Abstract
We evaluated whether bacteria with higher cell-specific nucleic acid content (HNA) or an active electron transport system, i.e., positive for reduction of 5-cyano-2,3-ditolyl tetrazolium chloride (CTC), were responsible for the bulk of bacterioplankton metabolic activity. We also examined whether the phylogenetic diversity of HNA and CTC-positive cells differed from the diversity of Bacteria with low nucleic acid content (LNA). Bacterial assemblages were sampled both in eutrophic shelf waters and in mesotrophic offshore waters in the Oregon coastal upwelling region. Cytometrically sorted HNA, LNA, and CTC-positive cells were assayed for their cell-specific [3H]leucine incorporation rates. Phylogenetic diversity in sorted non-radioactively labeled samples was assayed using denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA genes. Cell-specific rates of leucine incorporation of HNA and CTC-positive cells were on average only slightly greater than the cell-specific rates of LNA cells. HNA cells accounted for most bacterioplankton substrate incorporation due to high abundances, while the low abundances of CTC-positive cells resulted in only a small contribution by these cells to total bacterial activity. The proportion of the total bacterial leucine incorporation attributable to LNA cells was higher in offshore regions than in shelf waters. Sequence data obtained from DGGE bands showed broadly similar phylogenetic diversity across HNA, LNA, and CTC-positive cells, with between-sample and between-region variability in the distribution of phylotypes. Our results suggest that LNA bacteria are not substantially different from HNA bacteria in either cell-specific rates of substrate incorporation or phylogenetic composition and that they can be significant contributors to bacterial metabolism in the sea.
Marine bacteria are responsible for most respiration of organic matter in the sea and play important roles in marine biogeochemical cycles (16, 36, 37). Understanding how bacterial activity varies in time and space is thus vital to understanding the functioning of marine ecosystems. Recent studies support the idea that marine bacterial cells are not uniformly active; rather, subsets of the bacterial assemblage appear to be more or less active at any one time (9, 12, 67). Flow cytometric analysis of marine bacterioplankton routinely shows the presence of two major clusters of bacterial cells based on cell-specific nucleic acid content: high nucleic acid content (HNA) and low-nucleic-acid-content (LNA) cells (22, 29, 39). HNA cells have been proposed to be active cells and LNA cells to be inactive or dead cells (23, 29, 39) or cells with lower cell-specific metabolic activity than HNA cells (67, 77).
The relationship between the incorporation rate of radioactively labeled substrates and the cell-specific nucleic acid content has been examined by flow cytometric sorting of HNA and LNA cells (3, 35, 58, 59, 77). For both in situ samples and samples from a mesocosm enriched with nitrogen and phosphorus, HNA cells have been found to be responsible for a larger fraction of bacterial leucine incorporation than were LNA cells (35, 58, 59). However, in a study in the Celtic Sea, Zubkov et al. (77) reported that when incorporation rates of 35S-methionine were scaled to cell-specific protein content, LNA cells had biomass-specific incorporation rates higher than those found for HNA cells in surface waters, although not in pycnocline or deeper waters.
The relative activity of bacterial cells has also been assessed using the redox dye 5-cyano-2,3-ditolyl tetrazolium chloride (CTC) (55, 68). The abundance of CTC-positive cells has been reported to be positively correlated with whole-water respiration rates (66) and with volumetric incorporation rates of [3H]leucine or [3H]thymidine (13, 61). CTC reduction has been hypothesized to indicate the most highly metabolically active cells in a bacterial assemblage (48, 61). However, this does not necessarily imply that CTC-positive cells are dominant in terms of biosynthetic activity, since CTC reduction is an indicator of respiration. If balanced growth is not occurring, the number of cells reducing CTC may not correlate with community or cell-specific leucine or thymidine incorporation rates. For example, Servais et al. (57) sorted CTC-positive cells preincubated with [3H]leucine and found that CTC-positive cells contributed <40% of the total bacterial substrate incorporation in two eutrophic coastal marine systems in which the percentages of CTC-positive cells were higher than typically found in open-ocean habitats.
The idea that HNA, LNA, and CTC-positive bacteria may be phylogenetically, as well as metabolically, variable compared to the aggregate bacterial assemblage has been explored in a few studies. Differences in the phylogenetic diversities of HNA and LNA cells have been reported for North Sea bacterioplankton (75, 77), although samples from the Mediterranean Sea did not reveal phylogenetic differences based on cell-specific nucleic acid content (58). Investigations of the phylogenetic diversity of CTC-positive bacteria from coastal Mediterranean seawater indicated that most of the bacterial phylotypes in the whole-seawater community were also present in the CTC-positive subgroup of cells (2, 4). However, the diversity of Bacteria in these studies may have been biased by the addition of yeast extract added in order to obtain sufficient CTC-positive cells for genetic analysis.
Since the California Current System provides a wide range of environmental conditions, with pelagic ecosystems ranging from highly eutrophic to oligotrophic (8, 28, 52), it is an excellent region in which to examine variability in the cell-specific metabolic activity and diversity of bacterioplankton in relation to the trophic state. Bacterial leucine incorporation rates in these waters have been shown to vary over 3 orders of magnitude, with rates decreasing from upwelling shelf surface waters to subsurface depths and with increasing distance from shore (62). In this study, we examined both the cell-specific activity and the phylogenetic diversity of heterotrophic bacteria in seawater samples collected off the Oregon coast in late April/early May 2002. Samples from three stations, shelf, slope, and open basin, were sorted to examine the variability in cell-specific leucine incorporation and to determine the phylogenetic diversity of cells in the domain Bacteria within sorted populations of HNA, LNA, and CTC-positive cells. Our null hypothesis was that the three flow cytometrically defined groups would contain similar diversities of Bacteria. Most of the bacterial phylotypes identified in this study were present in both the HNA and LNA assemblages and had, at times, an active electron transport system (i.e., they were able to reduce CTC). CTC-positive cells had high cell-specific rates of leucine incorporation but insufficient abundances to account for more than, on average, 7 to 14% of the total bacterial activity. Although the HNA cells were responsible for the bulk of bacterioplankton leucine incorporation rates in all three regions, LNA cells contributed a larger proportion of total activity at the open-basin station, at the edge of the North Pacific gyre.
MATERIALS AND METHODS
Sample collection and oceanographic parameters.
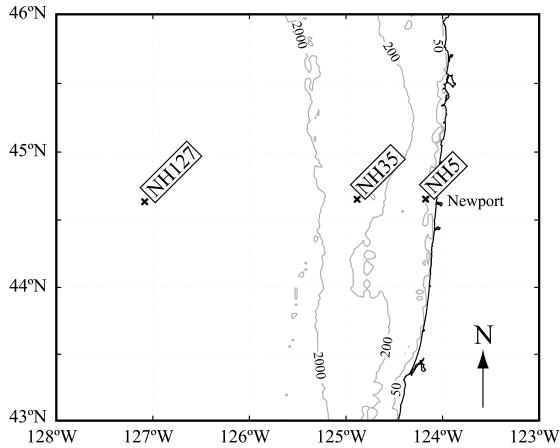
Samples were collected using General Oceanics 5-liter niskin bottles mounted on a rosette equipped with a SeaBird SBE 911+ CTD, a SeaTech fluorometer, a Biospherical PAR sensor, and a SeaTech 25-cm transmissometer. Three stations in different hydrographic regimes were chosen along the Newport hydrographic line extending westward from Newport, Oregon: a shelf station 9 km from shore (bottom depth, 60 m) (NH5; 44.65°N, 124.18°W), a slope station 65 km from shore (bottom depth, 450 m) (NH35; 44.65°N, 124.88°W), and a basin station above the abyssal plain 249 km from shore (bottom depth, 2,900 m) (NH127; 44.65°N, 127.1°W) (Fig. 1).
FIG. 1.
Regional map of the sampling area with contour lines indicating the 50-m, 200-m, and 2,000-m isobaths. Multiple casts were conducted at each of the three stations: a shelf station (NH5), a slope station (NH35), and a basin station above the abyssal plain (NH127).
Environmental parameters.
Chlorophyll concentration was used as a proxy for phytoplankton biomass. Discrete water samples were filtered through GF/F filters and kept frozen at −80°C for up to 1 month before being processed. Ninety percent high-performance liquid chromatography grade acetone was added to the filters and allowed to extract overnight at −20°C. Chlorophyll a and pheopigment concentrations were determined using a Turner Designs 10-AU fluorometer (69). Samples for nutrient analysis were stored frozen (−20°C) in 60-ml high-density polyethylene bottles. The analyses for phosphate, nitrate plus nitrite (N+N), nitrite, and silicic acid (silicate) were performed using a hybrid Technicon AutoAnalyzerII and Alpkem RFA300 system following protocols modified from Gordon et al. (26). Nitrate concentrations were determined by subtracting nitrite from the N+N value. The estimated precision for each element was as follows: PO4, ±0.008 μM; N+N, ±0.15 μM; NO2, ±0.01 μM; and silicic acid, ±0.3 μM.
