Abstract
We purified to homogeneity an enzyme from Citrobacter sp. strain KCTC 18061P capable of decolorizing triphenylmethane dyes. The native form of the enzyme was identified as a homodimer with a subunit molecular mass of about 31 kDa. It catalyzes the NADH-dependent reduction of triphenylmethane dyes, with remarkable substrate specificity related to dye structure. Maximal enzyme activity occurred at pH 9.0 and 60°C. The enzymatic reaction product of the triphenylmethane dye crystal violet was identified as its leuco form by UV-visible spectral changes and thin-layer chromatography. A gene encoding this enzyme was isolated based on its N-terminal and internal amino acid sequences. The nucleotide sequence of the gene has a single open reading frame encoding 287 amino acids with a predicted molecular mass of 30,954 Da. Although the deduced amino acid sequence displays 99% identity to the hypothetical protein from Listeria monocytogenes strain 4b H7858, it shows no overall functional similarity to any known protein in the public databases. At the N terminus, the amino acid sequence has high homology to sequences of NAD(P)H-dependent enzymes containing the dinucleotide-binding motif GXXGXXG. The enzyme was heterologously expressed in Escherichia coli, and the purified recombinant enzyme showed characteristics similar to those of the native enzyme. This is the first report of a triphenylmethane reductase characterized from any organism.
Triphenylmethane dyes are aromatic xenobiotic compounds that are used extensively in many industrial processes, such as textile dyeing, paper printing, and food and cosmetic manufacture (11). Studies of the biodegradation of triphenylmethane dyes have focused primarily on the decolorization of dyes via reduction reactions. Several triphenylmethane dye-decolorizing microorganisms have been reported and their characteristics reviewed (2). The biochemical mechanism underlying the decolorization of triphenylmethane dyes has been elucidated in fungi (2, 7, 20, 23) but not in bacteria. Triphenylmethane dyes are decolorized by lignin peroxidase of Phanerochaete chrysosporium (7). Laccase from the extracellular fluid of Cyathus bulleri (23) and peroxidase from Pleurotus ostreatus (20) also decolorize triphenylmethane dyes. The structural genes encoding lignin peroxidase and laccase have been cloned and characterized (8, 10). Although several triphenylmethane dye-decolorizing bacteria have been isolated (2), there are no reports of specific enzymes that decolorize these dyes. The decolorization of malachite green and crystal violet by intestinal microflora and several anaerobic bacteria proceeds through enzymatic reduction to their respective leuco derivatives (12, 16). However, the enzymes involved in this reduction have not yet been isolated or characterized in their purified forms. Their amino acid sequences and other biophysical parameters remain unknown.
We recently isolated a new bacterium, Citrobacter sp. strain KCTC 18061P, that has a higher decolorization capability than any microorganism reported to date, even at high concentrations of triphenylmethane dyes (1). We have biochemically purified from Citrobacter sp. strain KCTC 18061P and characterized an enzyme that decolorizes triphenylmethane dyes. This enzyme is designated triphenylmethane reductase (TMR) in this paper. We also report the cloning of the gene encoding this enzyme and its heterologous expression in Escherichia coli.
MATERIALS AND METHODS
Bacterial strains, media, and culture.
A Citrobacter sp. strain isolated in a previous study (1) was deposited in the Korean Collection for Type Cultures (KCTC) under accession number KCTC 18061P. This strain was aerobically grown at 37°C for 15 h in LB medium (1.0% tryptone, 0.5% yeast extract, and 1.0% NaCl, pH 7.0) containing streptomycin (50 μg/ml). Cells were harvested by centrifugation and washed with 20 mM sodium phosphate buffer (pH 7.0). The cell pellets were stored at −70°C until use. E. coli strains JM109 and BL21(DE3), which were used as hosts for recombinant plasmids, were cultured at 37°C in LB medium with the appropriate antibiotics.
Purification of TMR.
