Abstract
A sediment core spanning approximately 1,600 years was collected from a lake on Ardley Island, Antarctica. The sediment core had been greatly influenced by penguin guano. Using molecular methods, the chitinolytic bacterial community along the sediment core was studied over its entire length. Primers targeting conserved sequences of the catalytic domains of family 18 subgroup A chitinases detected group A chitinases from a wide taxonomic range of bacteria. Using quantitative competitive PCR (QC-PCR), chitinase gene copies in each 1-cm section of the whole sediment column were quantified. QC-PCR determination of the chitinase gene copies indicated significant correlation with phosphorus and total organic carbon concentration, suggesting a historical connection between chitinase gene copies and the amount of penguin guano input into the lake sediment. Most of the chitinase genes cloned from the historic sediment core were novel. Analysis of the chitinase gene diversity in selected sediment layers and in the fresh penguin deposits indicated frequent shifts in the chitinolytic bacterial community over time. Sequence analysis of the 16S rRNA genes of chitinolytic bacteria isolated from the lake sediment revealed that the isolates belonged to Janthinobacterium species, Stenotrophomonas species of γ-Proteobacteria, Cytophaga species of the Cytophaga-Flexibacter-Bacteroides group, and Streptomyces and Norcardiopsis species of Actinobacteria. Chitinase gene fragments were cloned and sequenced from these cultivated chitinolytic bacteria. The phylogeny of the chitinase genes obtained from the isolates did not correspond well to that of the isolates, suggesting acquisition via horizontal gene transfer.
More pristine ecosystems are found in Antarctica than elsewhere on our planet (6). Particular animals, plants, and microbes have been investigated in several Antarctic environments (7, 14, 23). However, the relationships among the organisms have rarely been described, in part because of the complexity of scientific investigation in this extreme environment or because of a lack of suitable study systems. Ornithogenic soils are the richest source of organic matter in Antarctica(19), from which many microbes, including new bacterial species, have been isolated and described (2, 18). The penguins on Ardley Island (62°13′S, 58°54′W) within the South Shetland Islands discharge large amounts of chitin-containing droppings onto the land which are transferred by ice or snowmelt water, and some of them are deposited in the Y2 lake (20). Penguin guano is the main source of chitin in this environment, as penguins consume krill as the main portion of their diet (5). Previous research has shown that the artificial addition of chitin into the soil stimulated growth of chitinolytic actinobacteria and changed the bacterial community (16, 24). But the impact of the long-term natural input of chitin into the lake sediment on the bacterial community in Antarctica has never been studied. We hypothesized that the input of penguin guano into the lake sediment would have a great impact on both the chitinolytic bacterial number and diversity.
In nature, chitin is recycled mainly through the chitinase pathway. Many organisms may produce chitinases to hydrolyze chitin for different purposes (3). Microorganisms use chitinases mainly to digest chitin as a nutrient source for growth, and chitinases play the most important role in chitin recycling in nature. Chitinases (EC 3.2.1.14) are quite versatile. They are classified into families 18 and 19 of the glycoside hydrolase classification system based on amino acid similarities within their catalytic domains (10, 12). Family 18 chitinases, subdivided into groups A, B, and C (22), are mainly from bacteria, whereas the majority of family 19 chitinases come from plants (11). Family 18 group A chitinases are most frequently detected in different environments. Thus, group A chitinases have been used to detect the diversity and distribution of chitinolytic bacteria in marine environments as well as in an upland pasture (4, 16, 17).
In this study, a 59.5-cm sediment core collected from the Y2 lake of Ardley Island, Antarctica, was studied. Both culture-independent and culture-dependent approaches were used to examine the chitinolytic bacterial communities, reflected by their chitinase genes, in order to reveal the impact of input penguin guano into the lake sediment on the bacterial community.
MATERIALS AND METHODS
Sample collection and handling.
