Abstract
A microbial community analysis of forest soil from Jindong Valley, Korea, revealed that the most abundant rRNA genes were related to Acidobacteria, a major taxon with few cultured representatives. To access the microbial genetic resources of this forest soil, metagenomic libraries were constructed in fosmids, with an average DNA insert size of more than 35 kb. We constructed 80,500 clones from Yuseong and 33,200 clones from Jindong Valley forest soils. The double-agar-layer method allowed us to select two antibacterial clones by screening the constructed libraries using Bacillus subtilis as a target organism. Several clones produced purple or brown colonies. One of the selected antibacterial clones, pJEC5, produced purple colonies. Structural analysis of the purified pigments demonstrated that the metagenomic clone produced both the pigment indirubin and its isomer, indigo blue, resulting in purple colonies. In vitro mutational and subclonal analyses revealed that two open reading frames (ORFs) are responsible for the pigment production and antibacterial activity. The ORFs encode an oxygenase-like protein and a putative transcriptional regulator. Mutations of the gene encoding the oxygenase canceled both pigment production and antibacterial activity, whereas a subclone carrying the two ORFs retained pigment production and antibacterial activity. This finding suggests that these forest soil microbial genes are responsible for producing the pigment with antibacterial activity.
The discovery of microbial products has depended primarily on the screening of cultured microbial species for desirable activity. However, the rediscovery rate of known microbial products derived from this classical approach is increasing, while the probability of obtaining novel resources is decreasing (21). A recently developed metagenomic approach clones the total microbial genome (the metagenome), which is directly isolated from natural environments, in culturable bacteria such as Escherichia coli (3, 21, 45, 46) to discover novel microbial resources (20). The metagenomic approach originated from the molecular analysis of microbial communities, which revealed that the majority of microorganisms in nature were not cultivable by standard culturing techniques (4, 6, 26, 36, 41). Therefore, most microorganisms in nature have not been characterized. Similarly, a microbial biomass study concluded that prokaryotes are the dominant organisms on Earth (60). A recent review (57) on the microbial diversity in various soil environments and sediments suggested that microbial diversity is higher in forest and pasture soils than in arable soils. Moreover, each gram of forest soil most probably contains several thousand bacterial species (54, 55). Thus, we focused on forest soil environments to explore the resources of soil microbes by using a metagenomic approach.
The difficulties in cultivating microorganisms exclude the majority of the microbial soil community from a functional analysis of their genes and the subsequent use of the microbial gene products (1, 27). Therefore, there is a high probability of finding novel microbial products, such as antibiotics and enzymes, in uncharacterized soil bacteria. Initial studies have demonstrated that expressing foreign genes in a heterologous host is a technically feasible approach for exploring microbial resources (45, 47). Several studies have shown that the metagenomic approach permits searches for various biocatalysts, such as lipase/esterase, protease, oxidoreductase, and nitrilase, in varied soils and microbial habitats (15, 18, 24, 32, 33, 35, 38, 45, 56, 58). Recent metagenome studies also have demonstrated the efficacy of the approach in searching for novel microbial bioactivities and anticancer drugs (20, 42). Several antibiotics and novel secondary metabolites have been identified from metagenomic studies, together with the genes responsible for them, including long-chain acyl amino acid and turbomycin antibiotics, and genes encoding polyketide synthases (7, 8, 14, 17, 39, 43, 51, 59).
In addition, the combination of phylogenetic-marker screening of metagenomic libraries and genomics could reveal the physiology of as-yet-uncultured microorganisms that have been identified only by culture-independent studies (37, 44). Ultimately, the isolation of novel soil microorganisms (28, 29, 52) can be advanced by the genetic and physiological information gained from analyses of the metagenomes of uncultured bacteria (20).
Our analysis of the microbial community of forest soils by this culture-independent method revealed that representatives of the Acidobacteria group were the numerically dominant phylotype. Two metagenomic libraries were constructed from two forest topsoils to explore their microbial genetic resources. Among several pigmented metagenomic clones, one produced indirubin as a major compound and indigo blue as a minor pigment. The corresponding genes were identified and characterized, although we found no evidence that the clone originated from the Acidobacteria, the major uncultured phylum in the forest soils.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
E. coli cultures were grown at 37°C on Luria-Bertani (LB) agar or in LB broth supplemented with the appropriate antibiotics (40). Bacillus subtilis strain JH642 carrying pBSK185 (a chloramphenicol-resistant plasmid) was kindly provided by Seung Hwan Park at the Korea Research Institute of Bioscience and Biotechnology. B. subtilis was routinely grown at 30°C on LB agar or in LB broth containing chloramphenicol. The following antibiotic concentrations were used for E. coli strains and B. subtilis: chloramphenicol, 25 μg/ml; ampicillin, 100 μg/ml; and kanamycin, and 25 μg/ml. E. coli strain CY15000 (61) was obtained from the E. coli Genetic Stock Center at Yale University.
DNA preparation from forest topsoil.
Forest topsoil samples were collected from Yuseong in Daejeon, Korea, where pine trees were planted about 40 years ago. Other forest topsoil samples were collected from Jindong Valley in Inje, Kangwon Province, Korea, from a site where oak and beech trees are dominant. The Yuseong and Jindong Valley forest soils were collected in January 2002 and October 2003, respectively. The soils from three different sites at each location were combined and used for further experiments. Table 1 lists each forest type and its soil characteristics. The soil type and chemical characteristics were analyzed at the National Institute of Agricultural Science and Technology, Rural Development Administration, Korea. Both soil samples were immediately stored at −80°C and were simultaneously subjected to metagenomic DNA isolation. The metagenomic DNA was extracted from bulk forest topsoil and the loosely attached soils removed from plant roots. The soils were sieved to remove plant debris and particulates larger than 2 mm. Soil DNA extraction was carried out as previously described with sodium dodecyl sulfate and proteinase K treatment (63). Further purification of the DNA for cloning into a fosmid was performed following the methods of Rondon et al. (45). The size of the extracted DNA was examined by pulsed-field gel electrophoresis (Bio-Rad CHEF-DR II, 1- to 6-s switch, 6 V/cm, 120° fixed angle, 5-h run time).