All purification procedures were carried out at 4°C in 20 mM sodium phosphate buffer (pH 7.0). Frozen cells were suspended in 20 mM sodium phosphate buffer (pH 7.0), disrupted by sonication on ice, and centrifuged at 15,000 × g for 20 min. The supernatant was used as the crude enzyme source for purification and was applied at 1 ml/min to a HiPrep 16/10 Q XL column (Amersham Bioscience, NJ) equilibrated with buffer. After the column was washed with buffer at 1 ml/min, proteins were eluted with a linear gradient of 0 to 0.5 M NaCl in buffer at a flow rate of 1 ml/min. The most enzymatically active fractions were pooled and dialyzed against the same buffer. After solid ammonium sulfate was added to a final concentration of 1 M, the pooled fraction was applied at 1 ml/min to a HiTrap phenyl Sepharose column (Amersham Bioscience) equilibrated with buffer containing 1 M ammonium sulfate. After the column was washed with the same buffer at 1 ml/min, proteins were eluted from the column at a flow rate of 0.5 ml/min with a decreasing linear gradient of 1.0 to 0 M ammonium sulfate. The most active fractions were pooled and dialyzed against the buffer, and the pooled fraction was applied at 1 ml/min to a fast protein liquid chromatography Mono Q HR 5/5 column (Amersham Bioscience) equilibrated with buffer. After the column was washed with buffer at 1 ml/min, proteins were eluted with a linear gradient of 0 to 0.5 M NaCl in buffer at a flow rate of 0.5 ml/min. Active enzyme fractions with a single protein peak were pooled and concentrated by ultrafiltration with a Centricon-YM10 filter (Millipore, MA). The purified enzyme was stored at −20°C until use.
Protein concentrations were determined using the Bio-Rad (Hercules, CA) protein assay kit according to the manufacturer's instructions. Bovine serum albumin was used as the protein standard for the calibration curve.
Enzyme assays.
The standard assay system for TMR comprised 20 mM sodium phosphate buffer (pH 7.0), 20 μM crystal violet, 0.1 mM NADH, and a suitable amount of the enzyme in a total volume of 1 ml. Each reaction was initiated by the addition of the enzyme, and the initial reaction rate was determined by monitoring the decrease in absorbance at 590 nm in the first 2 min in a temperature-controlled cuvette in a 1.0-cm light path at 40°C. Enzyme activity was a linear function of both incubation time and protein concentration. One unit of enzyme activity was defined as the amount that catalyzed the reduction of 1 μmol of crystal violet per min by using a molar absorption coefficient of 110,916 M−1 cm−1. The corresponding wavelengths and molar absorption coefficients were used when other triphenylmethane or azo dyes were tested in place of crystal violet (see Table 2). To determine the kinetic parameters of the triphenylmethanedyes, their concentrations were varied from 4 to 20 μM, while the concentration of NADH was kept constant at 100 μM. For kinetic studies of NADH and NADPH, their concentrations were varied from 40 to 240 μM. The concentration of basic fuchsin was set at 40 μM. Km and Vmax values were determined by fitting the data to Lineweaver-Burk plots using SWIFT II Applications software (Amersham Bioscience).
TABLE 2.
Substrate specificity and kinetic analysis of the purified TMRa
| Substrateb | Absorption maximum (nm) | Molar absorption coefficient (M−1 cm−1) | Relative specific activityc (%) | Km (μM) | Vmax (μmol/mg/min) |
|---|---|---|---|---|---|
| Malachite green | 616 | 137,000 | 100 | 0.53 | 330 |
| Crystal violet | 590 | 111,000 | 41 | 1.3 | 66 |
| Basic fuchsin | 544 | 116,000 | 35 | 3.1 | 61 |
| Brilliant green | 625 | 107,000 | ND | —d | — |
| Bromophenol blue | 589 | 60,500 | ND | — | — |
| Methyl red | 410 | 24,500 | ND | — | — |
| Congo red | 497 | 20,600 | ND | — | — |
| NADH | 340 | 6,220 | 100 | 0.03 | 600 |
| NADPH | 340 | 6,220 | 23 | 0.84 | 890 |
The reaction rates of the purified enzyme with different dyes were measured by the standard assay as described in Materials and Methods. The enzyme activities were determined at the absorption maxima of the respective dyes.
The concentrations of dye substrates in the enzyme assay for specific activity were 20 μM.
Expressed as a percentage of the specific activity of malachite green as the substrate, and of NADH as the cosubstrate, under the same conditions. ND, not detected.
—, not determined.
The optimal pH of the enzyme was determined over a pH range of 6.0 to 11.0 by using the following buffers: 20 mM sodium phosphate (pH 6.0 to 7.6), 20 mM Tris-HCl (pH 7.5 to 9.0), and 20 mM glycine-NaOH (pH 9.0 to 11.0). The optimal temperature for enzyme activity was determined over the range of 20 to 80°C by a standard assay.