A lake sediment core labeled Y2-4, collected near the location where the Y2 core was collected (21), was taken by driving a polyvinyl chloride pipe with a diameter of 12 cm into the soft substrate of the lake floor on Ardley Island during the 18th Chinese Antarctic Research Expedition. Core Y2-4 had a length of 59.5 cm. The sediment core was stored at −20°C for shipping and in the laboratory until being sectioned at 1.0-cm intervals on a clean bench. About 80% of each section was transferred to a separate sterile Falcon tube for DNA extraction. The remaining 20% was pulverized with an agate mortar and pestle, and the >200-mesh fraction was collected and analyzed. The phosphorus concentrations of all 59 subsamples were analyzed according to the method reported previously by Sun et al. (21). The total organic carbon (TOC) contents of the samples were determined by the method of Gaudette et al. (9).
Fresh deposits of droppings from Adélie and Gentoo penguins were collected during the 19th Chinese Antarctic Research Expedition. A soil sample not contaminated with penguin droppings from Antarctica was used for comparison.
Designing PCR primers and amplification of chitinase gene fragments.
The amino acid sequences of the chitinases encoded by chi69 of Janthinobacterium lividium (Swiss-Prot accession number Q48373), chiA of Stenotrophomonas maltophilia (accession number O30678), chiB of Arthrobacter spp. (accession number Q9REI6), and chi67 of Doohwaniella chitinasigens (accession number Q9RCG5) were aligned and compared. The forward primers chif1 (ATC/TTCGCTGGGT/CGGCTGG) and chif2 (GACGGCATCGACATCGATTGG) and a reverse primer, chir (CG/CGTCCAGCCGCGC/GCCG/ATA), were designed according to the conserved regions in the catalytic domain.
Primer pairs chif1-chir and chif2-chir were used to amplify chitinase gene fragments from the environmental DNA and from isolated total bacterial DNA, respectively. The PCR system included 5 μl 10× PCR buffer, 4 μl deoxynucleoside triphosphates (0.2 mM; Fermentas, MBI), 0.5 μl of each primer (10 μM), ∼20 ng template, and 2 U Taq polymerase (Fermentas, MBI). The thermal cycle (amplification) was performed in a T3 thermal cycler (Biometra) with PCR conditions consisting of an initial denaturation step at 95°C for 5 min followed by 35 cycles of denaturation at 94°C for 50 s, annealing at 65°C for 60 s, and extension at 72°C for 90 s and ending with an extension step at 72°C for 10 min.
Quantification of chitinase gene in the sediment core.
Sediment DNA was extracted as described previously (25, 27). Quantitative competitive PCR (QC-PCR) using the chif2-chir primers was used to determine the quantity of the original chitinase gene in the environments (13, 25). The extracted plasmid T-chi (QIAGEN plasmid kit), containing the cloned 439-bp chitinase gene fragment, was digested with BamHI and used as the positive standard fragment to evaluate the efficiency and accuracy of the competitor template DNA. The linear plasmid T-chi was digested with EcoRV to get a 200-bp deletion in the region between the PCR primer pair chif2-chir, self ligated, and then digested with BamHI. The linear plasmid T-chi Δ200bp was used as the competitive template DNA. The quantities of T-chi and T-chi Δ200bp were determined using a spectrophotometer (Ultrospec 2100; Amersham Pharmacia).
Chitinase gene library construction and RFLP analysis.
PCR products of chitinase gene fragments from samples were cloned into the pGEM-T vector by using a 2× rapid ligation kit according to the instructions of the manufacturer (Promega). Ligation mixtures were used to transform competent cells of Escherichia coli XL1-Blue according to the suggestions of the manufacturer (Promega). Around 40 clones from each sample library were subjected to restriction fragment length polymorphism (RFLP) analysis with restriction enzyme RsaI and visualized by electrophoresis on a 5% agarose gel containing ethidium bromide (0.5 mg/liter). The clones were grouped according to the different RFLP patterns, and one representative from each different RFLP pattern was selected for sequencing (Sangon Inc., Shanghai, China).