TABLE 1.
Soil chemical characteristics of two natural forest soils
| Site | Forest type | Soil type | pH | Organic C (%) | Phosphate (μg/ml) | Amt (mmol/kg) of:
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Ca | Mg | K | Na | ||||||
| Yuseong | Pine | Sandy loam | 5.4 | 2.9 | 10 | 32 | 8 | 0.9 | 2 |
| Jindong valley | Oakbeech | Sandy loam | 5.0 | 1.9 | 15 | 36 | 10 | 7.5 | 2.5 |
General DNA manipulation.
Plasmid preparation, restriction endonuclease digestion, DNA ligation, plasmid DNA transformation, agarose gel electrophoresis, and other standard recombinant DNA techniques were carried out by standard methods described by Sambrook et al. (49). DNA sequencing and primer synthesis were performed commercially at the DNA sequencing facility of GenoTech Corp. (Daejeon, Korea). DNA sequences were analyzed with a BLAST program provided by the National Center for Biotechnology Information.
PCR amplification of 16S rRNA genes.
Bacterial 16S rRNA genes were amplified with the two bacterial universal primers, 530F (5′-TGACTGACTGAGTGCCAGCMGCCGCGG-3′) and 1492R-1 (5′-TGACTGACTGAGGYTACCTTGTTACGMYTT-3′) (5), using metagenomic DNA obtained from Jindong Valley as the template. Before performing a PCR, the metagenomic DNA was further purified by using a Geneclean Turbo kit (Qbiogene, Montreal, Canada). The PCR amplification was carried out under the following conditions: 94°C for 2 min of initial denaturation; 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 68°C for 1 min, and a final extension at 68°C for 3 min. The reaction mixture (final volume, 50 μl) contained 1 μl of DNA (ca. 40 to 50 ng), 20 mM Tris-HCl (pH 8.0), 40 mM NaCl, 2 mM sodium phosphate, 0.1 mM EDTA, 1 mM dithiothreitol, 0.2 mM each deoxynucleoside triphosphate, 2 mM MgSO4, 0.25 μM each oligonucleotide primer, and 1 U of Platinum Taq high-fidelity polymerase (Invitrogen, Carlsbad, CA). The amplified products were visualized by agarose gel electrophoresis. After the amplified products were pooled, the band corresponding to the correctly sized product (∼1 kb) was excised, purified, and concentrated. The 16S rRNA gene library was generated with the purified 16S rRNAs ligated into the pGEM-Teasy vector (Promega, Madison, WI) and transformed into E. coli DH5α competent cells. White colonies were screened directly to confirm the presence of the inserts by performing colony PCR with T7 and SP6 primers. Subsequently, 100 randomly selected white colonies were used for nucleotide sequence analysis. The nucleotide sequencing and DNA oligonucleotide synthesis were performed commercially at the DNA sequencing facility of GenoTech Corp. (Daejeon, Korea).
Phylogenetic analysis.
Among the 100 colonies, 10 did not produce interpretable nucleotide sequences. The remaining 90 were analyzed by using BLAST (National Center for Biotechnology Information) and the Classifier and Sequence Match function with sequences in the Ribosomal Database Project II (RDP II [13]). Each sequence was submitted to the CHECK_CHIMERA program of the RDP II to search for the presence of chimeric artifacts. Among the 90 clones, four appeared to be chimeras and were excluded from the phylogenetic analyses. In addition, seven clones were redundant. Thus, 79 sequences were aligned with the CLUSTAL W program (53) and visually examined with the GENEDOC program. Phylogenetic trees were constructed by using DNADIST with the Jukes-Cantor model (30) and NEIGHBOR with the neighbor-joining method (48) in the PHYLIP (phylogeny inference package) programs, version 3.61. We generated 1,000 bootstrapped replicate resampling data sets with SEQBOOT (PHYLIP, version 3.61). We followed the standard protocol for the default settings of the computer programs used in this procedure.
Metagenomic library construction and selection of antibacterial clones.
Metagenomic libraries using the extracted DNA from the two forest soils were constructed using a commercial fosmid vector, pEpiFOS-5 (Epicenter, Madison, WI). The library was constructed by DNA size fractionation, clean-up of the metagenomic DNAs, and subsequent ligation into a fosmid vector as previously described (35). The ligation mixture was then packaged into lambda phages using MaxPlax Lambda Packaging Extracts (Epicenter, Madison, WI). The packaged library was transduced into E. coli EPI-100, and E. coli transformants were selected on LB agar supplemented with chloramphenicol. The presence of recombinant plasmids and the polymorphism of the insert DNA were examined by pulsed-field gel electrophoresis (Bio-Rad CHEF-DR II, 1- to 6-s switch, 6 V/cm, 120° fixed angle, 5-h run time) of a BamHI digestion of the purified plasmids from randomly selected E. coli transformants. The library clones were stored in cryotubes as clone pools, with ca. 500 clones per pool.
To select antibacterially active clones from the stored library, the library pool stocks were diluted in buffer (NaCl, 8.5 g/liter; KH2PO4, 0.3 g/liter; Na2HPO4, 0.6 g/liter; MgSO4, 0.2 g/liter; gelatin, 0.1 g/liter), and metagenomic clones in E. coli were cultured on LB agar supplemented with chloramphenicol for 3 days at 37°C. The number of clones per plate was adjusted to approximately 500 by dilution of the library stock, and at least fivefold the number per initial stock were bioassayed by using the procedure described by Rondon et al. (45). Briefly, the precultured E. coli clones were overlaid with 5 ml of LB top agar containing 0.5 ml of B. subtilis culture with an optical density at 600 nm of 0.2 to 0.3. The plates were incubated overnight at 28°C and scored for activity by looking for zones of growth inhibition in the bacterial lawn. During the library storage process, we had selected metagenomic clones that altered the phenotype of E. coli colonies, including pigmentation. From the bioassay, two antibacterially active clones were selected, one of which produced purple pigments. The pigments were extracted with ethyl acetate and fractionated. Two pigments appeared to be simultaneously produced from one clone, with one of the pigments being responsible for the antibacterial activity. Therefore, we further analyzed this clone, designated pJEC5, from the Jindong Valley library by subcloning the responsible genes and identifying the pigments.