Analysis of reaction products.
The presumed product formed from crystal violet or malachite green by purified TMR was analyzed using an Ultrospec 2100 pro spectrophotometer (Amersham Bioscience). An assay mixture containing 20 mM sodium phosphate buffer (pH 7.0), 50 μM crystal violet or malachite green, 0.1 mM NADH, and 0.5 μg of the purified enzyme was incubated at 40°C for various periods. It was extracted with an equal volume of dichloromethane, and the reaction products in the dichloromethane extract were analyzed spectrophotometrically. Product formation was detected by measuring the change in absorbance at the visible wavelength maximum for each dye. The UV-visible spectra of the reaction products were compared with those of authentic leuco crystal violet or leucomalachite green dissolved in dichloromethane and used as standards. Reaction products were also analyzed by thin-layer chromatography (TLC) with Kieselgel 60 F254 (Merck, Darmstadt, Germany), to which 40 μl of dichloromethane extract was applied in a line 2 cm from the bottom of the TLC plate and developed with n-hexane-ethyl acetate (4:6, vol/vol) or propanol-water-glacial acetic acid (90:9:1, vol/vol/vol).
Other methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 12% polyacrylamide gels according to the method of Laemmli (13). Native PAGE was performed using the same method as for SDS-PAGE, but without SDS. Low-range protein standards and Precision Plus protein standards (Bio-Rad) were used as molecular mass markers for SDS-PAGE and native PAGE, respectively. Activity staining was performed using a crystal violet solution containing NADH. After native PAGE, the gel was placed in a substrate solution containing 73.5 μM crystal violet and 0.1 mM NADH, and this mixture was incubated at 37°C until a clear band developed in the background of the dye-stained gel.
The native molecular mass of the purified enzyme was determined by size exclusion chromatography on a Superdex 200 HR 10/30 column (Amersham Bioscience) that had been equilibrated with 20 mM sodium phosphate buffer (pH 7.0) containing 0.15 M NaCl. The column was calibrated with the following proteins as standards: alcohol dehydrogenase (150 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa).
Digestion of the purified enzyme by trypsin (Promega, Madison, WI) and the subsequent separation of tryptic fragments by reversed-phase high-performance liquid chromatography (RP-HPLC) were performed as described previously (21). N-terminal and internal amino acid sequences of the purified enzyme were determined with an Applied Biosystems model 492 Procise protein-sequencing system at the Korea Basic Science Institute (Daejon, Korea).
Cloning and DNA sequencing of the TMR gene.
Chromosomal DNA, isolated from Citrobacter sp. strain KCTC 18061P as described previously (15), was used as the template for PCR. PCR conditions were five cycles at 94°C for 40 s, 37°C for 40 s, and 72°C for 1 min. This was followed by 30 cycles at 94°C for 40 s, 55°C for 40 s, and 72°C for 1 min. PCR products were subcloned into the pGEM-T Easy vector (Promega), and their nucleotide sequences were determined. Hybridization and detection were performed using the DIG DNA Labeling and Detection kit (Roche Applied Science, IN) according to the manufacturer's instructions. Clones showing a significant reaction to the probe were selected. Plasmid DNA prepared from these clones was subcloned into the corresponding sites of the pUC119 or pBluescript SK(+) vector and then sequenced. Analysis of sequence data and sequence similarity searches were performed using the BLAST(N) program of the National Center for Biotechnology Information (NCBI).
Expression of the TMR gene in E. coli.
The entire open reading frame (ORF) of TMR was amplified by PCR using Pyrobest DNA polymerase (Takara Biomedicals, Tokyo, Japan). PCR was undertaken with a sense P1 primer that contained a unique NdeI restriction site (underlined) and an ATG initiation codon (boldfaced) (5′-CTCATATGTCAATTGCGGTTACAGGTGCTAC-3′) and an antisense P3 primer that contained a unique XhoI restriction site (underlined) (5′-CACTCGAGTTACATTTTCAGGGCTTGTTTTACGG-3′). The amplified gene fragment (0.9 kb) was inserted into the SmaI site of the pUC119 vector, yielding pUTMR. The nucleotide sequence of the inserted fragment was then confirmed by DNA sequencing. The NdeI/XhoI fragment of pUTMR was introduced into the corresponding sites of the pET-29a(+) plasmid, resulting in pET-TMR. Escherichia coli BL21(DE3) cells transformed with pET-TMR were grown in LB medium containing kanamycin (50 μg/ml) at 37°C with shaking. Protein production was induced by addition of 0.5 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) when the culture turbidity at 660 nm was 0.8.
