Abstract
Escherichia coli O157:H7 carried on plant surfaces, including alfalfa sprouts, has been implicated in food poisoning and outbreaks of disease in the United States. Adhesion to cell surfaces is a key component for bacterial establishment and colonization on many types of surfaces. Several E. coli O157:H7 surface proteins are thought to be important for adhesion and/or biofilm formation. Therefore, we examined whether mutations in several genes encoding potential adhesins and regulators of adherence have an effect on bacterial binding to plants and also examined the role of these genes during adhesion to Caco-2 cells and during biofilm formation on plastic in vitro. The genes tested included those encoding adhesins (cah, aidA1, and ompA) and mediators of hyperadherence (tdcA, yidE, waaI, and cadA) and those associated with fimbria formation (csgA, csgD, and lpfD2). The introduction of some of these genes (cah, aidA1, and csg loci) into an E. coli K-12 strain markedly increased its ability to bind to alfalfa sprouts and seed coats. The addition of more than one of these genes did not show an additive effect. In contrast, deletion of one or more of these genes in a strain of E. coli O157:H7 did not affect its ability to bind to alfalfa. Only the absence of the ompA gene had a significant effect on binding, and the plant-bacterium interaction was markedly reduced in a tdcA ompA double mutant. In contrast, the E. coli O157:H7 ompA and tdcA ompA mutant strains were only slightly affected in adhesion to Caco-2 cells and during biofilm formation. These findings suggest that some adhesins alone are sufficient to promote binding to alfalfa and that they may exist in E. coli O157:H7 as redundant systems, allowing it to compensate for the loss of one or more of these systems. Binding to the three types of surfaces appeared to be mediated by overlapping but distinct sets of genes. The only gene which appeared to be irreplaceable for binding to plant surfaces was ompA.
The majority of infectious diseases are initiated by the adhesion of pathogenic organisms to the tissues of the host. This is considered the first stage in any infectious process and is an important and critical step for colonization (30). Many human pathogens, in addition to adhering to different mammalian cells, are capable of interacting with other surfaces outside their host, i.e., plants and fruits, and can live in association with other bacteria while obtaining nutrients for their survival (22). Food-borne illnesses present a significant health problem throughout the world, and in the United States alone, the Centers for Disease Control and Prevention (CDC) estimates that there are 76 million illnesses and 5,000 deaths annually (29). In recent years, the reported incidence of food poisoning caused by food-borne pathogens reported to the CDC has decreased (8). However, further efforts are needed to sustain these declines, to reduce pathogens in food animal reservoirs, and to prevent the contamination of produce. Among the outbreaks associated with fresh produce consumption and for which the etiology has been determined, Salmonella enterica serovar Enteritidis accounted for the largest number of outbreaks, cases, and deaths, followed by Escherichia coli O157:H7, which was frequently found during multistate outbreaks (42).
E. coli O157:H7 is a human pathogen which was first identified during two independent outbreaks of hemorrhagic colitis in 1982 (23, 36). E. coli O157:H7 colonizes the intestine and can cause a diarrheal syndrome characterized by a copious bloody discharge. In pediatric and elderly patients, the diarrhea caused by this strain can be fatal due to acute kidney failure (hemolytic-uremic syndrome) (reviewed in reference 31). Although cattle are known to be a reservoir of E. coli O157:H7 (9, 51) and contaminated undercooked beef has been most frequently implicated as a vehicle for pathogen transmission (15), the organism has also been associated with unexpected vehicles of transmission, such as alfalfa sprouts (5), radish sprouts (20), lettuce (1), and unpasteurized apple juice (10). E. coli O157:H7 contamination of fruits and vegetables is a concern in the United States, and its presence in a variety of food products suggests that this pathogen has the ability to express one or more adhesive structures which allow binding to surfaces of many different types of food.
E. coli O157:H7 expresses multiple fimbrial and nonfimbrial adhesins which have no obvious role in pathogenesis and may be involved in adhesion to other surfaces (reviewed in reference 46). For example, curli-expressing thin aggregative fimbriae, which are rarely reported in E. coli O157:H7 compared with other pathogenic E. coli strains (47, 48), have been reported to bind eukaryotic extracellular matrix proteins as well as to enhance the formation of E. coli O157:H7 biofilms on inert surfaces (11, 38). Biofilm formation may increase E. coli O157:H7 survival and would likely result in protection against many environmental conditions. Recently, Cah (calcium binding antigen 43 homologue), a surface-expressed and heat-extractable protein that causes autoaggregation and changes in bacterial shape, was characterized in E. coli O157:H7 and found to participate in the formation of biofilms (45). Finally, multiple mediators of adherence were identified during the screening of an E. coli O157:H7 transposon-insertion mutant library for hyperadherent strains (44). This analysis involved the isolation of several transposon mutant strains which displayed different levels of adhesion to eukaryotic cells and allowed the identification of OmpA (outer membrane protein A) as a novel adhesin of E. coli O157:H7.