Whole-seawater volumetric leucine incorporation rates.
Incorporation rates of [3H]leucine by bacterioplankton present in whole-seawater samples were assayed following the protocol of Smith and Azam (65). Four 1.5-ml aliquots of each water sample were pipetted into copolymer microcentrifuge tubes made by Eppendorf containing [3H]leucine (Perkin-Elmer Life Science Products; specific activity, 170 Ci mmol−1) to yield a 20 nM final concentration. One of the four aliquots served as a killed control, with 5% (final concentration) trichloroacetic acid (TCA) immediately added to the tube. The aliquots were incubated for 1 hour in the dark at the in situ water temperature (5.1 to 11.3°C). The samples were then killed with 5% TCA (final concentration) and stored frozen at −20°C. The samples were returned to shore and processed no more than 3 weeks after the sampling date. Prior experiments with radioactively labeled samples frozen for up to 1 month showed no difference in leucine incorporation rates compared to unfrozen samples processed immediately (data not shown). Activity was determined using a Wallac 1141 liquid scintillation counter. The activity of the killed control was subtracted from the values for the three live aliquots.
Flow cytometric analysis of bacteria.
A Becton-Dickinson FACSCalibur flow cytometer was used for all cell enumeration and cell sorting. For enumeration and sorting for molecular analysis, water samples in 3-ml aliquots were fixed with 0.2% (wt/vol) paraformaldehyde (final concentration), stored in the dark for at least 10 min at room temperature to harden the cells, and quick frozen in liquid nitrogen. The samples were then stored at −80°C until sample processing was done on shore.
To identify cells with an active electron transport system, water samples were incubated with CTC (Polysciences, Warrington, PA) immediately following collection. Three-milliliter subsamples were incubated with 2.5 mM CTC (final concentration) for 1 hour in the dark at 10°C. Previous studies off the Oregon coast had shown no difference in flow cytometric detection and incorporation of 2.5 mM CTC compared to 5 mM CTC (data not shown), confirming the finding of Servais et al. (57), who also used 2.5 mM CTC in their study. Furthermore, maximum CTC-positive cell counts were reached with 60-min incubations (data not shown). Water samples used to determine rates of cell-specific leucine incorporation were incubated immediately after collection with 40 nM of [3H]leucine (specific activity, 170 Ci mmol−1) in the dark for 1 hour at 10°C. Following the initial 1-hour incubation, a subset of the sample tubes was incubated with 2.5 mM of CTC for an additional hour in the 10°C water bath. Samples for radioactively labeled sorting were then fixed and stored as described above.
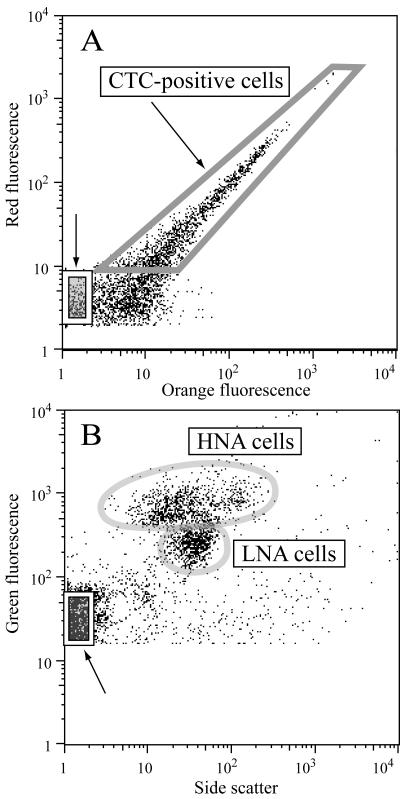
After being thawed, aliquots of samples collected for determination of bacterial abundance and cell-specific nucleic acid content were stained with a 1× working stock of SYBR Green I (Molecular Probes, Eugene, OR) and 25 mM potassium citrate for 15 min, following a protocol modified from Marie et al. (43). The abundance of cells was determined by calculating the flow rate with known concentrations of 1-μm fluorescent microspheres (Polysciences, Warrington, PA). Regions were established on cytogram plots of side scatter versus green fluorescence and adjusted for each sample to define HNA and LNA bacterial cells (Fig. 2B). The mean cell-specific SYBR fluorescences were obtained for HNA and LNA cells, along with the abundance of cells within each defined region. The fluorescence data from the HNA and LNA regions were normalized to the fluorescence of the beads prior to any further analysis in order to account for day-to-day machine variation. CTC-positive cell abundance was also determined via flow cytometry from cytogram plots of red versus orange fluorescence (13) (Fig. 2A). Prochlorococcus cells were not observed in any of the samples (63) and therefore did not interfere with CTC-positive cell counts. Synechococcus cells were easily distinguished from CTC-positive cells using cytograms of FL2 (orange fluorescence) versus FL3 (red fluorescence).
FIG. 2.
Cytograms indicating the sort regions for (A) the CTC-positive cells as defined on cytograms of orange versus red fluorescence and (B) the HNA and LNA regions as defined on the cytograms of side scatter versus green fluorescence. For both plots, the box marked with an arrow at the x-y origin of the cytogram contains the noncellular particles sorted to obtain each sample's background radioactivity level.
A total of 35 discrete water samples—15, 9, and 11 sorts at stations NH5, NH35, and NH127, respectively (see Table 2)—were sorted for determination of cell-specific leucine incorporation of total cells, HNA cells, LNA cells, and CTC-positive cells. Prior to the sorting, aliquots of each thawed sample were incubated in the dark for 15 min with a 2× working stock of SYBR Green I. Due to the length of the sorts (20 to 240 min), we used a higher concentration of the stain and placed a freshly stained aliquot of each sample on the flow cytometer every 15 min during a sort. All sorts were run on low flow using the single-cell option of the Becton-Dickinson CellQuest software. Bacterial cells were sorted using four different sort regions: an HNA cell region, an LNA cell region, and HNA plus LNA cells (i.e., a total bacterial region) in side scatter versus green-fluorescence cytograms and a CTC-positive cell region in red- versus orange-fluorescence cytograms. Cells were sorted onto 25-mm, 0.2-μm cellulose acetate membranes using the Becton-Dickinson cell concentrator unit attached to the flow cytometer's sort line. A total of 1.0 × 105 SYBR-stained cells were sorted for each region; at least 2 ×104 CTC-positive cells were sorted. As a control, 1.0× 105 low-fluorescent (noncell) particles at the x-y origin of the cytograms (Fig. 2) were sorted and processed for background radioactivity. There was no significant difference between the disintegrations per minute (DPM) obtained from the noncell region and the TCA-killed whole-seawater samples (paired t test, P = 0.17).
TABLE 2.
Water samples collected from three sampling stationsa
| Station | Depth (m) | Heterotrophic cell abundance (106 cells ml−1) | % HNA cells | % CTC-positive cells | No. of samples sorted
|
|
|---|---|---|---|---|---|---|
| Diversity | Leucine incorporation | |||||
| Shelf | 2-8 | 0.9-1.41 | 62-90 | 1.4-4.0 | 2 | 5 |
| 11-25 | 0.6-2.15 | 64-86 | 2.1-4.2 | 2 | 5 | |
| 32-50 | 0.48-0.86 | 63-82 | 1.2-5.1 | 2 | 5 | |
| Slope | 3-10 | 0.93-1.56 | 54-75 | 1.5-6.1 | 2 | 3 |
| 17-30 | 1.21-1.85 | 43-57 | 1.9-4.4 | 2 | 3 | |
| 40-100 | 0.35-1.16 | 45-70 | 1.7-10.3 | 2 | 3 | |
| Basin | 20-50 | 1.18-2.47 | 42-45 | 1.0-3.3 | 2 | 3 |
| 100-150 | 0.32-0.44 | 50-71 | 1.8-6.5 | 2 | 3 | |
| 300-500 | 0.07-0.24 | 53-70 | 1.6-24.4 | 2 | 5 | |
The table indicates the range of abundances of heterotrophic cells and the percentage of heterotrophic cells that were HNA or CTC-positive cells. Different numbers of samples were sorted to determine (i) the diversity of the HNA cells, LNA cells, and CTC-positive cells and (ii) cell-specific leucine incorporation rates of total cells, HNA cells, LNA cells, and CTC-positive cells.
After the desired number of cells had been collected, the filter was removed from the cell concentrator unit, transferred to a filter manifold, and processed following the method of Kirchman (33), which has previously been shown to result in values similar to those of the centrifugation method (65). The filter was washed twice with cold 5% TCA and twice with cold 80% ethanol. After being washed, the filter was folded into a 7-ml scintillation vial and allowed to dry. Ethyl acetate (0.5 ml) was added to the scintillation vial to dissolve the filter, followed by 3 ml of scintillation cocktail (Ultima Gold LLT; PerkinElmer). The samples were allowed to sit for 2 days before being counted on a Wallac 1141 liquid scintillation counter. For each sample, the DPM of the background filter (noncell sort) was subtracted from the DPM of the cell sorts for that sample. Cell-specific rates of leucine incorporation were obtained by dividing the molar leucine incorporation rate determined for each filter by the number of cells sorted for that sample. The volumetric leucine incorporation rate for each sorted group was calculated by multiplying the abundance of cells within each sorted region by the average cell-specific leucine incorporation rate for that group. The calculated volumetric leucine incorporation rates based on cell sorting are thus distinct from whole-seawater volumetric leucine incorporation rates obtained empirically from unsorted samples incubated with radioactively labeled leucine.