Chemicals.
Crystal violet, malachite green, basic fuchsin, brilliant green, bromophenol blue, methyl violet, Congo red, methyl red, leuco crystal violet, leucomalachite green, FAD, NAD, and NADPH were purchased from Sigma (St. Louis, MO). All other chemicals were of reagent grade and were also purchased from Sigma.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper was deposited in the GenBank database under accession number AY756172.
RESULTS AND DISCUSSION
Purification and characterization of TMR.
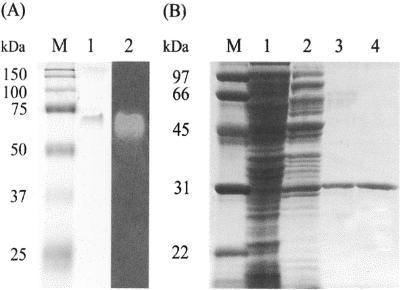
As shown in Table 1, the enzyme was purified 100-fold over the crude extract with an activity yield of 6.6%, and the specific activity of the final enzyme preparation was 50 U/mg. The purified enzyme was analyzed by native PAGE and SDS-PAGE. After native PAGE, the mobility of the active band, as determined by the staining of crystal violet-decolorizing activity, coincided with that of a single protein band with a molecular mass of 62 kDa when stained with Coomassie brilliant blue (Fig. 1A). The relative molecular mass of the pure enzyme under native conditions was determined to be 62 kDa by size exclusion chromatography (data not shown). However, SDS-PAGE of the purified enzyme produced a single band with a molecular mass of 31 kDa (Fig. 1B, lane 4). These results indicate that this enzyme has a homodimeric structure with a subunit size of about 31 kDa. This differs from the sizes of monomeric azoreductases that catalyze the NADPH-dependent reduction of azo dyes to aromatic amines (5, 6, 14, 17, 22).
TABLE 1.
Purification of TMR from Citrobacter sp. strain KCTC 18061P and recombinant E. coli TMR
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Citrobacter sp. TMR | |||||
| Crude extract | 1,300 | 610 | 0.5 | 100 | 1.0 |
| HiPrep Q XL | 200 | 550 | 2.8 | 90 | 5.6 |
| HiTrap phenyl Sepharose | 2.2 | 64 | 29 | 11 | 58 |
| Mono Q HR | 0.8 | 40 | 50 | 6.6 | 100 |
| Recombinant TMR | |||||
| Crude extract | 100 | 2,900 | 29 | 100 | 1.0 |
| HiPrep Q XL | 25 | 900 | 36 | 31 | 1.2 |
| HiTrap phenyl Sepharose | 1.7 | 110 | 65 | 3.8 | 2.2 |
| Mono Q HR | 1.0 | 77 | 77 | 2.7 | 2.7 |
FIG. 1.
Nondenaturing PAGE (A) and SDS-PAGE (B) of the protein obtained after purification of TMR from Citrobacter sp. strain KCTC 18061P. (A) The sample after the final Superdex 200 HR column chromatography analyzed by native PAGE (lane 1) and activity staining with crystal violet (lane 2). (B) Protein obtained from each purification step was analyzed by SDS-PAGE. Lane 1, 38 μg of protein from crude extract; lane 2, 16 μg of protein from the HiPrep Q XL column step; lane 3, 0.8 μg of protein from the HiTrap phenyl Sepharose column step; lane 4, 0.6 μg of protein from the Mono Q HR column step. Lanes M, marker proteins.