The adhesion of E. coli O157:H7 is a complex process, and it is unlikely that this organism relies on one mechanism for attachment to different surfaces (e.g., human or cattle intestinal cells, food products, or abiotic surfaces). In its most basic form, bacterial adhesion can be divided into the following two stages: the primary or docking stage and the secondary or locking phase (16). Primary adhesion results from a serendipitous meeting between a conditioned surface and a planktonic microorganism. This stage is reversible and is dictated by a number of physiochemical variables (electrostatic and hydrophobic interactions, temperature, hydrodynamic forces, etc.) that define the interaction between the bacterial cell surface and the conditioned surface of interest. Previous work has shown that E. coli O157:H7 expresses low surface hydrophobicity (24) and that the hydrophobic components on the surface can be masked by extracellular polymeric substances, such as the capsule, influencing the ability of E. coli O157:H7 to adhere to different surfaces (19). The second stage of adhesion employs molecularly mediated binding between specific adhesins and the surface. Once bacteria have irreversibly attached to a surface, the process of biofilm maturation begins (16). Since very little is known about the role of specific surface structures or regulatory proteins during binding of E. coli O157:H7 to food products, we tested whether a collection of E. coli O157:H7 strains carrying mutations in genes encoding proteins associated with binding to tissue culture cells also participate in the association with fresh produce. Furthermore, we determined the role of different mediators of adherence in the ability of E. coli O157:H7 to bind alfalfa sprouts and seed coats and attempted to correlate differences in adherence to plant surfaces with those observed on Caco-2 intestinal cells and during the formation of biofilms on plastic in vitro.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used for this study are listed in Table 1. Strains were routinely grown in Luria-Bertani (LB) broth or on L agar at 37°C (27). When indicated, the strains were grown in Dulbecco's modified Eagle's medium (Cellgro; Mediatech, Inc., Herndon, VA), M63 minimal glucose medium, or Congo red agar (Trypticase soy broth agar plus 0.01% Congo red dye) (35). Antibiotics (Sigma-Aldrich, Co., St. Louis, MO) were added to media at the following concentrations: kanamycin (Km), 50 μg/ml; ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 30 μg/ml; streptomycin (Sm), 100 μg/ml; tetracycline (Tc), 12.5 μg/ml; nalidixic acid (Nal), 30 μg/ml; and neomycin, 20 μg/ml in liquid and 60 μg/ml in solid media.
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| 86-24 | E. coli O157:H7 (41), Smr Nalr | A. G. Torres stock collection |
| AGT103 | 86-24 cah::cat cah::tet, Smr Cmr Tcr | 45 |
| AGT601 | 86-24 ompA::cat, Smr Cmr | 44 |
| AGT602 | 86-24 ompA::cat tdcA::Tnp, Smr Cmr Kmr | 44 |
| 86-24A | 86-24 csgA, Smr Apr | 21 |
| P9C8B1 | 86-24 csgD::Tnp, Smr Kmr | 44 |
| P10C9E1 | 86-24 yidE::Tnp, Smr Kmr | 44 |
| P9C8F2 | 86-24 tdcA::Tnp, Smr Kmr | 44 |
| P6C6E25 | 86-24 waaI::Tnp, Smr Kmr | 44 |
| P9C12D4 | 86-24 cadA::Tnp, Smr Kmr | 44 |
| P9C8F1 | 86-24 lpfD2::Tnp, Smr Kmr | 44 |
| AGT103A | 86-24 cah::cat cah::tet csgA, Smr Cmr Tcr Apr | 21 |
| DH5α pro | endA 1 hsdR17(rK− mK−) supE44 thi-1 recA 1 gyrA relA1 Δ(lacZYA-argF) U169deoR[f80dlac Δ(lacZ)M15] | Clontech |
| ER2267 | F′ proA+B+laclq Δ(lacZ)M15 zzf::mini-Tn10 (Kanr)/Δ(argF-lacZ)U169 glnV44 e14−(McrA−) rfbD1? recA1 relA1? endA1 spoT1? thi-1 Δ(mcrC-mrr)114::IS10, Kmr | New England Biolabs |
| Plasmids | ||
| pCAH100 | cah from E. coli O157:H7 strain EDL933 in EcoRV site of pACYC184, Tcr | 45 |
| pIB264 | 11.0-kb EcoRI fragment containing aidA gene in pBR322, Apr Tcr | 4 |
| pRMinv | 8.5-kb BamHI-SalI fragment containing csg cluster in pMMB33, Neor | 17 |
Recombinant DNA techniques.
Plasmid DNAs were isolated using a QIAGEN QIAprep plasmid preparation kit from 3 ml of overnight bacterial culture following the manufacturer's procedure. Plasmids were introduced by the transformation of RbCl2 chemically competent E. coli K-12 strains or by electroporation of enterohemorrhagic E. coli (EHEC) O157:H7 as described by Dower et al. (14). Standard molecular methods were used for restriction endonuclease analyses and the ligation and construction of recombinant plasmids (27).
Binding to alfalfa sprouts and seed coat assays.
Measurements of bacterial binding to alfalfa sprouts were carried out as previously described (21). Briefly, alfalfa sprouts were obtained by germinating seeds. In order to ensure that the alfalfa sprouts were axenic, seeds were surface sterilized by being soaked briefly in 80% ethanol followed by 20 min in a 2.6% sodium hypochlorite solution containing Tween 80 as a wetting agent. The seeds were then washed three times with sterile deionized water and germinated. After 1 day of germination, sprouts were placed in plastic dishes in groups of four with 5 ml of sterile deionized water. Bacteria were diluted and added to the germinated seeds to a final concentration of approximately 5 × 103 bacteria per ml. The inoculated sprouts were incubated at room temperature for 3 days. Sprouts and open seed coats were analyzed separately. Each was washed twice by vigorous inversions in 5 ml sterile deionized water. The sprouts or seed coats were then homogenized in 5 ml of washing buffer (26) in a Teflon glass homogenizer and plated to determine viable cell counts. The numbers given are the mean log10 values of the numbers of viable bacteria recovered from the homogenates of washed sprouts or seed coats ± standard deviations and were calculated per sprout or seed coat from a minimum of three experiments (26). The average sprout weighed about 50 mg, and the average opened seed coat weighed about 5 mg. In all cases, the number of bacteria recovered from the wash water (second wash), as measured by viable counts, was <1% of the number recovered from the sprout or seed coat. The statistical methods used are given above.
Bacterial adhesion to epithelial cells.
Caco-2 cells were seeded with 1 × 105 cells/well and incubated for 48 h at 37°C with 5% CO2 in 24-well plates (Corning, Inc., Corning, NY). The cell monolayers were washed twice with phosphate-buffered saline (pH 7.4), and the infection was carried out with wild-type bacteria and isogenic mutants as described previously (43). Briefly, bacterial strains were grown in LB broth overnight at 37°C, the monolayers were infected with 1 × 107 bacteria for 3 h, and adherence was evaluated qualitatively by Giemsa staining and quantitatively by plating adherent bacteria on L agar plates with an appropriate antibiotic. The results were performed in triplicate and repeated at least twice. P values were calculated using a paired t test.