Isolation and characterization of chitinolytic bacteria.
The sediment samples were diluted at a ratio of approximately 1:20 (wt/vol) in distilled water, and 100 μl containing 5 mg of mud (wet weight) of the suspension was spread onto basic M9 agar plates containing 1% colloidal chitin (derived from crab shell, treated by H3PO4). Plates were then incubated for 7 to 28 days at 10°C for retrieval of chitinolytic isolates. Colonies were subcultured for purification. Strains were selected on the basis of differences in colony morphology.
Genomic DNA of the isolated strains was extracted and purified by standard methods and PCR amplification, and cloning of the 16S rRNA gene fragments was performed according to methods described previously (26). DNA sequencing was carried out by the Sangong Company (Shanghai, China).
Phylogenetic analysis.
Deduced amino acid sequences of the chitinase genes retrieved from the library were searched in the NCBI databank. Related sequences were aligned by using the program DNAMAN (version 5.1; Lynnon Biosoft). A phylogenetic tree was constructed from a matrix of pairwise genetic distances by the maximum-parsimony algorithm and the neighbor-joining method using the DNAMAN program, and 1,000 trials of bootstrap analyses were used to provide confidence estimates for phylogenetic tree topologies.
Nucleotide sequence accession numbers.
The nucleic acid sequences of the chitinase genes in this study have been deposited in the GenBank/EMBL/DDBJ databases under accession numbers AY523556 to AY523563, AJ812554, AJ812562, and AJ968654 to AJ968656, and the 16S rRNA gene sequences of the isolated strains have been deposited under accession numbers AJ551147, AJ551150, AJ551168, and AM039885 to AM039887.
RESULTS
Analysis of sediment core structure and dating.
To evaluate the sediment structure, the core Y2-4 was examined by X ray. No inversions were observed, implying an absence of reworking during deposition (X-ray photo not shown).
The Y2-4 core was collected close to the Y2 core, which was well dated previously (21). The intermediate ages and bottom age of the Y2-4 core were obtained by comparing the pattern of its bioelements, such as phosphorus, with that in the Y2 core (21). The ages of the bottom layer (layer 59 [L59]) and intermediate layers L33 and L18 of the Y2-4 core were determined to be 1,600, 1,230, and 1,020 years before the present (BP), respectively (data not shown). The chronology of the whole core was then deduced on the basis of linear interpolation between the dated layers as described previously (15).
Designing and evaluating chitinase gene primers.
Primers for amplification of chitinase family 18 group A (chiA) gene fragments were designed as described in Materials and Methods. With primer pair chif2-chir, a specific DNA fragment of about 430 bp was obtained from all chitinolytic bacteria tested (data not shown). The tested chitinolytic bacteria spanned a broad range of taxa, including the gram-positive bacteria Streptomyces griesus, Streptomyces coelicolor, Nocardiopsis dassonvillei, and Arthrobacter spp.; the gram-negative γ-Proteobacteria Aeromonas caviae, Aeromonas hydrophila, Serratia marcescens, Stenotrophomonas spp., Janthinobacterium spp., and Shewanella benthica; and Cytophaga-Flexibacter-Bacteroides group bacteria including Cytophaga spp. The amplified band from each strain was confirmed to be group A chitinase by cloning and sequencing (data not shown). The results clearly demonstrated that the designed primer pair chif2-chir could be used to retrieve group A chitinases from a wide range of bacterial species.
Quantification of chitinase gene copies.
To determine the chiA gene copy number along the sediment core, quantitative PCRs were performed. The sensitivity and accuracy of the system were tested first. The results indicated that as few as 10 chitinase gene copies could be identified, and the quantity of the competitor DNA employed accurately reflected the amount of the standard tester DNA present (data not shown). The Antarctic ornithogenic core was determined to contain around 3.4 × 104 to 4.2 × 107 copies of chiA per gram of soil (Fig. 1). In addition, the chiA gene copy number in the deposits of fresh Gentoo and Adélie penguins, which are the major penguin species on Ardley Island, was also checked. The deposits of both Gentoo and Adélie penguins contained ∼1.25 × 105 to 2.5 × 105 copies of chiA per gram, whereas the soil sample without contamination of penguin droppings contained 2.5 × 103 copies of chiA per gram, around 100 times fewer than those in the penguin droppings.