Preliminary tests in our laboratory and recently published data (18) indicate that flow cytometry can be used to sort cells with high purity if flow rates are kept below 300 cells s−1. The low abundance of cells in our samples meant that sort rates averaged from 7 to 80 cells s−1. We used three metrics to examine the purity of sorting by the flow cytometer. First, the number of cells within the region of interest that were rejected by the flow cytometer was tested by sorting CTC-positive cells and using epifluorescence microscopy to count the number of CTC-positive cells that appeared in the waste tank. The average number of CTC-positive cells in the waste tank, which were therefore rejected by the flow cytometer, was less than 0.1% of the total CTC-positive cell count (n = 3). Second, samples were sorted within a region of interest; the sorted cells were then rerun through the flow cytometer to check for the appearance of cells within the region of interest. In the initial samples, cells within the sort region averaged 22% of the SYBR-stained cells; after sorting and rerunning the samples on the flow cytometer, the cells within the sort region averaged 76% of the SYBR-stained cells (n = 2). Finally, the whole-seawater volumetric leucine incorporation rates, which were independently obtained, agreed well with the total leucine incorporation rates (details below), indicating that while the flow cytometer may not sort 100% of the cells within the region of interest, the carryover is small enough not to impact the final rates.
Molecular analysis of bacterial diversity.
At each of the three stations, samples were collected for molecular analysis from three depths ranging from 5 to 300 m from two separate casts, for a total of 18 discrete water samples (see Table 2). For each sample, three sorts were conducted: HNA cells, LNA cells, and CTC-positive cells. To minimize bacterial contamination, Milli-Q water was filtered through a 0.1-μm Supor filter (Pall Corporation, East Hills, NY) and autoclaved prior to use as a sheath fluid. Each day, the sheath tank was rinsed once with 100% bleach and four times with filter-sterilized, autoclaved Milli-Q water. The sort line was cleaned at the beginning of each day by running 5% bleach for 15 min, followed by filter-sterilized, autoclaved Milli-Q water for 20 min. To prevent carryover between samples, the flow cytometer's sort line was also cleaned after each sample run with a solution of 5% bleach (10 min), followed by filter-sterilized, autoclaved Milli-Q water (20 min). The sort line was tested daily for the presence of contaminants by sorting only sheath fluid and collecting the outflow from the sort line on a filter; this filter was then treated as a sample. If the negative sort indicated the presence of DNA, the sorts from the entire day were repeated. Once the cleaning protocol had been optimized, only 1 day's sorts (three samples) had to be repeated because of contamination. At least 2 × 104 cells per sample were sorted for the CTC-positive samples, and at least 5 × 104 cells were sorted for HNA and LNA regions of SYBR-stained samples.
For each water sample, DNA was extracted from the sorted cells and from 3 ml of whole seawater collected on a filter. An extraction method modified from that of Giovannoni et al. (25) was chosen after several extraction methods were tested on paraformaldehyde-fixed cells, and it was used for both the whole-seawater samples and the sorted samples. Cells were lysed with a lysis buffer (40 mM EDTA, 50 mM Tris, 400 mM NaCl, 0.75 M sucrose, pH 8.0). Sodium dodecyl sulfate (final concentration, 0.5%) and proteinase K (final concentration, 500 μg ml−1) were added, and the cells were incubated overnight at 42°C. The solutions were then extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1). DNA was precipitated with 2 volumes ice-cold 96% ethanol for 6 hours at −20°C and then spun at 15,000 × g for 45 min at 4°C. After a wash with 70% ethanol and an additional 30-min centrifugation at 15,000 × g, the DNA was dried and resuspended in 10 mM Tris (pH 8.0). 16S rRNA genes in the extracted DNA were amplified using PCR with the following reaction conditions: 1× Promega PCR buffer, 200 μM deoxynucleoside triphosphates, 400 nM of each primer, 2 U of Promega Taq, and 2.5 mM MgCl2. PCR primers were 907RA (56) and 341F-GC (45), which has a 40-nucleotide GC-rich section for the denaturing gradient gel electrophoresis (DGGE) analysis. Conditions for the PCR were an initial denaturation at 94°C for 5 min; 30 cycles of 94°C for 30 s, 48°C for 30 s, and 72°C for 90 s; and a final 10 min at 72°C.
PCR products of the V3-V4-V5 variable regions (47) were analyzed using DGGE (46). Controls were run adjacent to the samples in order to facilitate comparison of the DGGE banding patterns between the different gels. After including positive controls on the DGGE gels, up to 12 samples were run per gel, resulting in six different gels. In addition, each sample was run on two different DGGE gels to facilitate comparison of the DGGE banding patterns between different samples and different sorts, and therefore a total of 12 DGGE gels were run. A 6% (wt/vol) acrylamide-bis-acrylamide gel was prepared in TAE buffer (40 mM Tris, 20 mM glacial acetic acid, 1 mM EDTA). A 30% to 70% urea-formamide denaturing gradient was used, where 100% is equal to 40% (vol/vol) formamide and 7 M urea. The gel was run in a DGGE-2001 system (CBS Scientific, Del Mar, CA) at 60°C and 100 V for 18 h. The acrylamide gel was stained with SYBR Green I for 30 min and then viewed on a UV light box. The gels were imaged with an AlphaImager 2200 Digital Imaging System (Alpha Innotech, San Leandro, CA). Multiple images were collected with different aperture and exposure settings in order to maximize detection of weak DGGE bands yet still allow separation of brighter DGGE bands.
The individual bands from the DGGE gel were removed by piercing the gel with a pipette tip and placing the tip in 20 μl of 10 mM Tris for 10 min at room temperature. Two to 5 μl of this mixture was used to reamplify the 16S rRNA genes using conditions as described above, except the forward primer lacked the GC-rich section. The PCR products were then purified with the Montage PCR filter kit (Millipore, Billerica, MA) following the manufacturer's instructions.
The reamplified DGGE bands were sequenced by cycle sequencing using fluorescent dideoxy terminators. The sequences were resolved using an ABI 3100 at Oregon State University's Central Services Laboratory. Sequences were checked manually using TraceViewer (CodonCode Corporation) and then were aligned in ARB (41), initially using the FastAligner, and then manually checked. The phylogenetic identities of the sequences were determined initially based on BLAST results. The sequences were then inserted into maximum-likelihood trees using the parsimony option in ARB (40). The phylogenetic affiliations of the DGGE sequences were determined based on which cluster the DGGE sequence grouped within in the maximum-likelihood trees.
Analysis of DGGE banding patterns.
The pattern of DGGE bands was reduced into a presence/absence matrix for each of the three sorts. Separate distance matrices for the HNA sorts, LNA sorts, and CTC-positive sorts were calculated using the Sorensen distance measure and compared in pairs using a Mantel test (38) on ranked distances as suggested by Dietz (15). The only significant difference between the three matrices was between the LNA DGGE band patterns and the CTC DGGE band patterns (Mantel test, P = 0.046; calculated from Monte Carlo randomization of 1,000 runs; all other comparisons resulted in P values of >0.05). Therefore, the matrices were combined for the cluster analysis; the resulting matrix included all the HNA bands, LNA bands, and CTC-positive bands. Since our primary interest was heterotrophic cells, we excluded the whole-seawater samples from these analyses in order to prevent analysis of geographical patterns related to the photoautotrophic cells. Cluster analysis was used to examine possible groups of samples based on the presence/absence matrix. The analysis was conducted with a Sorensen distance measure and the flexible beta linkage method (β = −0.25).
Statistical analysis.
Statistical analyses were conducted in Matlab 6.5 (Mathworks) or PC-ORD v. 4.19 (MjM Software Design). For the cluster analysis, the data were transformed using a square root transformation to reduce the skewness and coefficient of variation of the data. The other data were log transformed to improve normality. Analyses performed included Spearman rank correlations, paired t tests, Wilcoxon signed rank tests, Mantel tests, and Kruskal-Wallis tests. All relationships were significant at the P<0.01 level unless otherwise noted.
RESULTS
Environmental parameters.
Evidence of seasonal upwelling was seen in several of the measured environmental parameters (Table 1). The highest water temperatures were observed at the slope station; further inland at the shelf station, surface water temperatures were lower due to upwelling of cold, high-nutrient subsurface waters. There was evidence of a shallowing of the nutricline near shore due to upwelling; however, nutrient concentrations increased with depth at all three stations. The highest chlorophyll concentrations were observed at the surface of the shelf station, with lower values at the basin station. The slope station had middepth maxima in chlorophyll and slightly fresher water at the surface due to the influence of the Columbia River. Whole-seawater volumetric leucine incorporation rates also decreased with depth and with increasing distance from shore (Table 1).