Isolation of EHEC 86-24 mutant strains.
Mutants in csgA are described elsewhere (21). Briefly, the csgA gene was obtained by PCR and cloned into pUTgfpΔXho (28). The resulting plasmid was introduced into the E. coli O157:H7 strain by transformation, and cointegrates were selected using Apr. Transposon mutants of EHEC strain 86-24 (insertions in cadA, csgD, tdcA, lpf-2, waaI, and yidE) were generated with the kanamycin resistance-encoding transposome EZ::TN <R6Kγori/KAN-2> Tnp (Epicenter, Madison, WI), as previously described (44). Furthermore, disruption of the ompA gene was performed in the chromosomes of EHEC strains 86-24 (41) and P9C8F2 (Table 1) by the marker exchange approach known as the lambda red system, as described previously (44).
Biofilm assay.
The growth of biofilms in LB and M63 minimal glucose media was analyzed by the method optimized by Torres et al. (45). Bacterial strains were grown in polyvinyl chloride (PVC) 96-well plates (Falcon; Becton Dickinson, San Jose, CA) without shaking for 24 h in LB broth at 30°C. Next, the cells were diluted 1:100 into PVC plates containing either LB or M63 medium supplemented with 0.8% glucose. Strains were grown statically for an additional 24 h at 30°C, and the strains in M63 medium were diluted (1:100) again in fresh M63-glucose and incubated for 24 h at 30°C without shaking. The biofilms formed in LB or M63 minimal glucose medium were rinsed and stained with a solution of 1% (wt/vol) crystal violet. Biofilm assays with E. coli strains DH5α and ER2267 were performed with LB medium only because the DH5α strain displayed a growth-defective phenotype in M63 medium. To quantify the formation of biofilms, the crystal violet-stained cells were solubilized in dimethyl sulfoxide, and the samples were removed and transferred to a 96-well polystyrene plate. The optical density (570 nm) of each crystal violet-stained sample was determined using a microtiter plate reader. The values of at least six independently grown biofilms were used to calculate the average of each sample. P values were calculated by a paired t test.
RESULTS
E. coli adhesins mediate binding to alfalfa.
Since curli and the Cah protein have been associated with adhesion to inert surfaces in E. coli O157:H7 (11, 45) and since these proteins are either absent or not expressed in E. coli K-12 strains (6, 17, 21, 45), we examined whether these structures were able to mediate E. coli K-12 binding to alfalfa sprouts and seed coats. We first determined whether E. coli K-12 strains express curli or the Cah protein under the conditions used to infect alfalfa (incubating samples at room temperature for 2 to 3 days). The E. coli K-12 strains formed white colonies on Congo red agar plates, which was indicative of their inability to express curli. Furthermore, the K-12 strains tested do not carry the cah gene. Instead, they possess a homologue gene (agn-43). However, we have shown that Ag43 does not interfere with the function of Cah and seems to be repressed when Cah is overexpressed (45). In addition, Ag43 only contributes to E. coli biofilm formation when the bacteria are grown in minimal glucose medium and not when they are grown in Luria-Bertani broth (see below). As shown in Table 2, alfalfa was inoculated with the K-12 strains DH5α pro and ER2267 expressing either Cah (pCAH100) or curli (pRMinv). After incubation, bacteria were recovered from washed sprouts and seed coats. We found that the expression of Cah or curli enhanced binding to alfalfa, regardless of the E. coli K-12 strain used in the assay. In contrast, we were unable to recover bacteria from alfalfa inoculated with the DH5α pro or ER2267 strain alone. The Cah protein belongs to a family of autotransporters associated with adherence which includes Ag43 and AidA-I. Since AidA-I mediates bacterial attachment to a broad variety of human and other mammalian cells, participates in biofilm formation, and is only associated with some diarrheagenic E. coli strains (39), we determined whether AidA-I mediates binding to alfalfa to the same extent as Cah. We found that pIB264 (aidA+) also mediates binding to alfalfa sprouts and seed coats, but not as well as pCAH100 (cah+). The plasmid gene products encoded in plasmids pBI264 and pRMinv have previously been shown to be expressed in and to mediate the adhesion of K-12 strains to mammalian cells (4, 17). The next experiment was designed to determine the effect of a combination of plasmids expressing the different surface structures on the binding phenotype. As shown in Table 2, there were no significant differences in the total numbers of bacteria on seeds or sprouts for E. coli K-12 expressing either one or two adhesins, indicating that their expression did not have an additive effect on alfalfa binding. Since our data suggest that multiple factors influence the binding of E. coli K-12 to alfalfa, we determined whether these and other factors had an impact on the binding phenotype of E. coli O157:H7.
TABLE 2.
Binding to sprouts and seed coats of E. coli K-12 strains alone or with plasmids containing genes associated with adhesion and biofilm formation
| Bacterial strain [phenotype] | Mean log10 no. (± SD) of bacteria recovered per sprout or seed coat after 3 days of incubation
|
|
|---|---|---|
| Sprouts | Seed coats | |
| DH5α pro | 0a | 0a |
| DH5α pro(pCAH100) [cah1+] | 4.9 ± 0.6 | 4.7 ± 0.7 |
| DH5α pro(pIB264) [aidA1+] | 2.2 ± 0.3 | 2.0 ± 1.0 |
| DH5α pro(pRMinv) [csg+] | 3.7 ± 0.6 | 4.1 ± 0.5 |
| DH5α pro(pIB264)(pCAH100) [aidA1+cah1+] | 3.3 ± 0.2 | 3.7 ± 0.5 |
| DH5α pro(pIB264)(pRMinv) [aidA1+csg+] | 3.8 ± 0.4 | 4.8 ± 0.4 |
| DH5α pro(pCAH100)(pRMinv) [cah1+csg+] | 2.7 ± 0.6 | 2.8 ± 0.3 |
| ER2267 | 0a | 0a |
| ER2267(pCAH100) [cah1+] | 5.8 ± 0.4 | 5.9 ± 0.3 |
| ER2267(pIB264) [aidA1+] | 4.1 ± 1.5 | 3.9 ± 1.4 |
| ER2267(pRMinv) [csg+] | 5.6 ± 0.2 | 5.8 ± 0.3 |
No bacteria were recovered.