Subcloning and in vitro mutagenesis.
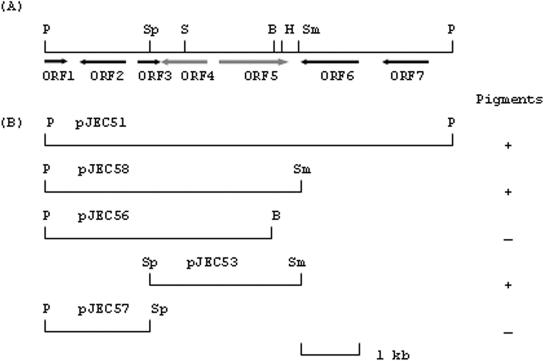
The selected clone, pJEC5, was analyzed with various restriction endonucleases in order to subclone the active gene for antibacterial activity and pigment production. The enzymes EcoRI, BamHI, PstI, and HindIII were each used to randomly digest pJEC5; the resulting fragments were ligated into pUC119 digested with the same enzyme. We selected E. coli transformants displaying purple pigments. Several dark-blue colonies also grew. We chose clone pJEC54, which carried a 7.5-kb PstI fragment. The PstI fragment was excised, blunt ended by treatment with Klenow fragment, and cloned into the original fosmid vector pEpiFOS-5 digested with PmlI to generate pJEC51. The E. coli cells carrying pJEC51 expressed the purple color of the initially selected clone, pJEC5. Therefore, pJEC54 was used to subclone various restriction fragments into the original fosmid vector to determine the genes involved in pigment production. The restriction-digested DNA fragments were blunt ended by treatment with Klenow DNA polymerase and were cloned into pEpiFOS-5 digested with PmlI to produce the subclones pJEC53, pJEC56, pJEC57, and pJEC58 (see Fig. 3).
FIG. 3.
Map of a 7.8-kb PstI fragment of pJEC5 carrying various ORFs involved in pigment production and their subclones. (A) Map of pJEC51 carrying ORF5 encoding indole dioxygenase and ORF4 encoding a putative protein similar to the TetR family response regulator. ORF1, partial gene similar to a transcriptional regulator gene; ORF2, similar to the TetR family response regulator (58% identical to ORF4); ORF6, similar to an outer membrane porin gene; ORF7, similar to an ABC transporter gene. (B) Pigment production by different subclones. The restriction endonuclease sites of the subclones are not indicated. “+” indicates pigmentation of E. coli colonies carrying the respective subclones, whereas “−” indicates a lack of pigmentation. Abbreviations: B, BamHI; H, HindIII; P, PstI; S, SalI; Sm, SmaI; Sp, SphI.
To confirm the presence of the pigmentation genes from pJEC5, in vitro transposon mutagenesis was carried out according to the mutagenesis protocol provided with the GPS-Mutagenesis System (GPS-M; New England BioLabs, Inc., Beverly, MA). Transformants without pigmented colonies were selected, and transposon insertion sites were determined by DNA sequencing and restriction analysis of the mutated subclones. The antibacterial activity of the mutated clones was determined by the procedure described above.
Isolation and spectral analysis of the pigments.
The E. coli carrying pJEC54 were grown in 100 ml of LB broth supplemented with ampicillin in a shaking incubator at 37°C for 2 days. The cells and pigment aggregates were harvested by centrifugation at 6,000 rpm for 10 min. The culture supernatant was extracted twice with an equal volume of ethyl acetate. The pellet was resuspended in 10 ml of 0.85% saline and sonicated to disrupt E. coli cells and aggregates. The solution was extracted twice with an equal volume of ethyl acetate. The two extracts were combined and concentrated to dryness. The residue was dissolved in chloroform and subsequently applied to a silica gel column (3.6 by 60 cm; Kiesel gel 60, 30 g, 230 to 400 mesh). The column was eluted with n-hexane and ethyl acetate (1:1 [vol/vol]). The eluted pigments were red and blue. Fractions containing the pigments were separately combined and concentrated to dryness. The purity of each individual compound was confirmed by high-performance liquid chromatography with a C18 column (μBondapak C18, 300 by 3.9 mm; Waters) using a linear gradient from 60% to 100% methanol. The compounds were detected by using a photo diode array detector (Waters).
The UV-visible absorption spectra of the two pigments were recorded on a UV/VIS spectrophotometer (Beckman). Mass and nuclear magnetic resonance (NMR) spectral analyses were performed to identify the molecular structures of the two compounds. Their electron impact mass spectra (EI-MS) were recorded on a double-focusing, high-resolution mass spectrophotometer (JEOL JMS-DX303; JEOL, Ltd., Tokyo, Japan). 1H-NMR spectra were examined in deuterochloroform on a Bruker AMX-500 (500 MHz) NMR spectrometer (Bruker Analytische Messtechnik Gmbh, Rheinstetten, Germany). The spectra were referenced to the tetramethylsilane (1H) signal.
Production of pigments.
We investigated the production of indigo and indirubin by mutated clones and various subclones. E. coli cells were grown in 10 ml of LB broth supplemented with chloramphenicol in a shaking incubator at 37°C for 4 days. The cells and pigment aggregates were harvested every 24 h by centrifugation at 12,000 rpm for 10 min. The pellet was resuspended with 1 ml of N,N′-dimethyl formamide and sonicated to disrupt the E. coli cells and aggregates. The solution was centrifuged at 14,000 rpm for 10 min to obtain a supernatant containing the pigments. The absorbance of the pigment solution was examined at 610 and 540 nm to monitor the relative production of indigo and indirubin, respectively.