TABLE 1.
Environmental parameters measured concurrently with sample collection summarized for the three sampling stationsa
| Station | Depth (m) | Temperature (°C) | Nitrate (μM) | Chlorophyll α (μg liter−1) | Volumetric leucine incorporation (pM Leu h−1) |
|---|---|---|---|---|---|
| Shelf | 0.5-8 | 9.5 (8.6-10.6) | 5.9 (0-25.8) | 16.8 (0.9-43.8) | 297 (102-543) |
| 10-25 | 8.5 (8.0-9.8) | 21.1 (2.8-29.3) | 5.0 (0.4-33.0) | 86 (6-515) | |
| 32-50 | 7.6 (7.2-7.9) | 29.6 (0.7-33.8) | 0.5 (0.4-1.0) | 10 (3-20) | |
| Slope | 1.3-10 | 11.1 (10.2-11.5) | 0.1 (0-0.9) | 0.6 (0.4-1.3) | 74 (21-109) |
| 13-30 | 9.9 (9.4-10.9) | 0.7 (0-2.5) | 1.9 (0.5-3.8) | 31 (17-67) | |
| 35-410 | 8.5 (5.5-9.4) | 14.1 (2.3-40.4) | 0.8 (0.3-1.6) | 6 (0-14) | |
| Basin | 1.2-50 | 9.3 (8.8-9.8) | 5.1 (0.1-6.0) | 0.7 (0.4-1.3) | 10 (6-16) |
| 60-150 | 8.2 (7.4-9.0) | 16.2 (6.3-28.9) | ND | 2 (1-3) | |
| 250-2,700 | 4.6 (1.7-7.1) | 40.8 (34.7-45.1) | ND | 1 (0-2) |
Values are mean values, with the range of measured values in parentheses. The volumetric leucine incorporation rate shown is the whole-seawater volumetric leucine incorporation rate. ND for the basin chlorophyll samples indicates that chlorophyll levels were below detection.
Heterotrophic-cell abundances were higher at the surfaces of the shelf and basin station samples and decreased with depth, while heterotrophic cell abundances reached maximum values in the pycnocline at the slope station (Table 2). The percentage of HNA cells reached maximum values at the surface of the shelf station, while the percentage of CTC-positive cells was always less than 25% of heterotrophic-cell abundances, and usually less than 10% (Table 2).
Cell-specific rates of leucine incorporation for total, HNA, and LNA cells.
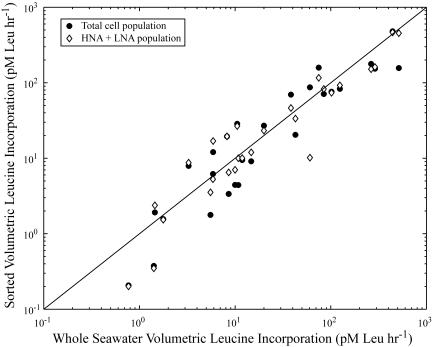
We checked whether cell sorting potentially resulted in underestimation of total bacterial leucine incorporation rates. The volumetric leucine incorporation rates of sorted total bacteria (HNA plus LNA cell regions) showed a strong, positive correlation around a 1:1 line to whole-seawater volumetric bacterial leucine incorporation rates (r = 0.92; Spearman rank correlation coefficient; P < 0.001) (Fig. 3). We also summed the calculated volumetric leucine incorporation rates for HNA and LNA groups sorted on the flow cytometer, obtained by multiplying the cell-specific leucine incorporation rate by the abundance of cells of each group in the sample. The summed sorted volumetric incorporation rates for the HNA and LNA cells were also significantly correlated with the whole-seawater volumetric leucine incorporation rate (r = 0.93; Spearman rank correlation coefficient; P < 0.001) (Fig. 3). Therefore, in general, the sum of the HNA and LNA cell incorporation rates was equivalent to the total bacterial cell incorporation rate. The good agreement found in these comparisons indicated that there was no detectable bias due to sorting with respect to evaluating the proportion of total bacterial activity rates due to specific subgroups of the bacterial assemblage.
FIG. 3.
Whole-seawater volumetric bacterial leucine incorporation rate compared to sorted volumetric leucine incorporation of the sorted total cells or to the sum of the sorted volumetric leucine incorporation by the HNA cells and LNA cells. The line marks a 1:1 relationship between whole-seawater volumetric leucine incorporation and the sorted volumetric incorporation rates. Sorted volumetric leucine incorporation is defined as the cell-specific leucine incorporation rate multiplied by the abundance of cells for the group.
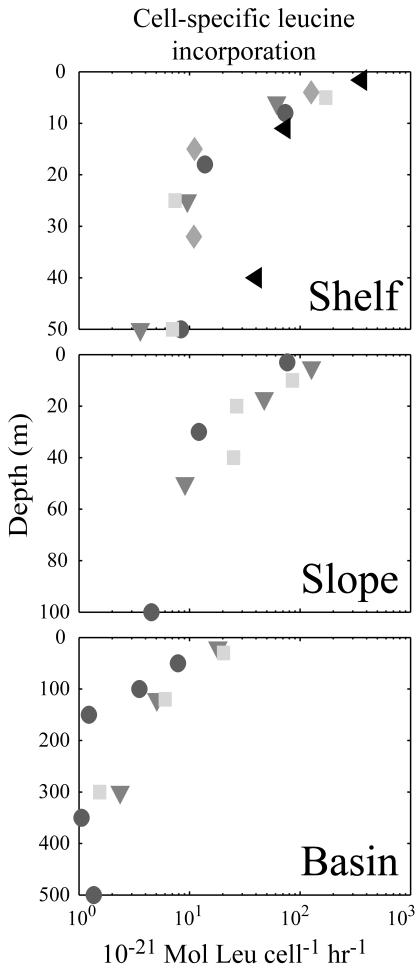
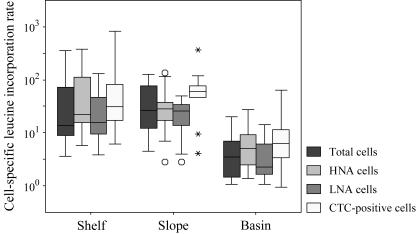
For all three regions sampled, cell-specific leucine incorporation rates decreased with depth (Fig. 4). The trend in cell-specific rates with depth shown for total cells (Fig. 4) was similar to the other sorted groups, although the range of cell-specific leucine incorporation rates varied for each group (Fig. 5). Cell-specific leucine incorporation rates were generally higher for HNA cells than for LNA cells (two-sided P < 0.001; paired t test). However, the average and range of cell-specific leucine incorporation rates for LNA cells were only marginally lower than for total and HNA cells. In four samples, the cell-specific leucine incorporation rate for LNA cells was actually greater than the cell-specific rate for HNA cells. Three of these samples, two from the basin station and one from the shelf, were from the deepest sample collected from the cast and may represent the lower limits of detection of the sorting method. The final sample was from the slope station at 10-m depth; the cell-specific leucine incorporation rate for the LNA cells was seven times greater than the HNA cell-specific leucine incorporation rate. This sample was sorted twice to confirm these numbers.
FIG. 4.
Variability in cell-specific leucine incorporation rates for total cells with depth. Each symbol denotes samples collected from a different cast.
FIG. 5.
Box plots of the cell-specific leucine incorporation rates across the three sampling stations. Four separate sorts were done for each sample: total cells, HNA cells, LNA cells, and CTC-positive cells. The boxes represent the middle 50% of the cell-specific leucine incorporation rates or the interquartile range (IQR) of the data. The bars extend to include data within 1.5 IQRs of the box. Outliers, marked with circles, are defined as cell-specific leucine incorporation rates between 1.5 and 3 IQRs away from the box; extreme outliers, marked with asterisks, are greater than 3 IQRs away from the box. The units for cell-specific leucine incorporation rates are pM leucine h−1.
The percentages of the sorted volumetric leucine incorporation rate attributable to the HNA and LNA groups were calculated by dividing the sorted volumetric leucine incorporation rate for each group by the sorted volumetric leucine incorporation rate of total bacterial cells (Table 3). HNA cells accounted for, on average, between 70% and >100%, while LNA cells accounted for between 25% and 38%, of total bacterial leucine incorporation. According to statistical analysis (Kruskal-Wallis test; P = 0.042; followed by a multiple comparison of mean ranks), the proportion of sorted volumetric leucine incorporation by LNA cells was higher at the basin station than at the shelf station. There were no other significant between-station differences.
TABLE 3.