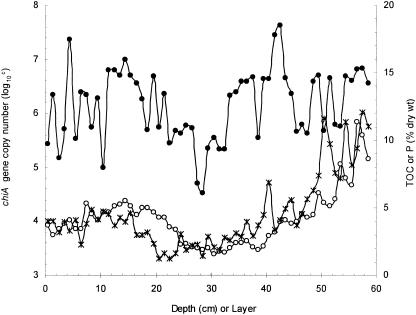
FIG. 1.
TOC contents, phosphorous concentrations, and chitinase gene copies in the Y2-4 core. The TOC contents and phosphorous concentrations in the Y2-4 core were determined according to previously published methods (9, 20). The chitinase gene copy number in each sediment layer was determined by QC-PCR as described in Materials and Methods. × represents TOC contents, ○ represents phosphorous concentrations, and • represents the chiA gene copy number.
The change in the chitinase gene abundance along the sediment column was compared with the geochemical data. As seen in Fig. 1, the variation in the chitinase gene copies through the column was similar to the bioelement P and to TOC. The Pearson correlation coefficient values between phosphorous concentration (P) and TOC, P and chiA gene copies, and TOC and chiA gene copies were 0.749, 0.369, and 0.308, respectively (P < 0.05; n = 59). The data clearly indicate that the chitinase gene copy, phosphorus concentration, and TOC content along the Y2-4 core were significantly correlated with one another.
Chitinase gene diversity.
Because the number of chitinase genes in L11 and L43 represent respective minimal and maximal values along the sediment core, we also selected two other sediment layers, L5 and L59, for analysis to represent chitinase gene numbers in the top and bottom of the sediment core and for indication of chitinase gene diversity in the environment. In addition, the chiA gene diversity in the excreta of fresh penguins and in soil not contaminated by penguin droppings was also analyzed.
Chitinase gene libraries of the four sediment layers (L5, L11, L43, and L59), fresh excreta of Gentoo and Adélie penguins, and the soil not contaminated with penguin droppings (labeled soil B) were constructed. More than 40 clones from each library were chosen randomly for RFLP analysis. In total, 12 RFLP types were found among the 295 clones of the seven libraries (Table 1), and a representative clone for each RFLP type was sequenced. Two RFLP types did not show any sequence similarities with those of chitinases, while all the other 10 RFLP types shared high sequence identities with those of chitinases (Table 1).
TABLE 1.
Distribution of the chitinase gene phylotypes in samples
| RFLP typea | Representative clone | No. of clones in library
|
Closest relatives from isolates (GenBank accession no., % identity) | Closest relatives from environmental clones (GenBank accession no., % identity) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bb | Gc | Ad | L5 | L11 | L43 | L59 | ||||
| 1 | B19 | 4 | 29 | 20 | 6 | J. lividium chi69 (U07025, 93) | controlC7S (AF484834, 95) | |||
| 2 | G37 | 1 | 5 | 17 | 18 | X. maltophilia chiA (AF014950, 89) | controlA2S (AF484819, 95) | |||
| 3 | G24 | 2 | S. coelicolor chiD (AB017011, 72) | sludgeC9S (AF484821, 65) | ||||||
| 4 | B5 | 5 | Vibrio vulnificus chi (AE016810, 57) | Nonee | ||||||
| 5 | L11-50 | 1 | Bacillus cereus chiCW (AF416570, 47) | None | ||||||
| 6 | L5-1 | 34 | Aphanocladium album chi1 (S81303, 47) | None | ||||||
| 7 | L5-18 | 3 | J. lividium chi69 (U07025, 51) | controlC7S (AF484834, 47) | ||||||
| 8 | L5-24 | 2 | 45 | 35 | B. thuringiensis chiCW (AF416570, 47) | None | ||||
| 9 | L59-15 | 1 | 3 | 1 | Aeromonas punctata chi1 (AJ534329, 99) | controlC8S (AF484827, 74) | ||||
| 10 | L59-28 | 2 | Halobacterium sp. chi (AE005023, 37) | Lime4 (AF455083, 39) | ||||||
Chitinase gene fragments cloned from the samples were subjected to RFLP analysis with restriction enzyme RsaI, clones were grouped according to the different RFLP patterns, and one representative from each different RFLP pattern was selected for sequencing.