The purified enzyme showed optimal catalytic activity at pH 9.0 and 60°C. The standard assay was linear at temperatures up to 50°C. TMR substrate specificity was investigated with the following substrates: crystal violet, malachite green, brilliant green, basic fuchsin, and bromophenol blue as triphenylmethanedyes; methyl red and Congo red as azo dyes; and NADH and NADPH as cosubstrates. As shown in Table 2, the specific activity of the enzyme was highest with malachite green, reaching 115 μmol decolorized mg of protein−1 min−1. Specific activities directed toward crystal violet and basic fuchsin were 41% and 35% of that for malachite green, respectively. The Vmax/Km value of the enzyme for malachite green was 623, whereas the values for crystal violet and basic fuchsin were 51 and 20, respectively. Brilliant green, which contains diethyl groups instead of dimethyl groups in the side chains, was not decolorized by the enzyme; nor was bromophenol blue, which contains no N-alkyl groups in the side chains. This substrate specificity indicates that the accessibility of triphenylmethane dyes to TMR is dependent on the chemical structures of the dyes. More-detailed biochemical and structural studies are required to understand these results. The enzyme also failed to decolorize methyl red and Congo red, which are azo dyes containing an azo group (—N=N—). Although TMR, like azoreductases, uses either NADPH or NADH as a cofactor, the specific activity of TMR for NADPH was only 23% of that observed for NADH, and its efficiency (Vmax/Km) for NADH was about 19-fold greater than that for NADPH (Table 2). Many reductases contain a tightly bound flavin prosthetic group, whereas others contain a bound pyridine nucleotide (24). The purified enzyme exhibited an absorption peak at 280 nm but no peaks in the range of 300 to 700 nm. This indicates that the enzyme does not contain any tightly bound flavin or heme cofactors, because both have absorption peaks in the 300-to-700-nm region (24). Taken together, these results indicate that TMR is a homodimeric flavin-free enzyme that preferentially uses NADH as a cofactor.
Identification of the reaction products.
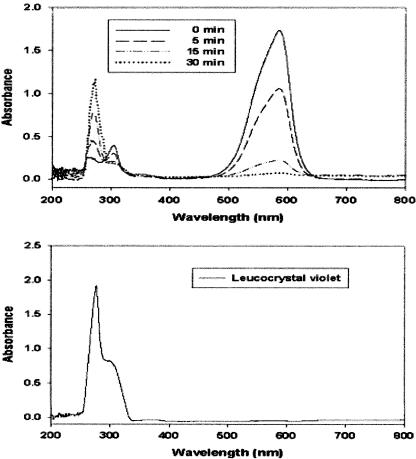
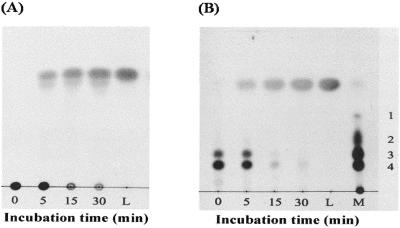
Triphenylmethane dyes such as malachite green and crystal violet are converted to their colorless leuco derivatives by intestinal microflora and several anaerobic bacteria (12, 16). UV-visible spectral analysis showed the disappearance of crystal violet, which has an absorbance maximum at 590 nm, and the concomitant appearance of a product with an absorbance maximum at 260 nm, which is characteristic of authentic leuco crystal violet (Fig. 2). TLC analysis of the reaction products showed that during incubation, the amount of crystal violet in the extract decreased whereas the amount of product increased (Fig. 3). This product comigrated with authentic leuco crystal violet. These results indicate that TMR catalyzes the reduction of crystal violet to leuco crystal violet in the presence of NADH as the electron donor. The reduction of malachite green to its leuco form by TMR was also detected by UV-visible spectral and TLC analyses (data not shown).
FIG. 2.
(Upper panel) Spectral changes observed during conversion of crystal violet to leuco crystal violet by the purified TMR. Spectral changes were recorded after enzymatic reactions for the indicated times (see the key). (Lower panel) The spectrum of authentic leuco crystal violet (20 μM) dissolved in dichloromethane is shown as the standard.
FIG. 3.
Thin-layer chromatogram of the reaction product of crystal violet with TMR. Crystal violet was incubated with the purified enzyme, and the reaction products were sampled at various time points and analyzed by TLC using n-hexane-ethyl acetate (4:6, vol/vol) (A) and propanol-water-glacial acetic acid (90:9:1, vol/vol/vol) (B) as solvents. Lanes L, authentic leuco crystal violet (the standard); lane M, methyl violet dissolved in methanol. Numbers on the right of panel B are as follows (7): 1, N,N′,N"-trimethylpararosaniline; 2, N,N,N′,N"-tetramethylpararosaniline; 3, N,N,N′,N′,N"-pentamethylpararosaniline; 4, N,N,N′,N′,N",N"-hexamethylpararosaniline (crystal violet).