Nucleotide sequence accession number.
The nucleotide sequence of the DNA insert of pJEC54 has been deposited in the GenBank database under accession number DQ000460. The nucleotide sequences of the 16S rRNA for phylogenetic analysis have been deposited in the GenBank database under accession numbers DQ004750 to DQ004828.
RESULTS
Microbial community profile of forest soil.
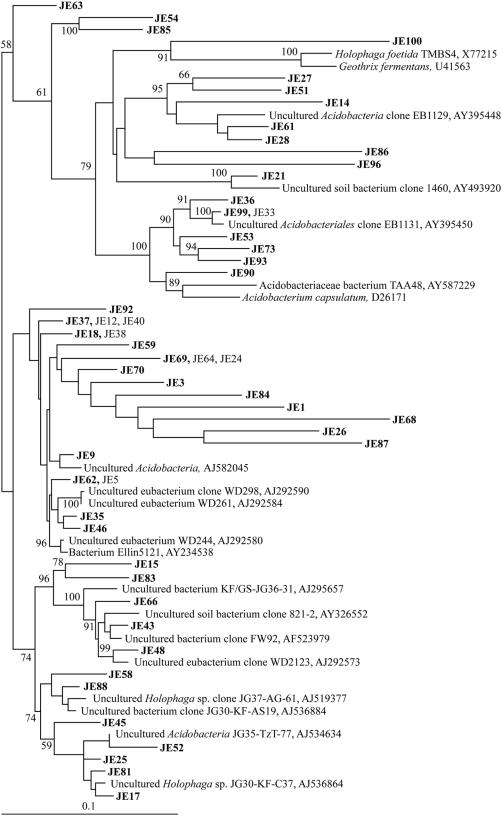
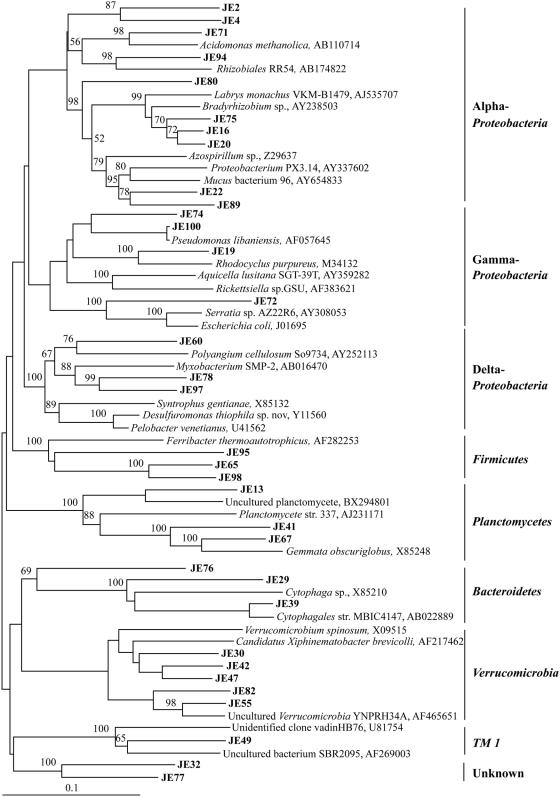
The chemical and physical characteristics of the two forest topsoils were similar, except that the Jindong Valley forest soils were more acidic and contained much more potassium ion (Table 1). The microbial community profile of the Jindong Valley forest topsoil was investigated by comparative sequence analysis of partial 16S rRNA genes. Of the 90 16S rRNA gene sequences, 4 appeared to be chimeras and 7 were redundant; these were excluded from the analysis. Therefore, a total of 79 clones underwent phylogenetic analysis. Most of the phylotypes identified were members of the phylum Acidobacteria (60%, 52 of 86) (Fig. 1), Proteobacteria (20%, 17 of 86), Verrucomicrobia (6%, 5 of 86), and others (14%, 12 of 86), including Plactomycetes, Firmicutes, and Bacteroidetes (2.43%) (Fig. 2). None of the clones belonged to β-Proteobacteria, and only three belonged to Firmicutes, which includes low G+C, gram-positive organisms. The phylogenetic analysis of the Acidobacteria group revealed a group of clones (JE59, JE69, JE70, JE3, JE84, JE1, JE68, JE26, and JE87; Fig. 1) that was deeply branching from other Acidobacteria clones. Many clones in the Acidobacteria group were unaffiliated with the Acidobacteria subgroup comprising cultured strains, such as A. capsulatum (25) and strain TAA48 (52).
FIG. 1.
Phylogenetic tree by 16S rRNA gene analysis of Acidobacteria from Jindong valley forest soil. The letters JE followed by a clone number indicates soil clones. Bootstrap values are shown for each node that had >50% support in a bootstrap analysis of 1,000 replicates. The scale bar indicates 0.1 change per nucleotide.
FIG. 2.
Phylogenetic tree by 16S rRNA gene analysis of Proteobacteria and other taxa from Jindong Valley forest soil. The letters JE followed by a clone number indicates soil clones. Bootstrap values are shown for each node that had >50% support in a bootstrap analysis of 1,000 replicates. The scale bar indicates 0.1 change per nucleotide.
Construction of a metagenomic library.
The quantity and quality of the prepared DNA to construct metagenomic library were almost identical to our previous study (35). However, the DNA solution prepared from the Jindong Valley soil was much darker than that from the Yuseong forest, indicating a higher contamination of humic substances. The enzymatic manipulation of the DNA to construct the library was difficult because of the humic substances, and purification did not completely remove them. The libraries constructed from the forest topsoils consisted of 80,500 Yuseong forest clones and 33,200 Jindong Valley clones and were maintained for activity-based screening. Since the metagenomic DNA from Jindong Valley was heavily contaminated with humic substances, its library construction was less efficient than that of Yuseong forest DNA. The average DNA insert size was estimated at more than 35 kb by analyzing randomly selected clones via preparative pulsed-field gel electrophoresis after BamHI digestion. Restriction analysis of the randomly selected 52 clones revealed various restriction digestion patterns without any redundant pattern, indicating a high diversity of cloned genes in a fosmid (data not shown).