Proportional contribution of sorted volumetric leucine incorporation rates of HNA cells, LNA cells, and CTC-positive cells to sorted volumetric leucine incorporation rate for total cellsa
| Sampling region | % of total volumetric leucine incorporation rate bya:
|
||
|---|---|---|---|
| HNA cells | LNA cells | CTC-positive cells | |
| Shelf | 116 (84-147) | 25 (14-36) | 7 (3-10) |
| Slope | 71 (44-99) | 25 (15-35) | 7 (3-12) |
| Basin | 82 (58-106) | 38 (26-51) | 14 (2-27) |
Values in parentheses are 95% confidence intervals. See the text for details on the calculations to determine the percent of volumetric leucine incorporation by the HNA, LNA, and CTC-positive populations.
Cell-specific leucine incorporation rates for CTC-positive cells.
As found for the SYBR-stained cells, the cell-specific leucine incorporation rates of CTC-positive cells were highest at the surface and decreased with depth. Cell-specific leucine incorporation rates for CTC-positive cells ranged over 3 orders of magnitude (0.9 × 10−21 to 840 × 10−21 M leucine cell−1 h−1). A point-to-point comparison showed that cell-specific leucine incorporation rates for CTC-positive cells were generally higher than rates found for total cells (Fig. 5) (two-sided P<0.0001; paired t test). The cell-specific leucine incorporation rate for the CTC-positive cells was not significantly greater than the cell-specific leucine incorporation rates for the HNA cells (two-sided P = 0.150; paired t test) but was significantly greater than the cell-specific leucine incorporation rate for the LNA cells (two-sided P < 0.0001; paired t test). In one-third of the samples, the cell-specific leucine incorporation rates for CTC-positive cells were 1% to 50% (average, 28%) lower than the rates for total cells. The percentage of the sorted volumetric leucine incorporation rate attributable to CTC-positive cells in individual samples ranged from 2% to 27% and on average accounted for only 7% to 14% of total activity (Table 3).
Phylogenetic analysis of sorted cell groups.
DNAs extracted from the sorted HNA, LNA, and CTC-positive cells were analyzed by running all three sorts from a single sample on the same DGGE gel. Bands from different sorts or from the whole-seawater sample that migrated to the same point in the DGGE gel were considered the same DGGE band. However, PCR products from different organisms can migrate to the same location in a DGGE gel (46), which would result in an underestimation of the diversity obtained from these samples. Conversely, the presence of heteroduplex molecules, in which strands of DNA from different PCR products reanneal, would artificially increase the number of DGGE bands, potentially overestimating sample diversity (46). Considering all the DGGE bands, only 7% of the bands were limited to the whole-seawater samples and were not obtained in any of the sorted assemblages. Since the samples were not prescreened to remove phytoplankton, some of these DGGE bands may have been due to photoautotrophs excluded from the later sorts. Even with the most stringent sorting criteria, there were DGGE bands that later were identified as phytoplankton or plastid DNA. After the sequence data revealed that some of the DGGE bands were chloroplasts or phytoplankton, the presence/absence matrix based on the DGGE gels was modified to consider only DGGE bands definitively identified as nonphytoplankton. Examination of all the DGGE bands combined revealed that 11% of the DGGE bands were observed in both the HNA and the LNA sorted assemblages, while 13% were present in all three assemblages: HNA, LNA, and CTC positive in the same sample. There was no significant difference between the proportion of DGGE bands observed in both the HNA and CTC-positive assemblages and the proportion of DGGE bands observed in both the LNA and CTC-positive assemblages in the same sample (6.7% and 5.7% of the DGGE bands, respectively; Wilcoxon signed rank test; P = 0.85).
Over 150 sequences were obtained from DGGE bands identified in the three sorted groups of cells, 146 in HNA sorts, 92 in LNA sorts, and 97 in CTC-positive sorts. Some of these sequences were observed only in either the HNA or LNA subset of the sorted assemblages. For example, sequences obtained only from the HNA assemblage included phylotypes of Bacteroidetes and Gammaproteobacteria (Table 4). Conversely, phylotypes of Gammaproteobacteria clustering with Marinobacterium and Pseudomonas, the one Methylobacteriaceae sequence, and the TM6 sequences were obtained only from the LNA assemblage. The single sequence from the SAR86 cluster within the Gammaproteobacteria, which occurred both in HNA and in LNA groups, was the only identified phylotype not found in the CTC-positive assemblage. Additional details concerning the sequence data are given by Longnecker (40).
TABLE 4.
Summary of phylogenetic information based on sequence data obtained from all DGGE bandsa
| Phylum | Class, order | Family, genus, or environmental cluster (reference[s]) | No. of sequences
|
|||
|---|---|---|---|---|---|---|
| HNA +/or LNA | CTC | HNA only | LNA only | |||
| Acidobacteria | 2 | 1 | ||||
| Actinobacteria | Marine gram-positive | 6 | 2 | |||
| Deinococcus-Thermus | Meiothermus | 3 | 2 | |||
| Bacteroidetes | Flavobacteria Flavobacteriales | Family Flavobacteriaceae, genus Polaribacter | 2 | 3 | ||
| Sphingobacteria Sphingobacteriales | Family Saprospiraceae | 3 | 1 | |||
| Environmental sequences | Delaware cluster 1 (34) | 5 | 1 | |||
| Delaware cluster 2 (34) | 1 | 1 | ||||
| ZD0203 from the North Sea (76) | 1 | 5 | ||||
| agg58 branch 2 (14, 49) | 13 | 5 | ||||
| Unaffiliated Cytophaga | 1 | 4 | ||||
| Proteobacteria | Alphaproteobacteria | |||||
| Rhodospiralles | 23 | 17 | ||||
| Rickettsiales | Family Rhodobacteraceae, genus Roseobacter | 18 | 6 | |||
| Environmental sequences | SAR11 cluster (5, 24, 44) | 17 | 7 | |||
| Rhizobiales | Family Bradyrhizobiaceae, genus Afipia | 5 | 4 | |||
| Family Methylobacteriaceae | 1 | 1 | ||||
| Betaproteobacteria | ||||||
| Burkholderiales | Family Burkholderiaceae, genus Burkholderia | 4 | 3 | |||
| Family Ralstoniaceae | 4 | 2 | ||||
| Family Comamonadaceae | 4 | 4 | ||||
| Hydrogenophilales | Family Hydrogenophilaceae | 2 | 1 | |||
| Rhodocyclales | Family Rhodocyclaceae | 1 | 1 | |||
| Deltaproteobacteria | Marine group B/SAR324 cluster (20, 73) | 9 | 5 | |||
| Gammaproteobacteria | Environmental sequences—Bano and Hollibaugh “Cluster A” (1) | 8 | 4 | |||
| Alteromonadales | Family Alteromonadaceae, genus Marinobacterium | 1 | 1 | |||
| Pseudomonadales | Family Moraxellaceae, genus Psychrobacter | 1 | 1 | |||
| Family Moraxellaceae, genus Acinetobacter | 2 | 2 | ||||
| Family Pseudomonadaceae, genus Pseudomonas | 2 | 2 | ||||
| Environmental sequences | SAR86/OCS5 cluster (21, 44) | 1 | ||||
| Arctic96B-16 cluster (1) | 4 | 4 | ||||
| OM60 cluster (54) | 4 | 3 | ||||
| SAR92 cluster (5, 24, 44) | 5 | 1 | ||||
| agg47/KTc1113 (14, 17) | 6 | 6 | ||||
| Unclassified | Candidate division | TM6 | 1 | 2 | ||
Phylogenetic groups were determined based on the insertion of the sequences from the DGGE bands into maximum-likelihood trees (40). The numbers indicate the number of sequences contained within the group for each of the four categories listed. The sequences noted in the HNA +/or LNA column were found either in both the HNA and LNA sorts in the same sample or in HNA and LNA sorts in different samples. The CTC column indicates bands from the CTC-positive sorts with a corresponding band in an HNA sort, an LNA sort, or both.
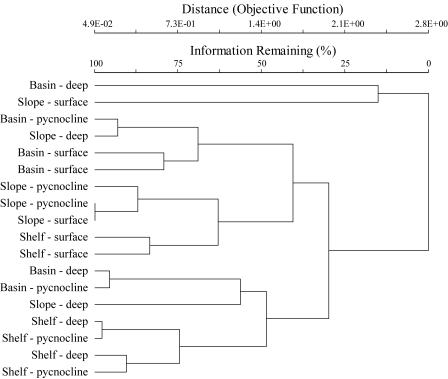
The cluster analysis revealed that there was some separation of the deep and pycnocline shelf station samples from the other samples. However, the two surface shelf station samples were more similar to the slope station samples (Fig. 6). While the surface samples from the basin station were also similar, the deep and pycnocline samples from the basin station were not as tightly clustered as the comparable samples from the shelf station.
FIG. 6.
Dendrogram from the cluster analysis conducted using the Sorensen distance measure and the flexible beta linkage method. Two samples from each of the three stations were sorted and are indicated as shelf, slope, or basin station. The sampling depth is noted as deep, pycnocline, or surface.
DISCUSSION
Validation of the sorting approach.
