. 84:285-297. [DOI] [PubMed] [Google Scholar]
- 23.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura, H., M. Hatanaka, K. Ochi, M. Nagao, J. Ogasawara, A. Hase, T. Kitase, K. Haruki, and Y. Nishikawa. 2004. Listeria monocytogenes isolated from cold-smoked fish products in Osaka City, Japan. Int. J. Food Microbiol. 94:323-328. [DOI] [PubMed] [Google Scholar]
- 25.Porto, A. C. S., L. Wonderling, J. E. Call, and J. B. Luchansky. 2003. Use of pulsed-field gel electrophoresis to monitor a five-strain mixture of Listeria monocytogenes in frankfurter packages. J. Food. Prot. 66:1465-1468. [DOI] [PubMed] [Google Scholar]
- 26.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 27.Revazishvili, T., M. Kotetishvili, O. C. Stine, A. S. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2004. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J. Clin. Microbiol. 42:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauders, B. D., K. Mangione, C. Vincent, J. Schermerhorn, C. M. Farchione, N. B. Dumas, D. Bopp, L. Kornstein, E. D. Fortes, K. Windham, and M. Wiedmann. 2004. Distribution of Listeria monocytogenes molecular subtypes among human and food isolates from New York State shows persistence of human disease-associated L. monocytogenes strains in retail environments. J. Food Prot. 67:1417-1428. [DOI] [PubMed] [Google Scholar]
- 29.Schuchat, A., B. Swaminathan, and C. V. Broome. 1991. Epidemiology of human listeriosis. Clin. Microbiol. Rev. 4:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smoot, L. M., and M. D. Pierson. 1997. Indicator microorganisms and microbiological criteria, p. 66-80. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers. ASM Press, Washington, D.C.
- 31.Vitas, A. I., and V. A. Garcia-Jalon. 2004. Occurrence of Listeria monocytogenes in fresh and processed foods in Navarra (Spain). Int. J. Food Microbiol. 90:349-356. [DOI] [PubMed] [Google Scholar]
- 32.Vogel, B. F., V. Fussing, B. Ojeniyi, L. Gram, and P. Ahrens. 2004. High resolution genotyping of Listeria monocytogenes by fluorescent amplified fragment length polymorphism analysis compared to pulsed-field gel electrophoresis, random amplified polymorphic DNA analysis, ribotyping, and PCR-restriction fragment length polymorphism analysis. J. Food Prot. 67:1656-1665. [DOI] [PubMed] [Google Scholar]
- 33.Wagner, M., and F. Allerberger. 2003. Characterization of Listeria monocytogenes recovered from 41 cases of sporadic listeriosis in Austria by serotyping and pulsed-field gel electrophoresis. FEMS Immunol. Med. Microbiol. 35:227-234. [DOI] [PubMed] [Google Scholar]
- 34.Wallace, F. M., J. E. Call, A. C. S. Porto, G. J. Cocoma, ERRC Special Projects Team, and J. B. Luchansky. 2003. Recovery rate of Listeria monocytogenes from commercially prepared frankfurters during extended refrigerated storage. J. Food Prot. 66:584-591. [DOI] [PubMed] [Google Scholar]
- 35.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yde, M., and A. Genicot. 2004. Use of PFGE to characterize clonal relationships among Belgian clinical isolates of Listeria monocytogenes. J. Med. Microbiol. 53:399-402. [DOI] [PubMed] [Google Scholar]