Selection of an antibacterial clone and identification of pigments.
Two antibacterially active clones were selected by the double-agar-layer method. One showed higher antibacterial activity against B. subtilis, as well as purple pigmentation of the E. coli colony. The clone (pJEC5) was further analyzed to identify the pigments and antibacterial activity. Crude culture and cell extracts of the E. coli carrying pJEC54 were partitioned through organic extraction, and two pigments were separated by silica gel column chromatography. These red and blue pigments were purified, and their purity was confirmed by high-pressure liquid chromatography analysis. The UV/VIS maxima were 241, 284, 336, and 610 nm for the blue pigment and 236, 289, 366, and 545 nm for the red pigment. Each pigment had a molecular weight of 262 atomic mass units, as determined by EI-MS. EI-MS analysis of the blue pigment showed major peaks at m/z 234, 205, 179, 158, 131, 104, 75, and 50, a profile identical to that of indigo blue. The red pigment showed major peaks at m/z 234, 205, 179, 158, 131, 103, 76, and 51, which is identical to the profile of indirubin. The NMR results also confirmed the identities of the pigments as indigo blue and indirubin (data not shown). Therefore, the purple coloration of the initial colony might result from the coproduction of two isomeric compounds: indigo blue and indirubin. Similar phenomena were previously observed by introducing the Rhodococcus gene into E. coli (22) and by soil metagenomic analysis (39).
Cloning and sequence analysis of genes for indirubin and indigo production.
In vitro insertion mutagenesis of pJEC5 generated five nonpigmented mutants lacking antibacterial activity. The insertion site of the transposon was determined by restriction digestion analysis, subcloning of the transposon flanking DNA, and sequence analysis. For the five mutants, the insertion site appeared to be in a single open reading frame (ORF), similar to that of genes encoding potential oxidoreductase. Thus, the ORF responsible for the antibacterial activity and pigmentation was defined, and subcloning experiments were performed. The random subcloning approach described in Materials and Methods was successful. It involved PstI or EcoRI digestion of pJEC5, the cloning of the digested DNA pools into pUC119, and the subsequent selection of E. coli transformants showing pigmentation. Since the PstI digest produced a smaller DNA insert fragment than did the EcoRI digest, the subclone carrying the PstI-digested insert (pJEC54) was used to further analyze the genes. Several subclones were constructed in the original fosmid vector to compare their pigment production with that of the original metagenomic clone, pJEC5 (Fig. 3B). Plasmid pJEC53, carrying two ORFs (ORF4 and ORF5), was selected as the subclone expressing an identical level of pigment production in E. coli and the equivalent antibacterial activity. The purified indirubin also showed antibacterial activity against B. subtilis in the growth inhibition assay (data not shown).
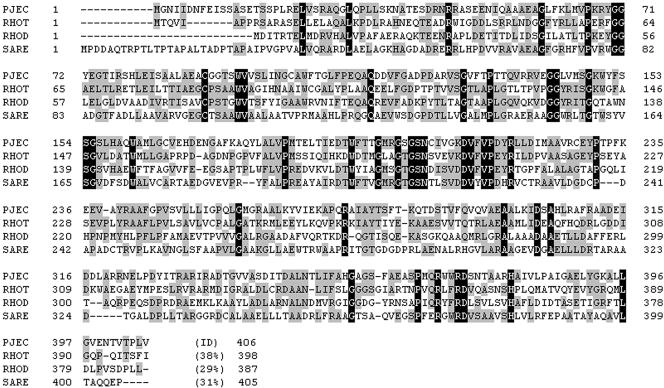
We determined the complete DNA sequence of plasmid pJEC51, a subclone of pJEC5, but not that of pJEC5, the original clone. We defined the potential ORFs and their organization by a BLAST search through the National Center for Biotechnology Information (Fig. 3A). The potential protein products encoded by each ORF are indicated in Fig. 3, based on the proteins from the GenBank database that are most similar. pJEC53, with its two ORFs, fully restored the pigmentation and antibacterial activity of the original clone, pJEC5. We described the extensive DNA sequence analysis earlier in this work. Based on the deduced amino acid sequence identity, it is likely that ORF4 and ORF5 encode a transcriptional regulatory protein and an oxygenase-like protein, respectively. ORF4 and ORF5 were divergently transcribed, revealing a 305-bp space between the ATG codons for ORF4 and ORF5, which should contain the promoter regions for the two transcriptional units. A comparable gene organization was found in a cultured Rhodococcus sp. (12), although the gene itself was not significantly similar. ORF4 encodes a protein composed of 240 amino acids, similar to the TetR family transcriptional regulator, which is known as a negative regulator. However, the level of similarity is 24 to 28% in the deduced amino acid sequences. ORF5 encodes a protein of 406 amino acids, similar to several oxidoreductases. When similar proteins in the database were aligned with the deduced amino acid sequence of ORF5 (Fig. 4), the sequences were 29 to 38% identical. Interestingly, the deduced amino acid sequence of ORF5 was virtually identical to the conserved domain of naphthocyclinone hydroxylase (NcnH), which is involved in the biosynthesis of the aromatic polyketide antibiotic naphthocyclinone in Streptomyces arenae (9).
FIG. 4.
Alignment of the putative indole oxygenase (pJEC) of pJEC5 with the oxygenases, including a flavon-containing monooxygenase of Rhodococcus sp. strain T104 (RHOT, accession AF494423 [12]), an indole oxygenase of Rhodococcus sp. (RHOD, accession M55641 [22, 23]), and a naphthocyclinone hydroxylase of Streptomyces arenae (SARE, accession AF218066 [9]). Identical bases between any two sequences are marked in light gray, and identical bases in all of the proteins are shown as white letters on a dark background. The identities (ID) between the amino acid sequences of oxygenases and pJEC5 are indicated.