Several previous studies have used flow cytometric sorting to determine cell-specific substrate incorporation rates for cytometrically identified bacterial cell groups (3, 35, 58, 59, 77). In this study, we directly compared volumetric estimates of leucine incorporation rates of bacteria in sorted and whole-water samples. We were thus able to validate this approach with the observation that the sorted leucine incorporation rates were in fact significantly correlated with whole-seawater bacterial leucine incorporation rates around a 1:1 line. Thus, the process of sorting radioactively labeled bacterial cells does not appear to introduce significant biases in estimating the contributions of the different sorted populations to total bacterial activity.
Variability in cell-specific leucine incorporation rates.
HNA cells have been generally viewed as the actively growing component of bacterial assemblages, while LNA cells have been thought to be less active or dead (22, 23, 29, 35, 60, 70, 72). It has also been suggested that cells with high respiration activity, i.e., CTC-positive cells, could account for a large part of the bacterial metabolism in marine pelagic systems (61, 66). We sought to test these ideas by determining the cell-specific activities of HNA cells, LNA cells, and CTC-positive cells across a range of trophic states in a coastal upwelling system.
We found that decrease in phytoplankton biomass with distance from shore was accompanied by decreasing average cell-specific leucine incorporation rates for HNA, LNA, and CTC-positive cells from shelf to basin stations (Fig. 5), as previously observed for bacterial cell-specific activity scaled to total bacterioplankton in this region (61). As has been reported in previous studies, HNA cells on average had higher cell-specific rates of leucine incorporation than LNA cells (Fig. 5); these higher specific rates combined with higher cell abundances resulted in HNA cells accounting for the largest fraction of total bacterial activity (Table 3). However, the average cell-specific incorporation rates for LNA cells were only marginally lower than the cell-specific rates of HNA cells, and the ranges found for cells in these two groups showed substantial overlap (Fig. 5). LNA cells contributed on average 25% of the total bacterial leucine incorporation rates in shelf and slope waters and a significantly larger fraction (average, 38%, and up to 51%) of the overall incorporation rates in basin waters.
While LNA cells have been shown to have high specific activity rates when methionine incorporation was normalized to biomass at three sampling stations in the Celtic Sea (77), this study is the first to link changes in the metabolic activities of the LNA assemblage with differences in the trophic states of a marine ecosystem over a range of sampling depths. LNA cells were responsible for a larger fraction of total bacterial leucine incorporation rates in low-chlorophyll basin waters than in high-chlorophyll shelf waters. Increased proportional abundance of LNA cells in open-ocean systems compared to coastal systems has been reported (7, 30, 39, 71). LNA cells in eutrophic coastal systems may indeed be inactive or dead cells (23, 29, 39), while LNA cells in oligotrophic systems may be cells with somewhat lower cell-specific metabolic activity (67) but which play a larger role in the heterotrophic processes of oligotrophic systems.
Contribution of CTC-positive cells to bacterial activity.
We made three important observations regarding CTC-positive cells in this study. First, the cell-specific leucine incorporation rate of CTC-positive cells had a range as large as that of the total bacterial community (Fig. 5). On average, CTC-positive cells were more active than the total heterotrophic-cell population and for the most part also had higher rates of cell-specific activity than the HNA cells. This result supports the hypothesis that CTC-positive cells are in general the most metabolically active cells within a bacterial assemblage (48, 61). The second observation was that, in some samples, the cell-specific leucine incorporation rate of the CTC-positive cells was lower than the cell-specific incorporation rate for total cells. Thus, the CTC-positive cells may have been less biosynthetically active at those times than the average bacterial cell. Some of the “CTC-positive cells” might actually have been free formazan granules released from CTC-reducing cells (J.Gasol, personal communication). These granules would not be labeled with [3H]leucine but would be counted as cells and so would lower estimates of average leucine incorporation rates. Finally, the low abundances of the CTC-positive cells meant they were responsible only for a mean of 7% to 14% of volumetric bacterial leucine incorporation (Table 3), so they were clearly not driving bacterial-community production in this system.
Cell-specific rates of CTC-positive cells and their proportional contribution to overall substrate incorporation by assemblages of marine bacteria have been examined in only one previous study (57), in which CTC-positive bacteria in two eutrophic coastal systems along the Mediterranean coast of France were found to have cell-specific leucine incorporation rates of 300 × 10−21 and 350 × 10−21 mol cell−1 h−1. These rates were 25% higher than cell-specific incorporation rates of total bacterial cells in a harbor and sixfold higher than specific rates for total bacteria in a lagoon. Estimates of the fractional contribution of CTC-positive bacteria to the total leucine incorporation rate were 37% and 27% in the harbor and lagoon, respectively (57). The lower proportional contribution of CTC-positive cells found in our study may reflect differences in the ecosystems studied. The highest fractional contribution of CTC-positive cells to total bacterial activity we encountered (27%) was in a basin sample.
Since differences in cell-specific nucleic acid content may not always be closely correlated with the respiration rate, patterns of CTC reduction by bacterial cells may reflect temporal or spatial disconnects between biosynthesis and respiration. For example, in experimental incubation of two marine bacterial isolates, one of the strains maintained high cell-specific ribosome concentrations even after 100 h in stationary growth (50). Thus, cells which are in the HNA assemblage but are CTC negative could represent cells maintaining a high cell-specific nucleic acid content in stationary growth and hence have low respiration rates and therefore not be identifiable as CTC-positive cells. CTC-positive cells may also be present in the LNA assemblage. Grégori et al. (27) used a combination of propidium iodide and SYBR to show that a fraction of LNA cells were able to exclude propidium iodide and therefore would be considered “live” cells. In addition, high biomass-specific rates of substrate uptake by LNA cells have been reported in an Arctic lake (6) and in the Celtic Sea (77), as well as in this study. Thus, LNA cells are not all inactive or dead cells, and some are likely to be able to respire at levels sufficient to be detected in the CTC-positive assemblage.
Phylogenetic diversity of HNA, LNA, and CTC-positive cells.
There was a high degree of uniformity in the phylogenetic compositions of bacterial assemblages in the three regions and in HNA, LNA, and CTC-positive cell groups in each region. The cluster analysis revealed a limited separation of the samples based on sampling regions and depth. Based on the DGGE band patterns and sequence data obtained from the individual DGGE bands, most of the bacterial phylogenetic groups identified in this study could be found in both the HNA and LNA cell assemblages, with some exceptions. Almost one-quarter of all the DGGE bands were present in both the HNA and LNA assemblages in the same sample. Therefore, at the level of taxonomic resolution employed here, there was little phylogenetic distinction during the period of sampling between these two flow cytometrically defined groups of bacteria. These results mirror conclusions reached during a similar analysis of samples collected in the Mediterranean (58). This suggests that cells may be present in both the HNA and LNA assemblages at the same time and that their ability to switch between the two assemblages is not a function of their phylogenetic diversity.
Use of microautoradiography combined with fluorescence in situ hybridization (FISH) has also revealed that no single phylogenetic group dominates the active proportion of the marine bacterioplankton (9, 11). Thus, our results support other findings that, in general, most phylotypes of Bacteria may occur in both HNA and LNA cell clusters, and also in active and inactive components of the marine bacterioplankton.
There were interesting exceptions to the observation that most of the phylogenetic groups could be present in either the HNA or the LNA assemblage. Several different phylotypes within the Bacteroidetes were identified only in the HNA sorts and were not identified in any LNA sorts. Reported FISH results have also confirmed the presence of strains affiliated with Bacteroidetes, more specifically, the members of the Cytophaga-Flavobacterium cluster, which bind to the CF319a probe, in the HNA assemblage but not the LNA assemblage in the North Sea (75, 77). Studies from the Delaware River estuary have indicated Bacteroidetes are involved in the breakdown of high-molecular-weight organic matter (10). Perhaps the metabolic processes used for the consumption of high-molecular-weight organic matter require the organisms to maintain larger genomes or multiple copies of the genome within their cells. Furthermore, contradictory evidence regarding the growth rates of the Bacteroidetes in aquatic systems (as reviewed in reference 32) indicate that they can have higher (31) or lower (19, 64) growth rates than other groups. If they have higher growth rates, this may also explain their presence within the HNA assemblage. Given the diversity within the Bacteroidetes and the observation that even within this study not all of them were limited to the HNA assemblage, more research is needed to examine whether some Bacteroidetes are physiologically or morphologically limited to the HNA assemblage or whether this was due to the environmental conditions observed during this study.
A lower total number of sequences were obtained from the LNA assemblage than from the HNA assemblage. Since the LNA cells have lower cell-specific DNA content, the lower number of sequences observed in this category could represent our inability to obtain sufficient DNA to detect these cells. Alternatively, those sequences obtained only from the LNA assemblage could represent groups that were not physiologically capable of maintaining higher cell-specific nucleic acid content but that still might have been metabolically active.