B represents a soil sample not contaminated by penguin droppings.
G represents the droppings of Gentoo penguins.
A represents the droppings of Adélie penguins.
None, no similar sequences were found.
The seven samples checked for chiA gene diversity showed different chiA composition, as shown in Table 1. Among the 10 RFLP types, 6 showed relatively low amino acid sequence similarities (less than 60% identity) with those of known chitinases (clone types 4 to 8 and 10). These new chitinases may be from uncultured bacteria. The dominant clone types in the seven samples were also different: type 2 was dominant in soil B (55%), type 1 and type 2 were each predominant in the droppings of Gentoo and Adélie penguins, respectively (71% and 100% each), type 6 constituted 85% of the RFLP groups in layer 5, types 1 and 2 each occupied 48% and 43% of the RFLP groups in layer 11, and type 8 occupied 100% and 78% of the RFLP groups in layers 43 and 59, respectively (Table 1). The phylogenetic relationship of the cloned chitinases with related chitinases in the databases is shown in Fig. 2.
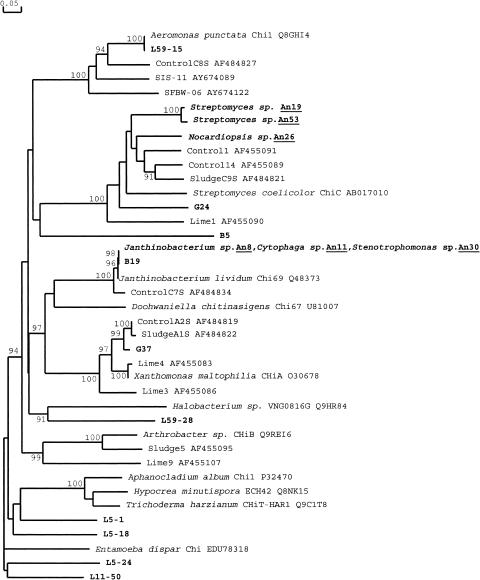
FIG. 2.
Phylogeny of cloned chitinase sequences. Chitinases cloned in this study and known chitinases taken from GenBank were aligned by DNAMAN. The phylogenetic tree was constructed from a matrix of pairwise genetic distances by the maximum-parsimony algorithm and the neighbor-joining method using the DNAMAN program. Bootstrap values above 40 from 1,000 replicates are shown. The scale bar represents 0.05 substitutions per amino acid site. The boldface type represents chitinases cloned in this study, and those from cultivated chitinolytic bacteria are underlined.
Chitinolytic isolates and their chitinase genes.
Some chitinolytic bacteria were isolated from sediment core samples by plating. Six morphologically different chitinolytic strains, named An8, An11, An19, An26, An30, and An53, were selected from plates with chitin-containing medium. Fragments of about 1.5 kb from 16S rRNA genes were amplified from the isolates and sequenced. The 16S rRNA gene sequence from each of the different isolates had high identity with that of one of the following: Janthinobacterium lividium (99%), Cytophaga spp. (97%), Streptomyces samponii (99%), Nocardiopsis Antarctica (99%), Stenotrophomonas maltophilia (99%), or Streptomyces griseus (99%). These species are ubiquitous soil bacteria on earth.