N-terminal amino acid sequencing.
To determine the N-terminal amino acid sequence of purified TMR, a sample of the enzyme was subjected to SDS-PAGE and blotted onto a polyvinylidene difluoride membrane. The N-terminal fragment was sequenced by automated Edman degradation, and the first 12 amino acids were identified (Table 3). To obtain additional information about the sequence, the purified protein was digested with trypsin and the internal peptide fragments were separated by RP-HPLC. The sequences of three fragments were thus determined (Table 3). They showed no homology to the sequences of any proteins of known function in the protein database.
TABLE 3.
Sequences of the N terminus, tryptic peptides, and deduced primers
| Peptide | Amino acid sequencea | Deduced primer sequenceb |
|---|---|---|
| N terminus | SIAVTGATGOLG | Primer 1, 5′-GCIGTNACNGGIGCIACNGGNCA(A/G)CT-3′ |
| 13 | HIAYTGYAFAE | Primer 2, 5′-AAIGC(A/G)TAICCNGT(A/G)TANGCNAT(A/G)TG-3′ |
| 24 | ASTLADOGVEVR | Primer 3, 5′-AC(C/T)TCIACICC(C/T)TG(A/G)TCNGCNAGNGT-3′ |
| 35 | VPASOIIAIVR | Primer 4, 5′-ACIATIGCIATIAT(C/T)TG(A/G)CTNGCNGG-3′ |
The underlined sequences of the N terminus and tryptic peptides from the purified TMR were used for the design of oligonucleotides for PCR.
I and N indicate inosine residues and each of four nucleotides, respectively.
Cloning and sequencing of the TMR gene.
To further characterize the structure and function of TMR, we cloned and sequenced the structural gene for TMR. Oligonucleotide primers (Table 3) designed from the N-terminal amino acid sequence of the purified enzyme and internal peptide fragments were used in PCR to amplify a portion of the TMR gene from Citrobacter sp. strain KCTC 18061P. Because the order of the internal peptides in the enzyme was unknown, it was necessary to test all possible combinations of sense primers from the N-terminal sequence with antisense primers from the internal peptides in PCR. PCR products amplified by primer combinations were subcloned and sequenced. An estimated 0.3-kb fragment amplified by a combination of primers 1 and 2 included the sequences of two peptides located in the N-terminal region of the enzyme, indicating that this PCR product was a portion of the TMR gene. This 320-bp fragment was labeled using digoxigenin and used as a probe to isolate the complete TMR gene. Southern hybridization of total genomic DNA digested with restriction enzymes using this probe gave hybridized bands ranging from 3.0 kb to16.0 kb (data not shown). An estimated 3.0-kb PstI fragment was eluted from a gel slice, ligated to the PstI site of pBluescript SK(+), and further screened by colony hybridization. A single positive colony containing a plasmid (pTMR) with a 3.0-kb insert was isolated and then subcloned into the pBluescript SK(+) vector for DNA sequencing. Nucleotide sequence analysis revealed an ORF starting with an ATG codon at nucleotide 241 and ending at a TAA codon at nucleotide 1102, thus encoding a protein of 287 amino acids (data not shown). A potential ribosome-binding sequence, AAGGAG, similar to that found in E. coli, was found 8 bp upstream from the ATG initiation codon. The deduced amino acid sequence extending from this initiation codon showed good agreement with the N-terminal amino acid sequence determined from purified native TMR, described above. Three internal tryptic peptides derived from the purified enzyme were also encoded by the deduced amino acid sequence of the ORF. The molecular mass of the deduced polypeptide was calculated to be 30,954 Da, which agrees closely with the 31 kDa determined by SDS-PAGE of the purified enzyme (Fig. 1B). These results indicate that the entire nucleotide sequence of the TMR gene was isolated.