Effects of mutations in genes encoding putative adhesins on binding of E. coli O157:H7 to alfalfa.
To analyze the role of Cah or curli in the binding of E. coli O157:H7 to alfalfa, sprouts were inoculated with the E. coli O157:H7 wild type or its isogenic mutant strains. As shown in Table 3, the isogenic strains AGT103 (cah1 cah2) and 86-24A (csgA) bound to alfalfa sprouts or seeds as well as the wild-type strain (86-24), indicating that single or double mutations in these genes did not have an effect on binding. Therefore, a triple mutant (AGT103A; cah1 cah2 csgA) was created, and the binding assay was repeated. Our results showed no differences in binding to sprouts and seed coats compared with the wild-type strain, suggesting that additional adhesins encoded in the genome of E. coli O157:H7 might be responsible for the binding phenotype of these mutant strains.
TABLE 3.
Binding of E. coli O157:H7 strain 86-24 and its mutants to alfalfa sprouts and seed coats
| Bacterial strain | Mutation or genotype | Mean log10 no. (± SD) of bacteria recovered per sprout or seed coat
|
|
|---|---|---|---|
| Sprouts | Seed coats | ||
| 86-24 | Wild type | 4.7 ± 0.6 | 5.6 ± 0.2 |
| AGT103 | cah1 cah2 | 3.8 ± 0.8 | 5.4 ± 0.5 |
| AGT601 | ompA | 3.2 ± 0.5 | 3.9 ± 1.5 |
| AGT602 | ompA tdcA | 0a | 0a |
| 86-24A | csgA | 4.1 ± 0.2 | 5.7 ± 0.2 |
| P9C8B1 | csgD | 5.8 ± 0.7 | 5.3 ± 0.5 |
| P10C9E1 | yidE | 5.4 ± 0.4 | 5.4 ± 0.1 |
| P9C8F2 | tdcA | 5.9 ± 0.1 | 5.9 ± 0.5 |
| P6C6E25 | waaI | 5.7 ± 0.1 | 5.4 ± 0.5 |
| P9C12D4 | cadA | 6.0 ± 0.3 | 6.1 ± 0.3 |
| P9C8F1 | lpfD2 | 6.2 ± 0.2 | 6.4 ± 0.4 |
| AGT103A | cah1 cah2 csgA | 4.7 ± 0.6 | 6.0 ± 0.2 |
No bacteria were recovered.
The expression of adhesion factors in E. coli O157:H7 appears to be tightly regulated under in vitro conditions (44). For this reason, we looked for factors increasing adhesion and recently isolated several mutants that adhere at higher levels than the wild-type strain to tissue culture cells (HeLa and Caco-2) (44). We tested this collection of mutants in the alfalfa binding assay, and as shown in Table 3, mutations in the mediators of hyperadherence encoded by tdcA, cadA, waaI, and lpfD2 caused a slight increase in binding to sprouts compared with that of the wild-type strain (mutations in yidE and csgD had no significant effect). None of the mutants was affected in binding to seed coats. cadA and lpfD2 mutants were recovered in the largest numbers from sprouts. Since we had previously shown that OmpA is an adhesin of E. coli O157:H7 which is differentially expressed during hyperadherence (44), the ompA mutant was assayed, and we found that strain AGT601 bound to sprouts at only about 10% the level of the wild-type strain (Table 3). This result suggests that OmpA has a role during binding to alfalfa. To confirm this observation, we tested the AGT602 strain containing mutations in the hyperadherence regulator TdcA and in OmpA, which has previously been shown to display reduced adherence to tissue culture cells (44). To our surprise, strain AGT602 showed no detectable binding to sprouts or seed coats, strongly indicating that OmpA and TdcA participate in the bacterium-plant interaction.
Role of hyperadherent mutants on binding of E. coli O157:H7 to Caco-2 cells.
It was previously shown that OmpA acts as an E. coli O157 surface protein mediating binding to different human-derived tissue culture cell lines (44). However, our previous studies to identify and characterize the other mediators of hyperadherence were performed with HeLa cells, an epithelial cell line obtained from a cervix adenocarcinoma that has been used extensively in the study of adherence patterns of E. coli strains, but which does not represent the actual epithelial cells that E. coli O157:H7 encounters during human infection. Since E. coli O157:H7 is a pathogen colonizing the large intestine, we were interested in determining whether these regulators play a role in adherence to intestinal epithelial cells. Therefore, we repeated the adherence experiments using Caco-2 cells, a colorectal adenocarcinomic cell line, and used this information to compare the abilities of these isogenic E. coli O157:H7 strains to mediate binding to alfalfa and to mammalian cells (Table 4). Our results confirmed our previous assays performed with HeLa cells indicating that transposon mutants with mutations in the mediators of adherence encoded by cadA, csgD, tdcA, yidE, waaI, and lpfD2 displayed an increase in adherence compared with the wild-type strain (the increase varied from 22% to 73% depending on the mutation; P < 0.05). In contrast, a slight reduction of adherence with no statistical significance was observed with the ompA and ompA tdcA mutants (the adherence of AGT601 was reduced 13.5% and that of AGT602 was reduced 17.3% compared with that of the wild-type strain; P > 0.05). Mutations in the cah genes did not have any effect on the adherence phenotype (P > 0.05). Our results are in accordance with our previous data for HeLa cells showing that E. coli O157:H7 carrying mutations in the mediators of adherence displayed increase adherence. The slight reduction in adherence of the ompA and ompA tdcA mutants during infections of Caco-2 cells did not reflect the striking reduction in binding of the E. coli O157:H7 ompA tdcA double mutant to alfalfa, which strongly suggests that OmpA acts as an accessory adhesin during binding to human-derived tissue culture cells but that OmpA and TdcA play an important role during binding to alfalfa sprouts and seed coats. Further experimentation is required to fully understand the role of OmpA and TdcA during binding to plant surfaces.