SAR11 sequences were obtained from both the HNA and LNA components of the heterotrophic community. In culture, “Candidatus Pelagibacter ubique” is a small organism with low growth rates (53) and therefore might be expected to be found exclusively in the LNA assemblage. However, we found SAR11 sequences in the HNA assemblage as well. Either the small size and low growth rates of SAR11 are a manifestation of the culture conditions, or low growth rates are characteristic of this group and not necessarily associated with lower cell-specific nucleic acid content. In the North Atlantic, bacterioplankton cells labeled with a suite of probes targeting the SAR11 group were similar in size to other Bacteria (42). Thus, the “Candidatus Pelagibacter ubique” culture from the Oregon coast may have an intrinsically smaller cell size than other Pacific SAR11 cells not yet cultured or isolated.
The observation of identical DGGE bands in both the LNA and CTC-positive assemblages could represent cells which have functioning electron transport systems but either do not have a constitutively high cell-specific nucleic acid content or have higher levels of respiration but not higher levels of biosynthetic activity. One question raised about the CTC method is whether all heterotrophic cells are able to reduce CTC (74). The diversity data indicated that the same phylotypes present in the HNA and LNA components of bacterioplankton were also found in CTC-positive cells. The only group we did not identify in sorts of CTC-positive cells was the SAR86 cluster within the Gammaproteobacteria. This may suggest that the environmental conditions we encountered were not conducive for the SAR86 group to reduce CTC sufficiently to be detectable as CTC positive. As only one SAR86 sequence was obtained in this study, strains of the SAR86 group may not have been an ecologically relevant component of the bacterioplankton at the beginning of upwelling season. In other systems and/or seasons, this group of Bacteria may be more important. For example, based on bromodeoxyuridine incorporation, SAR86 was a dominant component of the DNA-synthesizing population in the fall in the North Sea, when primary production levels were declining (51). To date, no phylotype in the SAR86 group has been cultured.
The diversity of Bacteria identified in the different flow cytometrically defined groups in this study was broader than that obtained from sorted populations originating from samples collected in the North Sea (75, 77). Some of the differences between the North Sea and the Oregon coast data are methodological, since the identification of the sorted populations in the North Sea partially relied on FISH using probes targeting broad phylogenetic groups. However, the Deltaproteobacteria, Actinobacteria, Acidobacteria, Meiothermus, and TM6 sequences obtained from the Oregon coast samples were all absent from the subset of sequences obtained from cloned 16S rRNA genes from the North Sea (75). They could represent regional or temporal differences in the metabolically “active” community (58). Now that it is possible to obtain DNA from sorted populations, future research can address the causes of temporal and spatial variability in the Bacteria present in the HNA or LNA assemblage in more detail, either by examination of the whole community or by focusing on specific phylogenetic groups.
Conclusion.
Our data on the cell-specific activity and diversity of HNA, LNA, and CTC-positive cells indicate that there were differences in the cell-specific activity for each of the three groups. However, regional variability in cell-specific activity was not directly correlated with variability in the phylogenetic diversity of the HNA, LNA, and CTC-positive cells. Thus, factors other than large-scale changes in the diversity of prokaryotes in the Oregon upwelling system are driving changes in their cell-specific activity.
Acknowledgments
We thank R. Condon, D. Homen, J. Fleischbein, J. Jennings, M. Lichtenberg, and the R/V Wecoma's captain, crew, and marine technicians for technical assistance. The other cruise participants provided thoughtful discussions during and after the cruise. The comments of the anonymous reviewers and D. Kirchman greatly improved this paper. The DNA sequences from the Center for Gene Research and Biotechnology at Oregon State University were outstanding.
This research was funded by NSF grants OCE-0002236 and OCE-0240785 to B.F.S. and E.B.S.
REFERENCES
- 1.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard, L., C. Courties, C. Duperray, H. Schäfer, G. Muyzer, and P. Lebaron. 2001. A new approach to determine the genetic diversity of viable and active bacteria in aquatic ecosystems. Cytometry 43:314-321. [PubMed] [Google Scholar]
- 3.Bernard, L., C. Courties, P. Servais, M. Troussellier, M. Petit, and P. Lebaron. 2000. Relationships among bacterial cell size, productivity, and genetic diversity in aquatic environments using cell sorting and flow cytometry. Microb. Ecol. 40:148-158. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, L., H. Schäfer, F. Joux, C. Courties, G. Muyzer, and P. Lebaron. 2000. Genetic diversity of total, active and culturable marine bacteria in coastal seawater. Aquat. Microb. Ecol. 23:1-11. [Google Scholar]
- 5.Britschgi, T. B., and S. J. Giovannoni. 1990. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 57:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Button, D. K., and B. R. Robertson. 2001. Determination of DNA content of aquatic bacteria by flow cytometry. Appl. Environ. Microbiol. 67:1636-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casotti, R., C. Brunet, B. Aronne, and M. Ribera d'Alcala. 2000. Mesoscale features of phytoplankton and planktonic bacteria in a coastal area as induced by external water masses. Mar. Ecol. Prog. Ser. 195:15-27. [Google Scholar]
- 8.Chavez, F. P., J. T. Pennington, C. G. Castro, J. P. Ryan, R. P. Michisaki, B. Schlining, P. Walz, K. R. Buck, A. McFadyen, and C. A. Collins. 2002. Biological and chemical consequences of the 1997-1998 El Niño in central California waters. Prog. Oceanogr. 54:205-232. [Google Scholar]
- 9.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 10.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottrell, M. T., and D. L. Kirchman. 2004. Single-cell analysis of bacterial growth, cell size and community structure in the Delaware estuary. Aquat. Microb. Ecol. 34:139-149. [Google Scholar]
- 12.del Giorgio, P. A., and T. C. Bouvier. 2002. Linking the physiologic and phylogenetic successions in free-living bacterial communities along an estuarine salinity gradient. Limnol. Oceanogr. 47:471-486. [Google Scholar]
- 13.del Giorgio, P. A., Y. T. Prairie, and D. F. Bird. 1997. Coupling between rates of bacterial production and the number of metabolically active cells in lake bacterioplankton, measured using CTC reduction and flow cytometry. Microb. Ecol. 34:144-154. [DOI] [PubMed] [Google Scholar]
- 14.DeLong, E. F., D. G. Franks, and A. L. Alldredge. 1993. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38:924-934. [Google Scholar]
- 15.Dietz, E. J. 1983. Permutation tests for association between two distance matrices. Syst. Zool. 32:21-26. [Google Scholar]
- 16.Ducklow, H. W. 1999. The bacterial component of the oceanic euphotic zone. FEMS Microbiol. Ecol. 30:1-10. [Google Scholar]
- 17.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari, B. C., G. Oregaard, and S. J. Sørensen. 2004. Recovery of GFP-labeled bacteria for culturing and molecular analysis after cell sorting using a benchtop flow cytometer. Microb. Ecol. 48:239-245. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs, B. M., M. V. Zubkov, K. Sahm, P. H. Burkill, and R. Amann. 2000. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ. Microbiol. 2:191-201. [DOI] [PubMed] [Google Scholar]
- 20.Fuhrman, J. A., and A. A. Davis. 1997. Widespread Archaea and novel Bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar. Ecol. Prog. Ser. 150:275-285. [Google Scholar]
- 21.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1993. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl. Environ. Microbiol. 59:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasol, J. M., and P. A. del Giorgio. 2000. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64:197-224. [Google Scholar]
- 23.Gasol, J. M., U. L. Zweifel, F. Peters, J. A. Fuhrman, and Å. Hagström. 1999. Significance of size and nucleic acid content in heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl. Environ. Microbiol. 65:4475-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 25.Giovannoni, S. J., E. F. DeLong, T. M. Schmidt, and N. R. Pace. 1990. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl. Environ. Microbiol. 56:2572-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon, L. I., J. C. Jennings, Jr., A. A. Ross, and J. M. Krest. 1994. A suggested protocol for continuous flow automated analysis of seawater nutrients (phosphate, nitrate, nitrite and silicic acid) in the WOCE Hydrographic Program and the Joint Global Ocean Fluxes Study. In WOCE operations manual. WOCE report no. 68/91, revision 1. WHP Office Report WHPO91-1. Woods Hole Oceanographic Institution, Woods Hole, Mass.
- 27.Grégori, G., S. Citterio, A. Ghiani, M. Labra, S. Sgorbati, S. Brown, and M. Denis. 2001. Resolution of viable and membrane-compromised bacteria in freshwater and marine waters based on analytical flow cytometry and nucleic acid double staining. Appl. Environ. Microbiol. 67:4662-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huyer, A., R. L. Smith, and J. Fleischbein. 2002. The coastal ocean off Oregon and northern California during the 1997-8 El Niño. Prog. Oceanogr. 54:311-341. [Google Scholar]
- 29.Jellett, J. F., W. K. W. Li, P. M. Dickie, A. Boraie, and P. E. Kepkay. 1996. Metabolic activity of bacterioplankton communities assessed by flow cytometry and single carbon substrate utilization. Mar. Ecol. Prog. Ser. 136:213-225. [Google Scholar]
- 30.Jochem, F. J. 2001. Morphology and DNA content of bacterioplankton in the northern Gulf of Mexico: analysis by epifluorescence microscopy and flow cytometry. Aquat. Microb. Ecol. 25:179-194. [Google Scholar]
- 31.Jürgens, K., and H. Güde. 1994. The potential importance of grazing-resistant bacteria in planktonic systems. Mar. Ecol. Prog. Ser. 112:169-188. [Google Scholar]
- 32.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 33.Kirchman, D. L. 1993. Leucine incorporation as a measure of biomass production by heterotrophic bacteria, p. 509-512. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Current methods in aquatic microbial ecology. Lewis Publishing, New York, N.Y.