Comparative sequence analyses using the BLAST program showed that TMR has 99% identity with the conserved hypothetical protein encoded in the genome sequence of Listeria monocytogenes strain 4b H7858 deposited in the NCBI database (GenBank accession no. ZP_00230411), even though it has no overall sequence similarity to any known protein, including other bacterial reductases. However, the amino acid sequence of the N-terminal region of TMR shows significant similarity to those of pyridine nucleotide-dependent enzymes in that it contains the characteristic motif of three conserved glycines and six conserved hydrophobic residues. The TMR sequence fulfils the 11 criteria for a nucleotide-binding sequence established by Wierenga et al. (25). An exception is the additional residue between the first and second glycines in the fingerprint sequence (GXXGXXG). This additional residue is also present in malate dehydrogenase, UDP-galactose 4-epimerase, and dihydrodipicolinate reductase, the tertiary structures of which are known to contain the classic NADP-binding βαβ-fold (3, 4, 19). This motif (GXXGXXG) is also found near the N termini of NADPH-dependent enzymes such as azoreductase (5), flavin reductase (18), and phenylcoumaran benzylic ether reductase (9). Moreover, a search of the NCBI conserved-domain database showed that this motif, which corresponds to amino acid residues 7 to 13 in TMR, is fully conserved in the isoflavone reductase family (pfam02716), the predicted nucleotide diphosphate-sugar epimerase family (COG0702), and the putative NADH-flavin reductase family (COG2910). These findings suggest that this motif of TMR is involved in NADP binding to facilitate enzyme activity.
Expression and characterization of TMR in E. coli.
When E. coli BL21(DE3) cells carrying pET-TMR were induced for 3 h with 0.5 mM IPTG at 37°C, TMR activity was detected in the soluble fraction. However, it was not detected in the extracellular or insoluble fractions. Furthermore, a predominant band corresponding to the expected size (31 kDa) of the recombinant enzyme was also observed in the soluble fraction of induced cells (data not shown). The expressed enzyme constituted about 30% of the total soluble proteins in the intracellular fraction. These results indicate that the recombinant enzyme was efficiently overexpressed in a soluble and active form in the cytoplasm of E. coli. The recombinant enzyme was purified from the intracellular fraction of E. coli by using a protocol similar to that used for the native enzyme and was characterized biochemically. The process resulted in 2.7-fold purification over the sonicated extract with 2.7% recovery of total activity; the final specific activity of the purified enzyme was 77 U/mg (Table 1). The recombinant enzyme displayed physical characteristics and kinetic parameters similar to those observed for the native enzyme.
In conclusion, we have described both the first TMR to be characterized from any organism and the first biochemical characterization of an enzyme responsible for the decolorization of triphenylmethane dyes in bacteria. We also report for the first time the molecular cloning and heterogeneous expression of the TMR gene. Moreover, we have succeeded in inducing the overexpression of recombinant TMR with enzymatic properties similar to those of the native enzyme. The availability of abundant quantities of recombinant TMR should permit more-detailed studies of the structure and function of this enzyme. X-ray crystallographic analysis of this enzyme is now in progress using the purified recombinant protein.
Acknowledgments
This work was supported by the 21C Frontier Microbial Genomics and Applications Center Program, Ministry of Science and Technology (grant MG02-0301-005-2-2-0 to Y.-C. Lee), Republic of Korea. Y.-M. Lee received a fellowship from the Brain Busan 21 Project.