TABLE 4.
Binding of E. coli O157:H7 strain 86-24 and its mutants to Caco-2 cells
| Bacterial strain | Mutation(s) | % Bacteria recovered after 3 h of Caco-2 infection (mean ± SD) |
|---|---|---|
| 86-24 | None (wild type) | 100 ± 2.9 |
| AGT103 | cah1 cah2 | 97.8 ± 19.2a |
| AGT601 | ompA | 86.5 ± 15.9a |
| AGT602 | ompA tdcA | 82.7 ± 16.2a |
| P9C8B1 | csgD | 131.1 ± 4.9b |
| P10C9E1 | yidE | 146.1 ± 1.6b |
| P9C8F2 | tdcA | 147.8 ± 1.03b |
| P6C6E25 | waaI | 172.9 ± 2.9b |
| P9C12D4 | cadA | 144.8 ± 1.4b |
| P9C8F1 | lpfD2 | 122.0 ± 9c |
P > 0.05.
P < 0.01.
P < 0.05.
Multiple mediators of adherence participate in the formation of biofilms.
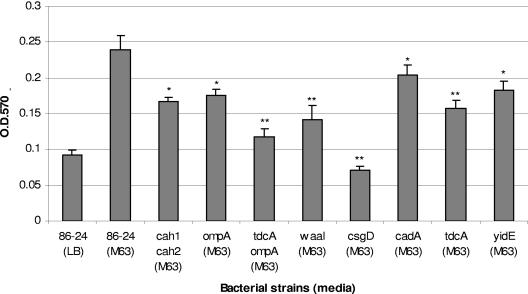
We determined whether the mediators of adherence, Cah, or the OmpA adhesin participates in the formation of E. coli O157:H7 biofilms when grown in minimal media. The E. coli wild-type strain 86-24 and several isogenic mutants with mutations in genes previously associated with adherence were tested using a crystal violet assay (12, 45). As previously shown, the wild-type strain 86-24 was able to form biofilms on PVC when grown in minimal glucose medium but was unable to form biofilms in LB broth (Fig. 1). We then compared the wild type and its isogenic mutants for the ability to form biofilms after growth in LB broth and found that none of the mutants tested showed any difference in biofilm formation compared to the wild-type strain (Fig. 1). We have previously shown that E. coli O157:H7 strains carrying mutations in either the flagellum apparatus or the quorum-sensing mechanism (both processes are important for biofilm formation in E. coli and other organisms) (50) did not show any difference in biofilm formation compared with the wild type when the strains were grown in Luria-Bertani broth (45). When the strains were grown in minimal glucose medium, we obtained significant differences between the biofilm formed by the wild type and those formed by the isogenic mutant strains. We have previously shown that an E. coli O157:H7 cah double mutant, AGT103, had a reduced ability to form biofilms compared with strain 86-24, and we proposed that the residual biofilm obtained with the mutant strain suggests that other proteins contribute to biofilm formation (Fig. 1) (45). To address this issue, we first determined whether mutations in the regulators of adherence had an effect on biofilm formation. All of the mutant strains tested (cadA, csgD, tdcA, waaI, and yidE) displayed an overall reduction in the ability to form biofilms. The csgD mutant (regulator of curli) had the largest effect on biofilm formation on PVC plates. We also observed that the ompA mutant strain (AGT601) had a reduced ability to establish biofilms compared to the AGT103 strain. Introduction of the ompA mutation into E. coli O157:H7 tdcA::Tnp (AGT602) further reduced the ability of this strain to form biofilms.
FIG. 1.
Quantification of crystal violet staining of biofilms formed by E. coli O157:H7 strain 86-24 and its isogenic mutants in minimal glucose medium. Strain 86-24 does not form biofilms in LB broth; therefore, this result was included as a negative control. *, P < 0.05; **, P < 0.01. OD, optical density.
To further establish the role of Cah and curli in biofilm formation, E. coli K-12 strains carrying plasmids with either the cah or csg gene were tested in the crystal violet assay. Neither of the strains tested was able to form biofilms in PVC plates (Table 5). Interestingly, when E. coli K-12 strain ER2267 containing the plasmid with the aidA1 gene was tested, we found that the strain was able to form a biofilm. Although this result is interesting, the nature of the interaction of the AidA-I protein with PVC plates is unknown. Overall, the results obtained with the crystal violet assay confirmed that (i) Cah participates in biofilm formation and (ii) curli expression is required for the formation of stable biofilms and suggest that (iii) OmpA plays a role in but is not the principal adhesin mediating biofilm formation in PVC plates.
TABLE 5.
Biofilm formation by DH5α pro and ER2267 strains grown in LB brotha
| Plasmid | Crystal violet staining of biofilm formed in PVC plates by E. coli K-12 strainc
|
|
|---|---|---|
| DH5α pro | ER2267 | |
| None (parent strain) | 0.007 ± 0.01 | 0.04 ± 0.03 |
| pCAH100 | 0.01 ± 0.01 | 0.06 ± 0.04 |
| pIB264 | 0.008 ± 0.01 | 0.23 ± 0.04b |
| pRMinv | 0.02 ± 0.04 | 0.04 ± 0.04 |
The assay was performed in LB medium due to the growth defect of DH5α in minimal glucose medium.
P < 0.01.
Values are indicated as optical densities at 570 nm.
DISCUSSION
In this study, we have demonstrated that the attachment of E. coli O157:H7 to plant tissues, human cell lines, and plastic appears to be mediated by overlapping but distinct sets of genes encoding multiple adhesins, some of which exist as redundant systems allowing the organism to compensate for the loss of one or more of them. It is possible that some of these systems may be selectively expressed or used under specific environmental conditions. The propensity for E. coli O157:H7 to colonize different surfaces is advantageous from an ecological standpoint because it allows the organism to persist in many different locations and may provide access to required nutrients or potential hosts. Our studies are important in increasing our understanding of the processes of surface adhesion and biofilm development by E. coli O157:H7, which will allow us to propose more effective approaches to treat contaminated plant, animal, or inert surfaces and thereby reduce the chance of human infection.