Characterization of indigo and indirubin production.
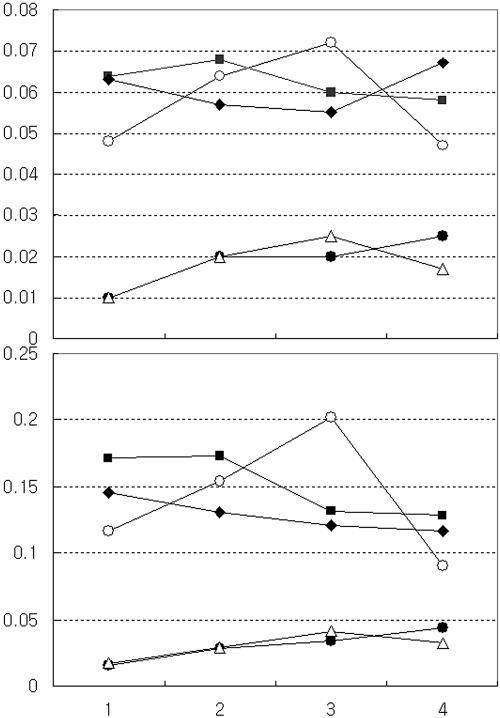
We found that the GPS-3 insertion mutation in ORF5 (plasmid pJEC5-1) completely shut down the level of pigment production to that of E. coli cells carrying an empty fosmid vector (Fig. 5). The original metagenomic clone, pJEC5, and its subclones, pJEC51 and pJEC53, produced almost equivalent amounts of both pigments. However, pigment production in E. coli carrying pJEC5 increased steadily for 3 days and then suddenly decreased, which may suggest growth stage-dependent regulation of the gene expression in pJEC5. In contrast, pigment production by the two subclones, pJEC51 and pJEC53, was relatively constant for 4 days. Our metagenomic clone produced more indirubin than indigo. We obtained approximately 6.4 mg of indirubin and 3.4 mg of indigo by culturing 100 ml of E. coli cells carrying pJEC54 (data not shown). To establish whether the pigments originated from indole accumulations in E. coli owing to the action of tryptophanase in releasing indole from tryptophan, we transformed E. coli CY15000 (61), which is tryptophanase minus, with pJEC5. The E. coli CY15000 carrying pJEC5 did not produce any detectable pigment because of the lack of indole generation. This confirmed that the gene product of ORF5 is most likely involved in indole oxidation to derive indirubin and indigo.
FIG. 5.
Production of indigo (upper panel) and indirubin (lower panel) pigments over time by various subclones and mutated clones. The y axis indicates the absorbance at 610 nm (A) and 540 nm (B) for indigo and indirubin, respectively. Symbols: •, fosmid vector only; ○, pJEC5; ▵, pJEC5-1; ▪, pJEC51; ⧫, pJEC53.
DISCUSSION
We performed a metagenomic analysis on a forest soil exhibiting high levels of acidobacteria rRNA genes. Phylum Acidobacteria was even more abundant than Proteobacteria. The Acidobacteria groups are phylogenetically diverse and may include novel classes (2). The prevalence of acidobacteria clones in our rRNA gene libraries suggests that acidobacteria may be the dominant microorganisms in Jindong Valley forest soil. The first cultured isolate in the Acidobacteria group was Acidobacteria capsulatum (25), and recently a few more acidobacteria isolates have been cultured (52). However, owing to the scarcity of cultured isolates, little is known about the physiology of acidobacteria. The ubiquity of the Acidobacteria in various types of soils suggests that they are probably metabolically versatile (2, 19). In natural forest soil, accumulated plant materials provide nutrients for potential use by microorganisms such as bacteria of the Acidobacteria group. Interestingly, none of the clones of β-Proteobacteria and only three clones of Firmicutes were detected from our culture-independent study, whereas Firmicutes and β-Proteobacteria groups have been frequently detected in soils by culture-independent microbial community analysis (5, 6, 10, 62). Our results were somewhat similar to those from Austrian forest soil (19) and arid U.S. southwestern soil (34) in that Acidobacteria was most abundant in both soils, whereas Firmicutes was scant.
Considering the diversity of soil bacteria, an immense number of metagenomic clones would be needed to completely assay microbial diversity using conventional cloning vectors. Depending on soil type, we usually obtained 30,000 to 100,000 clones in fosmids, using 1 μg of soil DNA with a 35-kb average insert (data not shown). Our libraries for the present study, consisting of 80,500 and 33,200 clones from each of two forest soils, represent almost 4 Gb of genomic DNA, which is equivalent to approximately 1,000 E. coli genomes. Fosmids are good vectors for constructing metagenomic libraries because of their high cloning efficiency, improved stability in E. coli, and optimum (40 kb) insert size (31). Given the low chance of finding desirable genes in a metagenomic library of diverse microbial genomes, cloning efficiency is important in constructing a large clone library, which should include most of the microbial DNA in the soil (38). The genomes of many rare microorganisms were probably not included in our library. An rRNA gene analysis from the Jindong Valley forest soil revealed Acidobacteria to be the most abundant clone type; therefore, our metagenomic library may consist largely of genes from this group.
The metagenomic clones of the forest topsoil microorganisms may be involved in the biotransformation of organic compounds originating from other forest organisms, mainly plants. The characterized metagenomic clone in our study carried genes involved in indigo and indirubin production. The gene for putative indole oxygenase (ORF5) is probably responsible for indoxyl production from indole, as noted in previous studies (16, 22). Indoxyl then spontaneously dimerizes to form indigo, which can be converted to indirubin. Since pJEC5 did not produce pigmentation when introduced into tryptophanase minus E. coli (CY15000), pigment production is likely dependent on indole generated from tryptophan catabolism (16). Our gene product was similar to several oxidoreductases, but the identity level was not particularly high. Nor was it well aligned to the gene products of Methylophaga sp. (11) and Ralstonia eutropha (16), which yielded indigo in E. coli. Our gene product was not at all similar to the gene for indirubin production from another soil metagenome (39), which produced red indirubin pigment and possessed antibacterial activity; however, the antibacterial activity was an effect of indirubin and an unknown unpigmented component. In contrast, the antibacterial activity of our clone was solely attributable to the antibiotic activity by the pigment. The role of the divergently transcribed putative transcriptional regulator (ORF4) during pigment production is not certain. It is also not clear whether the gene product is involved in regulating the gene expression of ORF5.