REFERENCES
- 1.An, S.-Y., S.-K. Min, I.-H. Cha, Y.-L. Choi, Y.-S. Cho, C.-H. Kim, and Y.-C. Lee. 2002. Decolorization of triphenylmethane and azo dyes by Citrobacter sp. Biotechnol. Lett. 24:1037-1040. [Google Scholar]
- 2.Azmi, W., R. K. Sani, and U. C. Banerjee. 1998. Biodegradation of triphenylmethane dyes. Enzyme Microb. Technol. 22:185-191. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, A. J., I. Rayment, P. A. Frey, and H. M. Holden. 1992. The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5 Å resolution. Proteins 12:372-381. [DOI] [PubMed] [Google Scholar]
- 4.Birktoft, J. J., G. Rhodes, and L. J. Banaszak. 1989. Refined crystal structure of cytoplasmic malate dehydrogenase at 2.5 Å resolution. Biochemistry 28:6065-6081. [DOI] [PubMed] [Google Scholar]
- 5.Blümel, S., H.-J. Knackmuss, and A. Stolz. 2002. Molecular cloning and characterization of the gene coding for the aerobic azoreductase from Xenophilus azovorans KF46F. Appl. Environ. Microbiol. 68:3948-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blümel, S., and A. Stolz. 2003. Cloning and characterization of the gene coding for the aerobic azoreductase from Pigmentiphaga kullae K24. Appl. Microbiol. Biotechnol. 62:186-190. [DOI] [PubMed] [Google Scholar]
- 7.Bumpus, J. A., and B. J. Brock. 1988. Biodegradation of crystal violet by the white rot fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 54:1143-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, D. 1997. Recent advances on the molecular genetics of ligninolytic fungi. J. Biotechnol. 53:273-289. [DOI] [PubMed] [Google Scholar]
- 9.Gang, D. R., H. Kasahara, Z.-Q. Xia, K. Vander Mijnsbrugge, G. Bauw, W. Boerjan, M. Van Montagu, L. B. Davin, and N. G. Lewis. 1999. Evolution of plant defense mechanisms: relationships of phenylcoumaran benzylic ether reductases to pinoresinol-lariciresinol and isoflavone reductases. J. Biol. Chem. 274:7516-7527. [DOI] [PubMed] [Google Scholar]
- 10.Gold, M. H., and M. Alic. 1993. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol. Rev. 57:605-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory, P. 1993. Dyes and dyes intermediates, p. 544-545. In J. I. Kroschwitz (ed.), Encyclopedia of chemical technology, vol. 8. John Wiley & Sons, New York, N.Y. [Google Scholar]
- 12.Henderson, A. L., T. C. Schmitt, T. M. Heinze, and C. E. Cerniglia. 1997. Reduction of malachite green to leucomalachite green by intestinal bacteria. Appl. Environ. Microbiol. 63:4099-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Maier, J., A. Kandelbauer, A. Erlacher, A. Cavaco-Paulo, and G. M. Gubitz. 2004. A new alkali-thermostable azoreductase from Bacillus sp. strain SF. Appl. Environ. Microbiol. 70:837-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 16.McDonald, J. J., and C. E. Cerniglia. 1984. Biotransformation of gentian violet to leucogentian violet by human, rat, and chicken intestinal microflora. Drug Metab. Dispos. 12:330-336. [PubMed] [Google Scholar]
- 17.Moutaouakkil, A., Y. Zeroual, F. Z. Dzayri, M. Talbi, K. Lee, and M. Blaghen. 2003. Purification and partial characterization of azoreductase from Enterobacter agglomerans. Arch. Biochem. Biophys. 413:139-146. [DOI] [PubMed] [Google Scholar]
- 18.Quandt, K. S., and D. E. Hultquist. 1994. Flavin reductase: sequence of cDNA from bovine liver and tissue distribution. Proc. Natl. Acad. Sci. USA 91:9322-9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scapin, G., J. S. Blanchard, and J. C. Sacchettini. 1995. Three-dimensional structure of Escherichia coli dihydrodipicolinate reductase. Biochemistry 34:3502-3512. [DOI] [PubMed] [Google Scholar]
- 20.Shin, K.-S., and C.-J. Kim. 1998. Decolorization of artificial dyes by peroxidase from the white-rot fungus Pleurotus ostreatus. Biotechnol. Lett. 20:569-572. [Google Scholar]
- 21.Stone, K. L., M. B. LoPresti, J. M. Crawford, R. DeAngelis, and K. R. Williams. 1989. Enzymatic digestion of proteins and HPLC peptide isolation, p. 31-47. In P. T. Matsudaira (ed.), A practical guide to protein and peptide purification for microsequencing. Academic Press, Inc., San Diego, Calif.
- 22.Suzuki, Y., T. Yoda, A. Ruhul, and W. Sugiura. 2001. Molecular cloning and characterization of the gene coding for azoreductase from Bacillus sp. OY1-2 isolated from soil. J. Biol. Chem. 276:9059-9065. [DOI] [PubMed] [Google Scholar]
- 23.Vasdev, K., R. C. Kuhad, and R. K. Saxena. 1995. Decolorization of triphenylmethane dyes by the birds nest fungus Cyathus bulleri. Curr. Microbiol. 30:269-272. [Google Scholar]
- 24.Walsh, C. 1979. Enzymatic reaction mechanisms. W. H. Freeman and Company, New York, N.Y.
- 25.Wierenga, R. K., P. Terpstra, and W. G. J. Hol. 1986. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 187:101-107. [DOI] [PubMed] [Google Scholar]