Attachment and biofilm formation by E. coli O157:H7 on food products and containers are a public health concern, and since E. coli O157:H7 has been shown to form biofilms on inert surfaces (37, 38, 45), our current study was intended to target adhesion to these surfaces. Thus, mutations in specific E. coli O157:H7 adhesins and/or mediators of adherence were analyzed. We removed unbound bacteria from plants, animal cells, and plastic surfaces before measuring the numbers of adherent organisms and thus would not have detected reversibly or loosely bound bacteria (see Materials and Methods). Our present results are in accordance with our previously published data indicating that bacteria which bound tightly to the surface included both those bacteria bound directly to the surface (bacterium-surface interactions) and those bacteria bound to other bacteria in a biofilm (bacterium-bacterium interactions) (21, 44).
The surfaces used in the current study are quite distinct, e.g., in their hydrophobicity, surface charge, and types of molecules expressed. The surfaces of plants are generally covered with polysaccharides and glycoproteins such as cellulose, pectins, xylans, and arabinogalactans, are hydrophilic, and have a net negative charge due to the presence of pectin and of acetyl groups on some sugars. The surfaces of animal cells, particularly of Caco-2 cells, are hydrophobic in nature, exhibiting dome formation and electrical properties similar to those of the intestinal epithelium. This cell line forms a polarized monolayer with the presence of tight junctions and regular microvilli and the presence of glycoproteins and hydrolases. Finally, the surface of the plastic used in our experiments, polyvinyl chloride, is relatively hydrophobic in character, with acid properties, and it is known to minimize bacterial adhesion. Therefore, we did not try to find a unique and overall mechanism that explains adherence to the three surfaces. Instead, we examined specific mediators of adherence of E. coli O157:H7 to establish their role during binding to alfalfa sprouts and seed coats and then showed that some of the differences in adherence correlate with those observed on Caco-2 intestinal cells and during biofilm formation on plastic.
The individual analysis of specific E. coli O157:H7 genes or gene clusters in an E. coli K-12 background showed that some of these adhesins participate in attachment, e.g., during binding to alfalfa sprouts and seed coats. Although these studies demonstrated that K-12 strains of E. coli expressing these proteins can display an increase in adherence, a decrease in attachment was not observed when a mutation in the same gene(s) was introduced into E. coli O157:H7. Our data generated with K-12 suggest that individual adhesins are sufficient to promote binding but that they may exist in E. coli O157:H7 as redundant systems allowing bacteria to compensate for the loss of one or more of these systems and to retain the ability to bind to plant and animal surfaces. The binding of K-12 strains to plant and mammalian cells was promoted by the addition of the cah, aidA1, or csg gene to the bacteria, suggesting that all of their gene products can participate in binding to both types of surfaces. While E. coli K-12 strains possess the genes for curli biosynthesis and a cah homologue gene (agn-43), it has been shown that the expression of their protein products is repressed or the genes are inactivated (6, 17, 21, 45), which renders them unable to interfere with the phenotypes observed.
Mutation in ompA and the ompA tdcA double mutant reduced the binding of O157 to plant cells, although these mutations had little effect on binding to Caco-2 cells. It is known that TdcA is a transcriptional activator of the tdcABC operon involved in the transport and metabolism of threonine and serine during anaerobic growth (18). In contrast, the nature of the genes regulated by TdcA which are required for binding (or alternatively, inhibiting binding) to plant surfaces is unknown, and characterization of the TdcA regulatory network is a current interest of our laboratory. OmpA, one of the major proteins in the outer membrane of E. coli, which forms small, nonspecific diffusion channels with a low permeability, is required for the structural integrity of the outer membrane and the generation of a normal cell shape (25). It is also involved in cell-cell interactions in laboratory strains of E. coli (2) and in the invasion of pathogenic E. coli K-1 strains associated with neonatal meningitis (33, 34). We have recently demonstrated that OmpA displays adhesive properties in E. coli O157:H7 strains and participates in bacterial binding to intestinal epithelial cells (44). The attachment of E. coli directly or indirectly triggers reactions in the bacterial cell, which cause major changes in the compositions of outer membrane proteins (32). It is feasible to speculate that the expression of OmpA is altered upon contact with different surfaces, and in addition to playing a multifunctional role in the outer membrane of E. coli O157:H7, OmpA acts as a critical adhesin for binding to plant surfaces. Another possibility is that TdcA controls the expression of other adhesins in E. coli O157:H7 but that their functions or location in the outer membrane is impaired by ompA mutation. This hypothesis is supported by previous data indicating that the C-terminal end of OmpA displays binding to peptidoglycan (13) and to the Pal protein (in the Tol-Pal system), which has a critical role in outer membrane integrity (7). Studies in progress will determine more precisely the change in OmpA expression and the specific role played by this protein during the adhesion of E. coli O157:H7 to abiotic, plant, and animal surfaces.
We also examined the functions of some of these genes during the adherence of E. coli O157:H7 to a plastic surface. Several genes appeared to inhibit binding to both plant and animal cells; these included lpfD2, waaI, cadA, and tdcA (as single mutants). Since some of these genes are not required for binding to the relatively hydrophobic plastic surface (e.g., cadA and tdcA), it is possible that they mediate interactions with putative receptors found on animal and plant surfaces which are absent from the plastic surface. In addition, a mutation in yidE which caused increased binding to Caco-2 cells did not have an effect on binding to plant or plastic surfaces. It is unclear how these genes inhibit binding so that mutants bind better than the wild type when specific surface conditions are encountered. Furthermore, we also tested the effect of the addition of the cah, aidA1, and csg genes on the ability of K-12 strains to form biofilms on plastic. Only the addition of the aidA1 gene to the ER2267 strain caused the bacteria to bind to plastic, suggesting that AidA-I can stimulate biofilm formation by this strain and that either the DH5α strain requires more than one adhesin for the formation of a biofilm or the regulatory mechanisms for the expression of AidA-I are different in the two strains.