- 34.Kirchman, D. L., L. Yu, and M. T. Cottrell. 2003. Diversity and abundance of uncultured Cytophaga-like bacteria in the Delaware estuary. Appl. Environ. Microbiol. 69:6587-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebaron, P., P. Servais, H. Agogué, C. Courties, and F. Joux. 2001. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl. Environ. Microbiol. 67:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legendre, L., and J. L. Fèvre. 1995. Microbial food webs and the export of biogenic carbon in oceans. Aquat. Microb. Ecol. 9:69-77. [Google Scholar]
- 37.Legendre, L., and F. Rassoulzadegan. 1996. Food-web mediated export of biogenic carbon in oceans: hydrodynamic control. Mar. Ecol. Prog. Ser. 145:179-193. [Google Scholar]
- 38.Legendre, P., and L. Legendre. 1988. Numerical ecology, 2nd English ed. Elsevier Science BV, Amsterdam, The Netherlands.
- 39.Li, W. K. W., J. F. Jellett, and P. M. Dickie. 1995. DNA distributions in planktonic bacteria stained with TOTO or TO-PRO. Limnol. Oceanogr. 40:1485-1495. [Google Scholar]
- 40.Longnecker, K. 2004. Bacterioplankton in the Oregon upwelling system: distribution, cell-specific leucine incorporation, and diversity. Ph.D. thesis. Oregon State University, Corvallis.
- 41.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malmstrom, R. R., R. P. Kiene, M. T. Cottrell, and D. L. Kirchman. 2004. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl. Environ. Microbiol. 70:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marie, D., F. Partensky, S. Jacquet, and D. Vaulot. 1997. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullins, T. D., T. B. Britschgi, R. L. Krest, and S. J. Giovannoni. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40:148-158. [Google Scholar]
- 45.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 47.Neefs, J.-M., Y. Van De Per, P. De Rijk, S. Chapelle, and R. De Wachter. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 21:3025-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen, J. L., M. Aquino de Muro, and P. H. Nielsen. 2003. Evaluation of the redox dye 5-cyano-2,3-tolyl-tetrazolium chloride for activity studies by simultaneous use of microautoradiography and fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:641-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Sullivan, L. A., K. E. Fuller, E. M. Thomas, C. M. Turley, J. C. Fry, and A. J. Weightman. 2004. Distribution and culturability of the uncultivated ′agg58′ cluster of the Bacteroidetes phylum in aquatic environments. FEMS Microbiol. Ecol. 47:359-370. [DOI] [PubMed] [Google Scholar]
- 50.Pernthaler, A., J. Pernthaler, H. Eilers, and R. Amann. 2001. Growth patterns of two marine isolates: adaptations to substrate patchiness. Appl. Environ. Microbiol. 67:4077-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pernthaler, A., J. Pernthaler, M. Schattenhofer, and R. Amann. 2002. Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton. Appl. Environ. Microbiol. 68:5728-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson, W. T., J. E. Keister, and L. R. Feinberg. 2002. The effects of the 1997-1999 El Niño/La Niña events on the hydrography and zooplankton off the central Oregon coast. Prog. Oceanogr. 54:381-398. [Google Scholar]
- 53.Rappé, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 54.Rappé, M. S., P. F. Kemp, and S. J. Giovannoni. 1997. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol. Oceanog. 42:811-826. [Google Scholar]
- 55.Rodriguez, G. G., D. Phipps, K. Ishiguro, and H. F. Ridgway. 1992. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl. Environ. Microbiol. 58:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schäfer, H., and G. Muyzer. 2001. Denaturing gradient gel electrophoresis in marine microbial ecology, p. 425-468. In J. H. Paul (ed.), Methods in microbiology, vol. 30. Academic Press, San Diego, Calif. [Google Scholar]
- 57.Servais, P., H. Agogué, C. Courties, F. Joux, and P. Lebaron. 2001. Are the actively respiring cells (CTC+) those responsible for bacterial production in aquatic environments? FEMS Microbiol. Ecol. 35:171-179. [DOI] [PubMed] [Google Scholar]
- 58.Servais, P., E. O. Casamayor, C. Courties, P. Catala, N. Parthuisot, and P. Lebaron. 2003. Activity and diversity of bacterial cells with high and low nucleic acid content. Aquat. Microb. Ecol. 33:41-51. [Google Scholar]
- 59.Servais, P., C. Courties, P. Lebaron, and M. Troussellier. 1999. Coupling bacterial activity measurements with cell sorting by flow cytometry. Microb. Ecol. 38:180-189. [DOI] [PubMed] [Google Scholar]
- 60.Seymour, J. R., J. G. Mitchell, and L. Seuront. 2004. Microscale heterogeneity in the activity of coastal bacterioplankton communities. Aquat. Microb. Ecol. 35:1-16. [Google Scholar]
- 61.Sherr, B. F., P. del Giorgio, and E. B. Sherr. 1999. Estimating abundance and single-cell characteristics of respiring bacteria via the redox dye CTC. Aquat. Microb. Ecol. 18:117-131. [Google Scholar]
- 62.Sherr, E. B., B. F. Sherr, and T. J. Cowles. 2001. Mesoscale variability in bacterial activity in the Northeast Pacific Ocean off Oregon, USA. Aquat. Microb. Ecol. 25:21-30. [Google Scholar]
- 63.Sherr, E. B., B. F. Sherr, and P. A. Wheeler. 2005. Distribution of coccoid cyanobacteria and small eukaryotic phytoplankton in the upwelling ecosystem off the Oregon coast during 2001 and 2002. Deep-Sea Res. II 52:317-330. [Google Scholar]
- 64.Šimek, K., J. Pernthaler, M. G. Weinbauer, K. Hornák, J. R. Dolan, J. Nedoma, M. Mašín, and R. Amann. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacterial protein synthesis rates in sea water using 3H-leucine. Mar. Microb. Food Webs 6:107-114. [Google Scholar]
- 66.Smith, E. M. 1998. Coherence of microbial respiration rate and cell-specific bacterial activity in a coastal planktonic community. Aquat. Microb. Ecol. 16:27-35. [Google Scholar]
- 67.Smith, E. M., and P. A. del Giorgio. 2003. Low fractions of active bacteria in natural aquatic communities? Aquat. Microb. Ecol. 31:203-208. [Google Scholar]
- 68.Smith, J. J., and G. A. McFeters. 1997. Mechanisms of INT (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl tetrazolium chloride), and CTC (5-cyano-2,3-ditolyl tetrazolium chloride) reduction in Escherichia coli K-12. J. Microbiol. Methods. 29:161-175. [DOI] [PubMed] [Google Scholar]
- 69.Strickland, J. D. H., and T. R. Parsons (ed.). 1972. A practical handbook of seawater analysis, 2nd ed., vol. 167. Fisheries Research Board of Canada, Ottawa, Canada.
- 70.Troussellier, M., C. Courties, P. Lebaron, and P. Servais. 1999. Flow cytometric discrimination of bacterial populations in seawater based on SYTO 13 staining of nucleic acids. FEMS Microbiol. Ecol. 29:319-330. [Google Scholar]
- 71.Vaqué, D., E. O. Casamayor, and J. M. Gasol. 2001. Dynamics of whole community bacterial production and grazing losses in seawater incubations as related to the changes in the proportions of bacteria with different DNA content. Aquat. Microb. Ecol. 25:163-177. [Google Scholar]
- 72.Wetz, M. S., and P. A. Wheeler. 2004. Response of bacteria to simulated upwelling phytoplankton blooms. Mar. Ecol. Prog. Ser. 272:49-57. [Google Scholar]
- 73.Wright, T. D., K. L. Vergin, P. W. Boyd, and S. J. Giovannoni. 1997. A novel δ-subdivision proteobacterial lineage from the lower ocean surface layer. Appl. Environ. Microbiol. 63:1441-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamaguchi, N., and M. Nasu. 1997. Flow cytometric analysis of bacterial respiratory and enzymatic activity in the natural aquatic environment. J. Appl. Microbiol. 83:43-52. [Google Scholar]
- 75.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]
- 76.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2002. Rapid turnover of dissolved DMS and DMSP by defined bacterioplankton communities in the stratified euphotic zone of the North Sea. Deep-Sea Res. II 49:3017-3038. [Google Scholar]
- 77.Zubkov, M. V., B. M. Fuchs, P. H. Burkill, and R. Amann. 2001. Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl. Environ. Microbiol. 67:5210-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]