Chitinase gene fragments were amplified, cloned, and sequenced from the six isolates using the chif2-chir primer pair. The chitinase genes cloned from the three different isolates An8, An11, and An30 were identical and shared 95% identity with chi69 of J. lividum, whereas the chiA fragment of the actinobacteria of An19, An26, and An53 had high similarity and shared ∼80% identity with chiC from Streptomyces coelicolor. The chitinases from the isolates were compared with those cloned directly from the sediments and from the reference sequences (Fig. 2).
DISCUSSION
Krill is the main food of penguins on Ardley Island, but squid is also an important food source (20). The shells of krill and squid are mainly composed of chitin; therefore, penguin droppings serve as a source of chitin, stimulating the growth of chitinolytic bacteria in the sediment. Six chitinolytic bacterial strains were isolated from the sediment core Y2-4, affiliated with the γ-Proteobacteria including Janthinobacterium and Stenotrophomonas species, Cytophaga species of the Cytophaga-Flexibacter-Bacteroides group, and the actinomycetes Streptomyces and Nocardiopsis species. All of the isolates resemble typical and ubiquitous soil bacteria. None of the isolates were psychrophilic; they had optimal growth temperatures above 20°C. However, they could grow relatively well at 4°C, indicating that they were psychrotolerant (data not shown). The relatively low diversity of bacteria isolated may be reflective of the medium used for isolation. Only two clusters of chiA were cloned from the isolates (Fig. 2); the data again indicated that cultivation-based microbiological approaches are of rather limited utility in assessing the microbial diversity in the environment. The phylogeny of the cloned chitinase genes (Fig. 2) did not correlate well with that of the species from which the chitinase genes were isolated. This suggests that horizontal transfer of the chitinase gene during the evolution of these microbes could have occurred. At present, conflicting opinions exist regarding the relationship between chiA and 16S rRNA phylogeny (4, 17). Like other analyses of functional genes, our results support the view that chiA sequences do not closely follow the bacterial 16S rRNA-based phylogeny.
It has been well documented that standard laboratory cultivation methods retrieve less than 1% of microbes from the environment (1). We therefore used molecular microbiological methods in our study, targeting the chitinase group A gene to quantify the chitinolytic bacteria and to reveal the diversity of the chitinolytic community over time in a sediment core. Using QC-PCR, we semiquantified the chitinase gene copy numbers in fresh penguin droppings from Gentoo and Adélie penguins, in the sediment that was uncontaminated with penguin droppings, and in an ornithogenic sediment core at 1-cm intervals. The sediment not contaminated with penguin droppings contained ∼103 copies/g of chiA, the two kinds of fresh penguin relics contained 105 copies/g of chiA, and the sediment core contained 104 to 107 copies/g of chiA. The data suggested that the input of penguin excrement in the sediment stimulated the number of chitinolytic bacteria. The significant correlation coefficient of chitinase gene copies with bioelement P and TOC concentration implied that the fluctuation of the chitinase gene copies along the column could be used together with the geochemical element data in inferring the size of the penguin population and/or climate change over time. The ages of the Y2-4 intermediate layers L18 and L33 and the bottom layer were determined to be around 1,020, 1,300, and 1,600 years BP, respectively. The age of each sediment layer was deduced by linear interpolation between the dated layers. According to the temporal profile of the chitinase gene numbers, the lower and higher values observed at layers 11 and 53 indicated a lower and a relatively higher number of krill-eating penguins, respectively. Layers 11 and 53 correspond to approximately 600 years BP, during a brief ice age, and before 1,400 years BP, the Medieval warm time period, respectively. The data again suggested that a colder climate may have reduced the penguin population during the past 1,600 years, which is consistent with our previous studies using geochemistry approaches (21). Our chitinase gene approach suggests that by combining molecular methods with other approaches such as a geochemistry approach, together with the proper selection of sampling sites in other locations such as Adelie Land, Ross Sea, East Antarctica, and elsewhere, researchers may be able to have an overall insight into the change in the entire penguin population of Antarctica and its correlation with climate changes in the last thousand years. All these thousand-year-scale studies may provide more convincing arguments for the biological and ecological change affected by the earth climate.