Another isogenic strain tested carried a mutation in the csgD gene, and this strain produced a hyperadherence phenotype in Caco-2 cells, was unable to form a stable biofilm on plastic, and did not seem to have a major impact on adherence to plant cells. We have previously shown that E. coli O157:H7 produces red colonies on Congo red plates, which is indicative of curli biosynthesis. CsgD is the regulator of curli biosynthesis: curli proteins are highly aggregated extracellular fibers expressed by E. coli and Salmonella spp. (40, 49). Both genera have curli genes; however, it has been reported that single-base-pair csgD promoter mutations leave ≥95% of E. coli O157:H7 cells unable to express curli (47). These mutations also cause E. coli O157:H7 to display increased virulence in mice and increased invasion of HEp-2 cells (48). We previously reported that E. coli O157:H7 is not an invasive organism and that a mutation in the csgD gene does not increase the invasion ability of the mutant compared to the E. coli O157:H7 wild-type strain (43). Although it has been reported that curli production by E. coli O157:H7 enhances biofilm formation on inert surfaces, i.e., stainless steel (37, 38), our data do not support these findings. In our hands, the csgD gene product of E. coli O157:H7 may be controlling the expression of an adhesive factor required for hyperadherence to tissue culture cells and/or a factor necessary for biofilm formation because the csgD mutant strain was unable to form biofilms on PVC plastic plates. These differences could be explained if we considered that bacteria are able to “sense” the surface structures and, through a regulatory mechanism, control the expression of specific adhesins. In the case of alfalfa sprouts and seed coats, curli may play a role in the attachment of those E. coli O157:H7 strains which are able to express these fimbriae, since curli fimbriae are induced in an environment similar to that in which sprouts are grown, i.e., a low temperature and low osmolarity (3). Current studies in the laboratory and our understanding of the biology of E. coli O157:H7 will allow us to examine the specificity of binding to the different surfaces and determine the specific roles of these putative redundant systems in the pathogenesis of this organism.
In summary, we were able to perform an initial analysis of multiple gene products associated, directly or indirectly, with the binding of E. coli O157:H7 to different surfaces and to demonstrate that only the absence of the ompA and tdcA genes had a significant effect on binding, particularly during plant-bacterium interactions. Furthermore, we showed that binding to the three types of surfaces appeared to be mediated by overlapping but distinct sets of genes. The definition and characterization of genes important in the adherence of E. coli O157:H7 will provide new and deeper insight into the colonization process of this pathogen on intestinal epithelial cells, plants, and inert surfaces and build up a foundation for developing novel approaches for the control of E. coli O157:H7 and potentially other pathogenic E. coli strains.
Acknowledgments
This work was supported by institutional funds from the UTMB John Sealy Memorial Endowment Fund for Biomedical Research to A.G.T. and by USDA grant 2003-35201-13693 to A.G.M.
REFERENCES
- 1.Ackers, M. L., B. E. Mahon, E. Leahy, B. Goode, T. Damrow, P. S. Hayes, W. F. Bibb, D. H. Rice, T. J. Barrett, L. Hutwagner, P. M. Griffin, and L. Slutsker. 1998. An outbreak of Escherichia coli O157:H7 infections associated with leaf lettuce consumption. J. Infect. Dis. 177:1588-1593. [DOI] [PubMed] [Google Scholar]
- 2.Anthony, K. G., C. Sherburne, R. Sherburne, and L. S. Frost. 1994. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol. Microbiol. 13:939-953. [DOI] [PubMed] [Google Scholar]
- 3.Barak, J. D., L. C. Whitehand, and A. O. Charkowski. 2002. Differences in attachment of Salmonella enterica serovars and Escherichia coli O157:H7 to alfalfa sprouts. Appl. Environ. Microbiol. 68:4758-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benz, I., and M. A. Schmidt. 1989. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breuer, T., D. H. Benkel, R. L. Shapiro, W. N. Hall, M. M. Winnett, M. J. Linn, J. Neimann, T. J. Barrett, S. Dietrich, F. P. Downes, D. M. Toney, J. L. Pearson, H. Rolka, L. Slutsker, P. M. Griffin, and I. Team. 2001. A multistate outbreak of Escherichia coli O157:H7 infections linked to alfalfa sprouts grown from contaminated seeds. Emerg. Infect. Dis. 7:977-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, P. K., C. M. Dozois, C. A. Nickerson, A. Zuppardo, J. Terlonge, and R. Curtiss III. 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol. Microbiol. 41:349-363. [DOI] [PubMed] [Google Scholar]
- 7.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2005. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 sites, United States, 2004. Morb. Mortal. Wkly. Rep. 54:352-356. [PubMed] [Google Scholar]
- 9.Chapman, P. A. 2001. Sources of Escherichia coli O157 and experiences over the past 15 years in Sheffield, UK. Symp. Ser. Soc. Appl. Microbiol. 29:51S-60S. [DOI] [PubMed] [Google Scholar]
- 10.Cody, S. H., M. K. Glynn, J. A. Farrar, K. L. Cairns, P. M. Griffin, J. Kobayashi, M. Fyfe, R. Hoffman, A. S. King, J. H. Lewis, B. Swaminathan, R. G. Bryant, and D. J. Vugia. 1999. An outbreak of Escherichia coli O157:H7 infection from unpasteurized commercial apple juice. Ann. Intern. Med. 30:202-209. [DOI] [PubMed] [Google Scholar]
- 11.Cookson, A. L., W. A. Cooley, and M. J. Woodward. 2002. The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int. J. Med. Microbiol. 292:195-205. [DOI] [PubMed] [Google Scholar]
- 12.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 13.De Mot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 14.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle, M. P. 2001. Escherichia coli O157:H7 and its significance in foods. Int. J. Food Microbiol. 12:289-301. [DOI] [PubMed] [Google Scholar]