Since the full-length original clone was not sequenced, we had no phylogenetic markers by which to study the origin of the individual clones. In addition, our attempts to amplify and clone 16S rRNA gene from the pJEC5 fosmid clone were not successful. Several strategies have been described for linking phylogenetic groups to their activity and function in soil (56), but defining the origin of a specific metagenomic clone remains a great challenge. It is likely that the most abundant phylotype is an Acidobacteria, and it will be interesting to determine whether our clone originated from the Acidobacteria. We plan to examine the origin of the clone by plate-wash PCR (52) with ORF5 gene-specific primers, using bacterial cultures from various culture conditions and media.
We describe here the microbial community composition of two forest soils and the subsequent construction of metagenomic libraries to select novel antibiotics. Surprisingly, our analysis of the small subunit rRNA gene library from the forest soil, from which we took the metagenomic DNA, showed that the most abundant microbial group was the major unculturable phylum Acidobacteria. Studying the library by expression-dependent or sequence homology-dependent methods would provide a unique opportunity to analyze the genome of the Acidobacteria. Our study has shown that metagenomic genes and their products can be obtained from these libraries, suggesting that these libraries could be used for many biotechnology applications (50).
Acknowledgments
We thank C. O. Jeon for valuable assistance in the phylogenetic analysis.
This research was supported by grant MG 05-0103-1-0 to S.W.L from the Microbial Genomics and Application Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology of the Republic of Korea.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Javanovich, R. A. Feldman, and E. F. Delong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. [DOI] [PubMed] [Google Scholar]
- 4.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of archaea from soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borneman, J., P. W. Skroch, K. M. O'Sullivan, J. A. Palus, N. G., Rumjanek, J. L. Jansen, J. Nienhuis, and E. W. Triplett. 1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 62:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brady, S. F., C. J. Chao, and J. Clardy. 2002. New natural product families from an environmental DNA (eDNA) gene cluster. J. Am. Chem. Soc. 124:9968-9969. [DOI] [PubMed] [Google Scholar]
- 8.Brady, S. F., and J. Clardy. 2000. Long-chain N-acyl amino acid antibiotics isolated from heterologously expressed environmental DNA. J. Am. Chem. Soc. 122:12903-12904. [Google Scholar]
- 9.Brünker, P., O. Sterner, J. E. Bailey, and W. Minas. 2001. Heterologous expression of the naphthocyclinone hydroxylase gene from Streptomyces arenaefor production of novel hybrid polyketides. Antonie Leeuwenhoek 79:235-245. [DOI] [PubMed] [Google Scholar]
- 10.Cho, J.-C., and S.-J. Kim. 2000. Increase in Bacterial community diversity in subsurface aquifers receiving livestock wastewater input. Appl. Environ. Microbiol. 66:956-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, H. S., J. K. Kim, E. H. Cho, Y. C. Kim, J. I. Kim, and S. W. Kim. 2003. A novel flavin-containing monooxygenase from Methylophaga sp. strain SK1 and its indigo synthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 306:930-936. [DOI] [PubMed] [Google Scholar]
- 12.Choi, K. Y., D. Kim, S.-C. Koh, J.-S. So, Y. M. Kim, and E. Kim. 2004. Molecular cloning and identification of a novel oxygenase gene specifically induced during the growth of Rhodococcus sp. strain T104 on limonene. J. Microbiol. 42:160-162. [PubMed] [Google Scholar]
- 13.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courtois, S., C. M. Cappellano, M. Ball, F.-X. Francou, P. Normand, G. Helynck, A. Martinez, S. J. Kolvek, J. Hopke, M. S. Osburne, P. R. August, R. Nalin, M. Guerineau, P. Jeannin, P. Simonet, and J.-L. Pernodet. 2003. Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl. Environ. Microbiol. 69:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeSantis, G., Z. Zhu, W. A. Greenberg, K. Wong, J. Chaplin, S. R. Hanson, B. Farwell, L. W. Nicholson, C. L. Randi, D. P. Weiner, D. E. Robertson, and M. J. Burk. 2002. An enzyme library approach to biocatalysis: development of nitrilases for enantioselective production of carboxylic acid derivatives. J. Am. Chem. Soc. 124:9024-9025. [DOI] [PubMed] [Google Scholar]
- 16.Drewlo, S., C. O. Brämer, M. Madkour, F. Mayer, and A. Steinbüchel. 2001. Cloning and expression of a Ralstonia eutropha HF39 gene mediating indigo formation in Escherichia coli. Appl. Environ. Microbiol. 67:1964-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillespie, D. E., S. F. Brady, A. D. Bettermann, N. P. Cianciotto, M. R. Liles, M. R., Rondon, J. Clardy, R. M. Goodman, and J. Handelsman. 2002. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 68:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta, R., Q. K. Beg, and P. Lorenz. 2002. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 59:15-32. [DOI] [PubMed] [Google Scholar]
- 19.Hackl, E., S. Zechmeister-Boltenstern, L. Bodrossy, and A. Sessitsch. 2004. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microbiol. 70:5057-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handelsman, J., M. R. Rondon, S. P. Brady, J. Clady, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-249. [DOI] [PubMed] [Google Scholar]
- 22.Hart, S., K. R. Koch, and D. R. Woods. 1992. Identification of indigo-related pigments produced by Escherichia coli containing a cloned Rhodococcus gene. J. Gen. Microbiol. 138:211-216. [DOI] [PubMed] [Google Scholar]
- 23.Hart, S., R. Kirby, and D. R. Woods. 1990. Structure of a Rhodococcus gene encoding pigment production in Escherichia coli. J. Gen. Microbiol. 136:1357-1363. [DOI] [PubMed] [Google Scholar]
- 24.Henne, A., R. A. Schmitz, M. Bomeke, G. Gottschalk, and R. Daniel. 2000. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity of Escherichia coli. Appl. Environ. Microbiol. 66:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiraishi, A., N. Kishimoto, Y. Kosako, N. Wakao, and T. Tano. 1995. Phylogenetic position of the menaquinone-containing acidophilic chemo-organotroph Acidobacterium capsulatum. FEMS Microbiol. Lett. 132:91-94. [DOI] [PubMed] [Google Scholar]
- 26.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hugenholtz, P., and N. R. Pace. 1996. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 14:190-197. [DOI] [PubMed] [Google Scholar]
- 28.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, N.Y.