The chitinase gene diversity in the sediment and in the fresh penguin droppings was analyzed. It was shown that the chitinase genes in the sediments were not of high diversity (Table 1).The highest diversity was observed in layers 5, 11, and 59, which had four RFLP types, while layer 43 and droppings of Adélie penguins only had one RFLP type. However, most of the chitinase sequences cloned from the sediments had low identities (<60%) with known chitinases present in sequence databases from both cultivated bacteria and environmental clones (Table 1). Chitinases are widespread in nearly every genus of the bacterial domain. These novel chitinases may be from uncultured bacteria or uncloned chitinases from cultured organisms. The reason for the chitinase diversity change along the column is not clear. However, the data suggest that the quantity of penguin guano input into the sediment has a profound influence on the growth of different chitinolytic bacteria. Another contribution to the frequent shift in the chitinolytic bacterial community in different sediment layers could be a diet shift for the penguins during different periods of time. A preliminary analysis of squid and fish remains discovered in abandoned colonies of Adélie penguins suggests that Adélie penguins may change their diets following warming and cooling cycles (8). Additional evidence of the influence of penguin diet on sediment bacteria comes from our analysis of the chitinase gene diversity in the droppings of Emperor penguins, whose diet consists mostly of small fish. Entirely different chitinase types are present in the droppings of Emperor penguins compared with those in the excreta of Adélie and Gentoo penguins (data not shown), suggesting that chitin from different sources could stimulate the growth of different kinds of chitinolytic bacteria. Among the five chitinase types from Emperor penguin droppings, only one type corresponding toRFLP type 2, which was also detected in the blank soil (Table1), was commonly detected in the three different penguins (data not shown). The RFLP type 2 chitinase may be widespread in Antarctica. Emperor penguins are not observed in the Ardley Island nowadays. The absence of most of the chitinase genes from the Emperor penguin droppings in the entire Y2-4 core is consistent with this fact. Furthermore, our data indicate that Emperor penguins may not live in Ardley Island, at least in the past 1,600 years.
Acknowledgments
We are grateful to Douglas Bartlett for help with editing of the manuscript. We give our particular thanks to the two anonymous reviewers for their great efforts in improving the manuscript. We thank Yinbao Gai for help in sediment DNA extraction.
This work is financially supported by China Natural Science Foundation grants 40376050 and 40476001, a COMRA fund grant (DY105-04-02-7), and the Chinese High Tech (863) program (grant 2004AA621010).
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detecting of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman, J. P., J. Cavanagh, J. J. Austin, and K. Sanderson. 1996. Novel psychrobacter species from Antarctic ornithogenic soils. Int. J. Syst. Bacteriol. 46:841-848. [DOI] [PubMed] [Google Scholar]
- 3.Cohen-Kupiec, R., and I. Chert. 1998. The molecular biology of chitin digestion. Curr. Opin. Biotechnol. 9:270-277. [DOI] [PubMed] [Google Scholar]
- 4.Cottrell, M. T., D. N. Wood, L. Y. Yu, and D. L. Kirchman. 2000. Selected chitinase genes in cultured and uncultured marine bacteria in the α- and γ-subclasses of the proteobacteria. Appl. Environ. Microbiol. 66:1195-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croll, D. A., and B. R. Tershy. 1998. Penguins, fur seals and fishing: prey requirements and potential competition in the South Shetland islands, Antarctica. Polar Biol. 6:365-374. [Google Scholar]
- 6.Croxall, J. P., P. N. Trathan, and E. J. Murphy. 2002. Environmental change and Antarctic seabird populations. Science 297:1510-1514. [DOI] [PubMed] [Google Scholar]
- 7.Croxall, J. R., and R. A. Prince. 1979. Antarctic seabird and seal monitoring studies. Polar Rec. 19:593-595. [Google Scholar]
- 8.Emslie, S. D., W. R. Fraser, R. C. Smith, and W. Walker. 1998. Abandoned penguin colonies and environmental change in the Palmer Station area, Anvers Island, Antarctic Peninsula. Antarc. Sci. 3:257-268. [Google Scholar]
- 9.Gaudette, H. E., W. R. Flight, L. Toner, and D. W. Folger. 1974. An inexpensive titration method for determination of organic carbon in recent sediments. J. Sedim. Petrol. 44:249-253. [Google Scholar]
- 10.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henrissat, B. 1999. Classification of chitinase modules, p. 137-156. In P. Jollès and R. A. A. Muzzarelli (ed.), Chitin and chitinases. Birkhäuser Verlag, Basel, Switzerland.