- 16.Dunne, W. M. J. 2002. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gophna, U., M. Barlev, R. Seijffers, T. A. Oelschlager, J. Hacker, and E. Z. Ron. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69:2659-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagewood, B. T., Y. L. Ganduri, and P. Datta. 1994. Functional analysis of the tdcABC promoter of Escherichia coli: roles of TdcA and TdcR. J. Bacteriol. 176:6214-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassan, A. N., and J. F. Frank. 2004. Attachment of Escherichia coli O157:H7 grown in tryptic soy broth and nutrient broth to apple and lettuce surfaces as related to cell hydrophobicity, surface charge, and capsule production. Int. J. Food Microbiol. 96:103-109. [DOI] [PubMed] [Google Scholar]
- 20.Itoh, Y., Y. Sugita-Konishi, F. Kasuga, M. Iwaki, Y. Hara-Kudo, N. Saito, Y. Noguchi, H. Konuma, and S. Kumagai. 1998. Enterohemorrhagic Escherichia coli O157:H7 present in radish sprouts. Appl. Environ. Microbiol. 64:1532-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeter, C., and A. G. Matthysse. 2005. Characterization of the binding of diarrheagenic strains of E. coli to plant surfaces and the role of curli in the interaction of the bacteria with alfalfa sprouts. Mol. Plant-Microbe Interact. 18:1235-1242. [DOI] [PubMed]
- 22.Ji, C., J. Smith-Becker, and N. T. Keen. 1998. Genetics of plant-pathogen interactions. Curr. Opin. Biotechnol. 9:202-207. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, W. M., H. Lior, and G. S. Bezanson. 1983. Cytotoxic Escherichia coli O157:H7 associated with haemorrhagic colitis in Canada. Lancet i:76. [DOI] [PubMed] [Google Scholar]
- 24.Junkins, A. D., and M. P. Doyle. 1989. Comparison of adherence properties of Escherichia coli O157:H7 and a 60-megadalton plasmid-cured derivative. Curr. Microbiol. 19:21-27. [Google Scholar]
- 25.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 26.Loper, J. E., T. V. Suslow, and M. N. Schroth. 1984. Lognormal distribution of bacterial populations in the rhizosphere. Phytopathology 74:1454-1460. [Google Scholar]
- 27.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Matthysse, A. G., S. Stretton, C. Dandie, N. C. McClure, and A. E. Goodman. 1996. Construction of GFP vectors for use in gram-negative bacteria other than Escherichia coli. FEMS Microbiol. Lett. 145:87-94. [DOI] [PubMed] [Google Scholar]
- 29.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mietzner, T. A., and S. A. Morse. 1994. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu. Rev. Nutr. 14:471-493. [DOI] [PubMed] [Google Scholar]
- 31.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otto, K., J. Norbeck, T. Larsson, K. A. Karlsson, and M. Hermansson. 2001. Adhesion of type 1-fimbriated Escherichia coli to abiotic surfaces leads to altered composition of outer membrane proteins. J. Bacteriol. 183:2445-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prasadarao, N. V. 2002. Identification of Escherichia coli outer membrane protein A receptor on human brain microvascular endothelial cells. Infect. Immun. 70:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prasadarao, N. V., C. A. Wass, J. N. Weiser, M. F. Stins, S. H. Huang, and K. S. Kim. 1996. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reeves, S. A., A. G. Torres, and S. M. Payne. 2000. TonB is required for intracellular growth and virulence of Shigella dysenteriae. Infect. Immun. 68:6329-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riley, L. W., R. S. Remis, S. D. Helgerson, et al. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 37.Ryu, J. H., and L. R. Beuchat. 2005. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and curli production on its resistance to chlorine. Appl. Environ. Microbiol. 71:247-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryu, J. H., H. Kim, J. F. Frank, and L. R. Beuchat. 2004. Attachment and biofilm formation on stainless steel by Escherichia coli O157:H7 as affected by curli production. Lett. Appl. Microbiol. 39:359-362. [DOI] [PubMed] [Google Scholar]
- 39.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186:8058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sukupolvi, S., R. G. Lorenz, J. I. Gordon, Z. Bian, J. D. Pfeifer, S. J. Normark, and M. Rhen. 1997. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect. Immun. 65:5320-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarr, P. I., M. A. Neill, C. R. Clausen, J. W. Newland, R. J. Neill, and S. L. Moseley. 1989. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington, 1984-1987. J. Infect. Dis. 159:344-347. [DOI] [PubMed] [Google Scholar]
- 42.Tauxe, R., H. Kruse, C. Hedberg, M. Potter, J. Madden, and K. Wachsmuth. 1997. Microbial hazards and emerging issues associated with produce: a preliminary report to the National Advisory Committee on Microbiologic Criteria for Foods. J. Food Prot. 60:1400-1408. [DOI] [PubMed] [Google Scholar]
- 43.Torres, A. G., K. J. Kanack, C. B. Tutt, V. Popov, and J. B. Kaper. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol. Lett. 238:333-344. [DOI] [PubMed] [Google Scholar]
- 44.Torres, A. G., and J. B. Kaper. 2003. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 71:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torres, A. G., N. T. Perna, V. Burland, A. Ruknudin, F. R. Blattner, and J. B. Kaper. 2002. Characterization of Cah, a calcium-binding and heat-extractable autotransporter protein of enterohaemorrhagic Escherichia coli. Mol. Microbiol. 45:951-966. [DOI] [PubMed] [Google Scholar]
- 46.Torres, A. G., X. Zhou, and J. B. Kaper. 2005. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73:18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uhlich, G. A., J. E. Keen, and R. O. Elder. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlich, G. A., J. E. Keen, and R. O. Elder. 2002. Variations in the csgD promoter of Escherichia coli O157:H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect. Immun. 70:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao, T., M. P. Doyle, J. Shere, and L. Garber. 1995. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 61:1290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]