- 31.Kim, U.-J., H. Shizuya, P. J. De Jong, B. Birren, and M. I. Simon. 1992. Stable propagation of cosmid-sized human DNA inserts in an F factor based vector. Nucleic Acids Res. 20:1083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knietsch, A., S. Bowien, G. Whited, G. Gottschalk, and R. Daniel. 2003. Identification and characterization of coenzyme B12-dependent glycerol dehydratase- and diol dehydratase-encoding genes from metagenomic DNA libraries derived from enrichment cultures. Appl. Environ. Microbiol. 69:3048-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knietsch, A., T. Waschkowitz, S. Bowien, A. Henne, and R. Daniel. 2003. Construction and screening of metagenomic libraries derived from enrichment cultures: generation of a gene bank for genes conferring alcohol oxidoreductase activity on Escherichia coli. Appl. Environ. Microbiol. 69:1408-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid Southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, S.-W., K. Won, H. K. Lim, J.-C. Kim, G. J. Choi, and K. Y. Cho. 2004. Screening for novel lipolytic enzymes from uncultured soil microorganisms. Appl. Microbiol. Biotechnol. 65:720-726. [DOI] [PubMed] [Google Scholar]
- 36.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenz, P., and C. Schleper. 2002. Metagenome—a challenging source of enzyme discovery. J. Mol. Catal. B-Enzym. 19-20:13-19. [Google Scholar]
- 39.MacNeil, I. A., C. L. Tiong, C. Minor, P. R. August, T. H. Grossman, K. A. Loiacono, B. A. Lynch, T. Phillips, S. Narula, R. Sundaramoorthi, A. Tyler, T. Aldredge, H. Long, M. Gilman, D. Holt, and M. S. Osburne. 2001. Expression and isolation of antimicrobial small molecules from soil DNA libraries. J. Mol. Microbiol. Biotechnol. 3:301-308. [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 42.Pettit, R. K. 2004. Soil DNA libraries for anticancer drug discovery. Cancer Chemother. Pharmacol. 54:1-6. [DOI] [PubMed] [Google Scholar]
- 43.Piel, J., D. Hui, N. Fusetani, and S. Matsunaga. 2004. Targeting modular polyketide synthases with iteratively acting acyltransferases from metagenome of uncultured bacterial consortia. Environ. Microbiol. 6:921-927. [DOI] [PubMed] [Google Scholar]
- 44.Quaiser, A., T. Ochsenreiter, H.-P. Klenk, A. Kletzin, A. H. Treusch, G. Meurer, J. Eck, C. W. Sensen, and C. Schleper. 2002. First insight into the genome of an uncultivated crenarchaeote from soil. Environ. Microbiol. 4:603-611. [DOI] [PubMed] [Google Scholar]
- 45.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rondon, M. R., R. M. Goodman, and J. Handelsman. 1999. The earth's bounty: assessing and accessing soil microbial diversity. Trends Biotechnol. 17:403-409. [DOI] [PubMed] [Google Scholar]
- 47.Rondon, M. R., S. J. Raffel, R. M. Goodman, and J. Handelsman. 1999. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc. Natl. Acad. Sci. USA 96:6451-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 50.Schloss, P. D., and J. Handelsman. 2003. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14:303-310. [DOI] [PubMed] [Google Scholar]
- 51.Seow, K.-T., G. Meurer, M. Gerlitz, E. Wendt-Pienkowski, C. R. Hutchinson, and J. Davies. 1997. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J. Bacteriol. 179:7360-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torsvik, V., F. L. Daae, R.-A. Sandaa, and L. Øvreås. 1998. Novel techniques for analyzing microbial diversity in natural and perturbed environments. J. Biotechnol. 64:53-62. [DOI] [PubMed] [Google Scholar]
- 55.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torsvik, V., and L. Øvreås. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 57.Torsvik, V., L. Øvreås, and T. F. Thingstad. 2002. Prokaryotic diversity: magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 58.Voget, S., C. Leggewie, A. Uesbeck, C. Raasch, K.-E. Jaeger, and W. R. Streit. 2003. Prospecting for novel biocatalysts in a soil metagenome. Appl. Environ. Microbiol. 69:6235-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, G.-Y.-S., E. Graziani, B. Waters, W. Pan, X. Li, J. McDermott, G. Meurer, G. Saxena, R. J. Anderson, and J. Davies. 2000. Novel natural products from soil DNA libraries in a streptomycete host. Org. Lett. 2:2401-2404. [DOI] [PubMed] [Google Scholar]
- 60.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yanofsky, C., and V. Horn. 1981. Rifampin resistance mutations that alter the efficiency of transcription termination at the tryptophan operon attenuator. J. Bacteriol. 145:1334-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou, J., B. Xia, H. Huang, A. V. Palumbo, and J. M. Tiedje. 2004. Microbial diversity and heterogeneity in sandy subsurface soils. Appl. Environ. Microbiol. 70:1723-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]