- 12.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurie, A. D., and G. Lloyd-Jones. 2000. Quantification of phnAc and nahAc in contaminated New Zealand soils by competitive PCR. Appl. Environ. Microbiol. 66:1814-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawley, B., S. Ripley, P. Bridge, and P. Convey. 2004. Molecular analysis of geographic patterns of eukaryotic diversity in Antarctic soils. Appl. Environ. Microbiol. 70:5963-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melles, M., S. R Verkulich, and W. D. Hermichen. 1994. Radiocarbon dating of lacustrine and marine sediments from the Bunger Hills, east Antarctica. Antarc. Sci. 6:375-378. [Google Scholar]
- 16.Metcalfe, A. C., M. Krsek, G. W. Gooday, J. I. Prosser, and E. M. H. Wellington. 2002. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl. Environ. Microbiol. 68:5042-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaiah, N., R. T. Hill, J. Chun, J. Ravel, M. H. Matte, W. L. Straube, and R. R. Colwell. 2000. Use of a chiA probe for detection of chitinase genes in bacteria from the Chesapeake Bay. FEMS Microbiol. Ecol. 34:63-71. [DOI] [PubMed] [Google Scholar]
- 18.Ramsay, A. J., and R. E. Stannard. 1986. Numbers and viability of bacteria in ornithogenic soils of Antarctica. Polar Biol. 5:195-198. [Google Scholar]
- 19.Staley, J. T., and R. P. Herwig. 1993. Degradation of particulate organic material in the Antarctic, p. 241-264. In E. I. Freidmann (ed.), Antarctic microbiology. Wiley-Liss, Inc., New York, N.Y.
- 20.Sun, L. G., and Z. Q. Xie. 2001. Relics: penguin population programs. Sci. Prog. 84:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun, L. G., Z. Q. Xie, and J. L. Zhao. 2000. A 3000-year record of penguin populations. Nature 407:858. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki, K., M. Taiyoji, N. Sugawara, N. Nikaidou, B. Henrissat, and T. Watanabe. 1999. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 343:587-596. [PMC free article] [PubMed] [Google Scholar]
- 23.Tindall, B. J. 2004. Prokaryotic diversity in the Antarctic: the tip of the iceberg. Microb. Ecol. 47:271-283. [DOI] [PubMed] [Google Scholar]
- 24.Vionis, A. P., F. Niemeyer, A. D. Karagouni, and H. Schrempf. 1996. Production and processing of a 59-kilodalton exochitinase during growth of Streptomyces lividans carrying pCHIO12 in soil microcosms amended with crab or fungal chitin. Appl. Environ. Microbiol. 62:1774-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, P., F. P. Wang, M. X. Xu, and X. Xiao. 2004. Molecular phylogeny of methylotrophs in a deep-sea sediment from tropical west Pacific warm pool. FEMS Microbiol. Ecol. 47:77-84. [DOI] [PubMed] [Google Scholar]
- 26.Zeng, X., X. Xiao, P. Wang, and F. P. Wang. 2004. Screening and characterization of psychrotrophic, lipolytic bacteria from deep-sea sediments. J. Microbiol. Biotechnol. 14:952-958. [Google Scholar]
- 27.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]